Abstract
Background
Metastasis to the cervical (neck) lymph nodes is one of the most significant clinical factors responsible for death from oral squamous cell carcinoma (SCC). Therefore, the lymph nodes are frequently removed when the tumor is excised (neck dissection), even though the majority of patients will not benefit from the extra surgery. Two subtypes of oral SCC distinguished by the presence of tumor genomic aberrations +3q, -8p, +8q and/or +20 differ in risk for metastasis – high for the 3q8pq20 subtype, harboring one or more of the aberrations and low for the non-3q8pq20 subtype, lacking these alterations. A prior analysis of the literature suggested genes differentially methylated in the two subtypes. Therefore, the goal of this study was to further investigate the methylation status of candidate biomarkers of the non-3q8pq20 subtype, and evaluate their utility for identifying patients at low risk for metastasis.
Methods
Methylation status of genes in a cohort of 52 oral SCC patients with at least five year follow up was determined by pyrosequencing. Gene expression levels were determined by quantitative RT-PCR. Growth following re-expression of HOXA9 in cultured oral SCC cells was assessed by proliferation and colony formation assays.
Results
A pilot study evaluating methylation levels of HOXA9, MT1A and HOXA11 promoters in DNA from 12 tumors (six each of the 3q8pq20 and non-3q8pq20 subtypes) revealed that only HOXA9 was differentially methylated. Significant differences in methylation levels of HOXA9 were observed amongst the 52 oral SCCs with respect to genomic subtype and nodal status (p = 0.014, and p = 0.024, respectively, Wilcoxon rank sum test). High levels of HOXA9 methylation and low levels of expression in oral SCC cell lines were observed compared to HaCaT, a non-tumorigenic keratinocyte cell line. Re-expression of HOXA9 in the SCC4 oral cancer cell line resulted in diminished proliferation and colony formation.
Conclusions
HOXA9 methylation is frequent in oral cancers and levels are higher in tumors with greater risk of metastasis. Expression of HOXA9 is low in cells with high levels of methylation and reduced expression appears to confer a growth advantage.
Keywords: Oral cancer, Metastasis, HOXA9, Methylation, Pyrosequencing
Background
Metastasis to the cervical (neck) lymph nodes is one of the most significant clinical factors responsible for death from oral squamous cell carcinoma (SCC). Currently, there are no satisfactory clinical, imaging, pathologic or molecular techniques that can reliably determine if neck metastases are present at the time of surgery to remove the primary tumor. Therefore, patients and physicians frequently elect to remove the cervical lymph nodes (neck dissection) at the time the tumor is excised if the chance of metastasis is > 20% based on current imperfect risk assessment capability. This surgical procedure is lengthy, complex, and risky with high morbidity due to functional deficits and disfigurement. Being able to determine which patients do not need such surgery would have a substantial, immediate clinical benefit.
We recently reported that two subtypes of oral SCC distinguished by tumor genomic aberrations differ in risk for metastasis [1]. One subtype, the 3q8pq20 subtype, is characterized by the presence of one or more of the recurrent copy number aberrations, +3q, -8p, +8q and/or +20 and has a substantial risk of metastasis (46%). The other subtype (non-3q8pq20) lacks these copy number alterations and is associated with a low risk of metastasis (7%). These initial studies, which were replicated in a small independent cohort, indicated that non-3q8pq20 status has 93% negative predictive value (NPV), i.e., ability to predict that these cases do not have neck metastases, and thus do not need neck dissection.
The non-3q8pq20 tumors lack chromosome level instability, which suggests that development of these tumors could be associated with other, copy number neutral, mechanisms, such as microsatellite instability or epigenetic alterations. Microsatellite instability is not common in oral SCC from western countries, whereas genome-wide alterations in methylation patterns are observed [2,3]. Analysis of a head and neck cancer patient cohort [2] for which both copy number and methylation measurements were available (NCBI GEO accession numbers GSE20939 and GSE20742, respectively) found 15 loci significantly differentially methylated in 3q8pq20 compared to non-3q8pq20 tumors and normal oral tissue [1]. To investigate whether these loci are a potential biomarker for distinguishing 3q8pq20 and non-3q8pq20 tumors, we investigated the methylation status of the loci in an oral SCC cohort in which 3q8pq20 status had been determined [1]. The overall goal was to develop a simple assay for 3q8pq20 status utilizing a panel of differentially methylated loci that could be performed on tissue samples obtained by a non-invasive technique prior to surgery and thus guide decisions regarding the need for neck dissection.
Methods
Patients and tissue samples
The study was approved by the Institutional Review Board of the University of California San Francisco (H7867-23910-05). Formalin fixed paraffin embedded (FFPE) SCC surgical resection specimens were available from 52 cases of the previously published SCC cohort#2 [1] and included oral cavity sites–tongue, gingiva, floor of mouth, retromolar trigone and buccal mucosa. Associated clinical data were obtained through the University of California San Francisco Oral Cancer Tissue Bank and Cancer Registry (Table 1 and Additional file 1: Table S1 of reference [1]). Patient consent was obtained for use of all specimens.
Table 1.
Patient characteristics
| Sample | Site | Nodal status | 3q8pq20 |
|---|---|---|---|
| AB003 |
Retromolar Region |
N0 |
Yes |
| AB004 |
Gingiva |
N0 |
Yes |
| AB007 |
Floor of Mouth |
N+ |
Yes |
| AB010 |
Tongue |
N0 |
Yes |
| AB011 |
Tongue |
N0 |
Yes |
| AB014 |
Retromolar Region |
N+ |
Yes |
| AB015 |
Tongue |
N+ |
Yes |
| AB017 |
Buccal Mucosa |
N+ |
Yes |
| AB018 |
Floor of Mouth |
N+ |
Yes |
| AB019 |
Floor of Mouth, tongue |
N+ |
Yes |
| AB020 |
Hard Palate |
N+ |
Yes |
| AB021 |
Tongue |
N0 |
no |
| AB023 |
Tongue |
N0 |
Yes |
| AB025 |
Gingiva |
N0 |
No |
| AB026 |
Retromolar Region |
N0 |
Yes |
| AB029 |
Floor of Mouth, tongue, buccal mucosa |
N0 |
Yes |
| AB031 |
Tongue |
N0 |
Yes |
| AB032 |
Buccal Mucosa |
N0 |
Yes |
| AB033 |
Retromolar Region |
N+ |
Yes |
| AB034 |
Buccal Mucosa |
N0 |
No |
| AB035 |
Tongue |
N0 |
No |
| AB039 |
Gingiva |
N0 |
Yes |
| AB041 |
Tongue |
N0 |
Yes |
| AB042 |
Tongue |
N0 |
Yes |
| AB045 |
Gingiva |
N0 |
No |
| AB048 |
Tongue |
N0 |
Yes |
| AB049 |
Tongue |
N0 |
No |
| AB051 |
Tongue |
N+ |
Yes |
| AB054 |
Buccal Mucosa |
N+ |
Yes |
| AB055 |
Floor of Mouth |
N0 |
Yes |
| AB056 |
Retromolar Region |
N+ |
Yes |
| AB059 |
Tongue |
N+ |
Yes |
| AB060 |
Tongue, Floor of Mouth |
N+ |
Yes |
| AB061 |
Buccal Mucosa |
N0 |
Yes |
| AB062 |
Gingiva |
N0 |
No |
| AB063 |
Tongue |
N0 |
Yes |
| AB064 |
Buccal Mucosa |
N0 |
Yes |
| AB066 |
Tongue |
N0 |
Yes |
| AB067 |
Floor of Mouth |
N0 |
Yes |
| AB068 |
Gingiva |
N0 |
No |
| AB070 |
Floor of Mouth |
N0 |
Yes |
| AB071 |
Hard Palate |
N0 |
Yes |
| AB073 |
Gingiva |
N+ |
No |
| AB077 |
Floor of Mouth |
N0 |
Yes |
| AB079 |
Tongue |
N0 |
No |
| AB080 |
Tongue |
N0 |
No |
| AB081 |
Gingiva |
N+ |
Yes |
| AB082 |
Floor of Mouth |
N+ |
Yes |
| AB083 |
Floor of Mouth, tongue, gingiva |
N+ |
Yes |
| AB084 |
Gingiva |
N+ |
Yes |
| AB085 |
Tongue |
N0 |
Yes |
| AB086 | Floor of Mouth | N0 | No |
Tumor cell lines
Human oral tongue SCC cell lines SCC4, SCC9, SCC15 and SCC25 were obtained from the American Type Culture Collection (Manassas, VA), BICR16, H357, H103, PE/CA-PJ15, and PE/CA-PJ49 from the Health Protection Agency Culture Collections (HPA, Salisbury, UK), CAL33 from Deutsche Sammlung von Mikroorganismen und Zellculturen GmbH (DSMZ, Braunschweig, Germany), and OSC20 from the Japanese Collection of Research Biosources (Osaka, Japan). The DOK cell line, derived from a human oral dysplasia was obtained from the Health Protection Agency Culture Collections (HPA, Salisbury, UK) and HaCaT, a skin keratinocyte line was from the Deutsche Sammlung von Mikroorganismen und Zellculturen GmbH (DSMZ, Braunschweig, Germany). Cells were propagated according to the methods recommended by the suppliers.
Bisulfite conversion and pyrosequencing
The DNA concentration was quantified by Quant-iT™ dsDNA BR Assay (Life Technologies, Grand Island, NY). A total of 200 ng of each DNA sample was bisulfite converted with the EZ DNA Methylation-Direct Kit (Zymo Research, Orange, CA). EpiTect Control DNA (QIAGEN, Germantown, MD) was used as methylated and unmethylated control DNA. PyroMark assays (QIAGEN, Germantown, MD) were used to determine methylation status of HOXA9 (Hs_HOXA9_05_PM) and MT1A (HS_MT1A_02_PM). A custom assay was designed for HOXA11 (Forward: biotin-5’-AGAGGTAGGTAGGGAAGATG-3’ , Reverse: 5’-CCCCTCCCATAAACTTACTCTAAA-3’ , Sequencing: 5’-ACACTCTCTCATTCATAATC-3’). Bisulfite PCR was performed using the PyroMark PCR kit (QIAGEN, Germantown, MD) and amplification was carried out by an initial incubation at 95°C for 15 min, followed by 45 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec. A final incubation was carried out at 72°C for 10 min. The biotinylated PCR product was purified and subjected to pyrosequencing using the PyroMark Q24 System and PyroMark Q24 gold reagents (QIAGEN, Germantown, MD). Data were analyzed by PyroMark Q24 2.0.6 software. The HOXA9 assay (Hs_HOXA9_05_PM) includes three CpG islands. Methylation level was assigned as the mean of the three sites.
RT-PCR
Total RNA was extracted from cell lines using TRIzol® Reagent (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. RNA quantity was determined with a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific Inc., Asheville NC, US) and RNA integrity was assessed with the Bioanalyzer™ (Agilent Technologies, Inc., average RIN for cell lines = 8.2). A fixed amount of total RNA (500 ng) per each sample was reverse transcribed with iScript™ Select cDNA Synthesis kit (Bio-Rad, Hercules CA, US).
Standard TaqMan qRT-PCR Gene Expression assays were conducted in triplicate to quantify HOXA9 expression levels relative to GUSB. Duplex PCR was performed with the FAM labeled Taqman assay for HOXA9 (Hs00365956_m1) and VIC labeled Taqman assay for GUSB (Hs00939627_m1). Reactions (10 μL per well) included 5 μL TaqMan Gene Expression Master Mix (Life Technologies, Grand Island, NY), 0.5 μL 20× Gene Expression Assay Mix, and 1 μL cDNA diluted to a final concentration of 10 ng/μL. Assay plates were run on an Applied Biosystems 7900HT detection system using standard settings (cycling program included 2 min incubation at 50°C and 10 min incubation at 95°C followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min). Data values (Cycle Threshold [Ct] values) were extracted from each assay with the SDS v2.0 software tool (Life Technologies, Grand Island, NY). Gene expression values were derived from the equation: ΔCt = (Ctgene–CtGUSB) and expressed as 2-ΔCt.
For end point PCR, the primer sequences used to amplify GUSB were 5’-TGCGCACAAGAGTGGTGCTGA-3′ and 5′-TCGACCCCATTCACCCACACGA-3′. The primers for BRCA1 have been reported previously [4]. Amplification reactions used the HotStarTaq Master Mix kit (QIAGEN, Germantown, MD). An initial incubation at 95°C for 15 min was followed by 45 cycles of 94°C for 30 sec, 55°C (BRCA1) or 60°C (GUSB) for 30 sec, 72°C for 30 sec and a final incubation at 72°C for 10 min. Nucleotide free water was used for the negative control and Universal Human Reference RNA (Agilent technologies, Santa Clara, CA) was used for the positive control.
Establishment of the SCC4 cell line with the HOXA9 inducible-expression construct
To generate stable, inducible cell lines expressing HOXA9 upon doxycycline induction, HOXA9 cDNA (BC10023, Open Biosystems) was subcloned into pLVX-tight-puro (Clontech, CA). SCC4 cells were transfected with pLVX-Tet-On and pLVX-HOXA9 using Lipofectamine LTX (Life Technologies, Grand Island, NY)) according to the manufacturer’s instructions. Three days after transfection, stable clones were selected by culturing for 2 weeks in medium supplemented with 1 μg/mL Puromycin (Sigma-Aldrich) and 300 μg/mL G418 (Roche). These SCC4-HOXA9 cells were cultured with 1 μg/mL doxycycline to induce HOXA9 expression, which was verified by qPCR and western blotting. Empty vector control SCC4 cells (SCC4-empty vector) were generated by transfecting with pLVX-Tet-On and pLVX-tight-puro and selection by culturing for 2 weeks in medium supplemented with 1 μg/mL puromycin and 300 μg/mL G418.
Proliferation and colony formation assays
To assess cell proliferation, 500 SCC4-HOXA9 and SCC4-empty vector cells were seeded in 96-well culture plates. After allowing cells to attach and grow for 24 hours, 100 μL of culture medium supplemented with 1 μg/mL doxycycline was added to the cultures. Plates containing six replicate wells of each cell type were harvested over a period of nine days and proliferation was measured using the CyQUANT® NF assay (Life Technologies, Grand Island, NY).
Colony formation assays were performed by seeding cells in six-well plates at a density of 1000 cells/well in 2 mL complete medium. After culturing for two weeks, colonies were stained with 0.5% crystal violet in 2% ethanol and colonies were counted.
Statistical analysis
A Wilcoxon rank sum test was used to assess differential methylation between groups. A T-test was used to assess differences in proliferation and colony formation assays.
Results
Pilot study evaluating methylation status of HOXA9, HOXA11 and MT1A
Methylation and tumor genome copy number data reported by Poage and colleagues [2] were accessioned and cases assigned to the 3q8pq20 or non-3q8pq20 subtypes as described previously [1]. Information on nodal status was not available for these cases. Methylation levels were compared between tumor subtypes, and this analysis revealed 15 loci that were differentially methylated between 3q8pq20 and non-3q8pq20 plus normal tissue (Additional file 2: Figure S1, Additional file 1: Table S1). HOXA9, HOXA11 and MT1A were selected for further study, because promoters of these genes were consistently highly methylated in 3q8pq20 cases. Methylation status of the CpG islands in these promoters was evaluated by pyrosequencing in 12 tumors (six 3q8pq20 and six non-3q8pq20 subtypes) from the previously reported SCC cohort#2 for which copy number and clinical follow up data were available [1]. In this pilot study, only HOXA9 appeared to be differentially methylated between the two subtypes (Figure 1).
Figure 1.
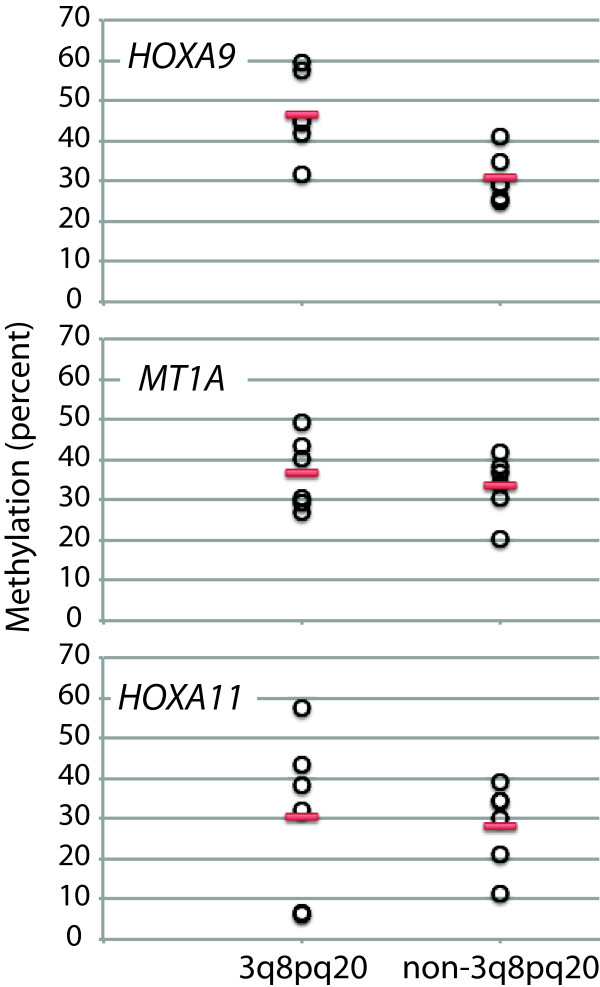
Pilot study evaluating methylation of HOXA9, MT1A and HOXA11 in 3q8pq20 and non-3q8pq20 tumors. Methylation levels of the gene promoters are shown for individual 3q8pq20 and non-3q8pq20 tumors (open circles). The mean level is indicated by the red bar. Differential methylation in the two oral cancer subtypes is evident for HOXA9.
Association of HOXA9 methylation with risk for metastasis
The methylation status of HOXA9 was further investigated in cell lines and 40 additional tumors from SCC cohort#2 [1] by pyrosequencing (Figure 2, Additional file 1: Table S2). The HOXA9 promoter is nearly completely methylated in oral cancer cell lines and to a slightly lesser extent in DOX, a cell line derived from dysplastic tissue (Figure 3a). By contrast, only 30% methylation of HOXA9 was observed in the non-tumorigenic HaCaT cells. The mean level of methylation of non-3q8pq20 tumors (31%) was similar to HaCaT cells and significantly less than that of 3q8pq20 tumors (45%) or clinically normal epithelium from four cancer patients (16%). We also observed a significant difference in methylation when comparing node positive (49%) and node negative cases (38%), irrespective of genomic subtype.
Figure 2.
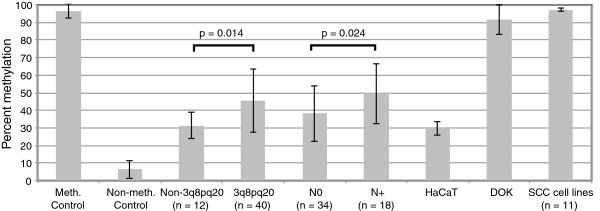
Methylation of HOXA9 in oral cancers and cell lines. Shown are the mean and standard deviation.
Figure 3.
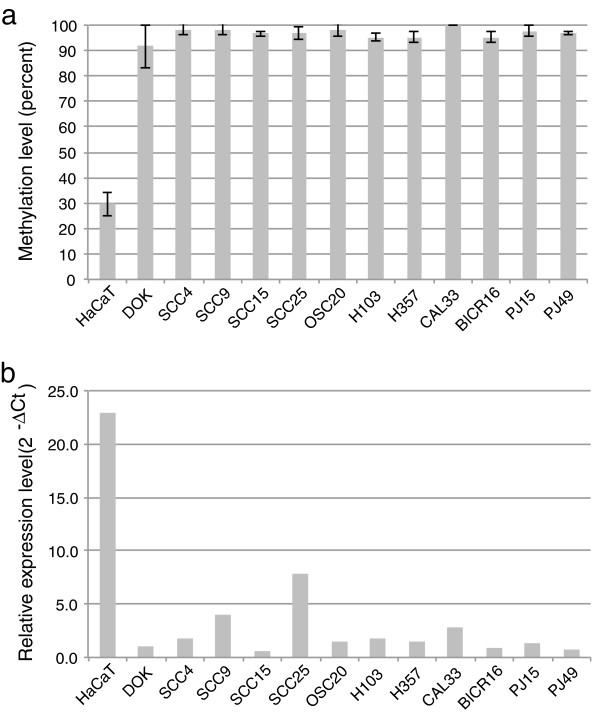
HOXA9 is methylated and expressed at low levels in oral cancer cell lines. a. Methylation levels (mean and standard deviation) as determined by pyrosequencing. b. Transcript levels determined by quantitative RT-PCR
Re-expression of HOXA9 inhibits growth
In oral cancer cell lines with nearly complete methylation of the HOXA9 promoter, HOXA9 expression levels are low (Figure 3). By contrast, HOXA9 expression levels are higher in HaCaT cells with only partial methylation of the promoter (Figure 3). These observations suggest that promoter methylation is frequently a mechanism whereby expression of HOXA9 expression can be repressed in oral cancer and pre-cancer cells.
To investigate the possible functional significance of reduced expression of HOXA9, we re-expressed HOXA9 in SCC4 tongue cancer cells (SCC4-HOXA9), which resulted in diminished proliferation compared to control SCC4 cells with an empty vector construct (SCC4-empty vector, Figure 4a). We also observed that fewer (p = 0.04), and smaller colonies were formed by SCC4-HOXA9 cells in colony formation assays compared to SCC4 cells with an empty vector construct (Figure 4b and Additional file 2: Figure S2). In breast cancer cells, HOXA9 has been shown to directly regulate BRCA1 expression and to suppress growth and survival [4]. Expression of HOXA9 in SCC4 cells also resulted in increased expression of BRCA1 (Figure 4c), suggesting that, in addition to promoting growth, reduced expression of HOXA9 may contribute to tumor genome instability.
Figure 4.
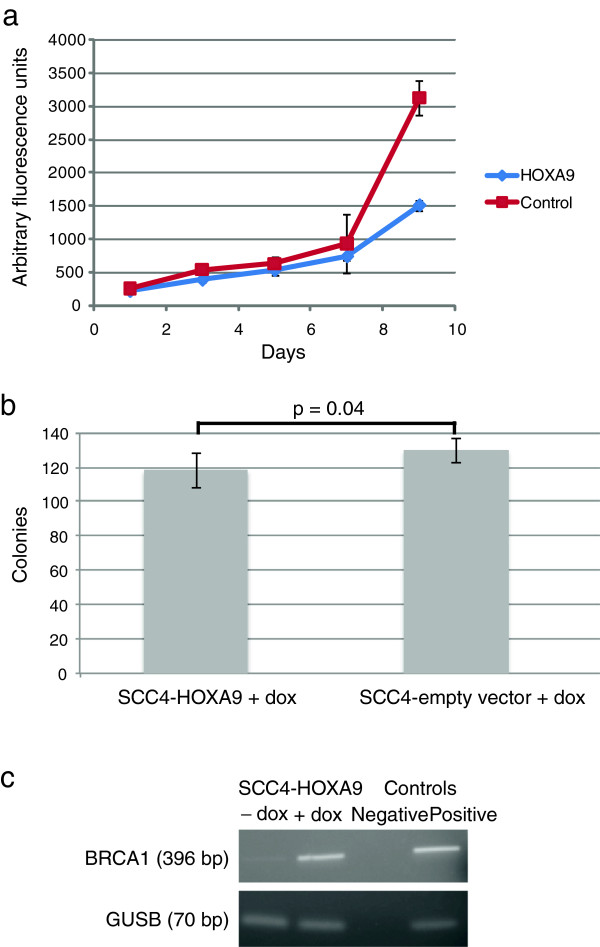
Re-expression of HOXA9 represses proliferation and colony formation and induces expression of BRCA1. a. Proliferation of SCC4-HOXA9 and SCC4-empty vector cells. b. Colony formation by SCC4-HOXA9 and SCC4-empty vector cells. c. Endpoint PCR analysis of BRCA1 expression.
Discussion
The risk of metastasis in oral cancer patients is associated with the status of copy number alterations at chromosome 3q, 8p, 8q and 20. The presence of any one of these aberrations (3q8pq20 genomic subtype) is associated with a substantial risk of metastasis (46%), while absence of alterations in all of these regions (non-3q8pq20 subtype) shows greater promise as a biomarker for low risk of metastasis (93% NPV). If validated in further larger clinical studies, determination of non-3q8pq20 status prior to surgery could identify those patients at low risk of metastasis who could be spared the extra surgery of an elective neck dissection. While non-3q8pq20 status could be determined prior to surgery by profiling tumor biopsies for copy number alterations, assignment of non-3q8pq20 status might be ambiguous if no copy number changes were present on other chromosome arms. Therefore, we investigated whether differential methylation of loci could act as a surrogate and identify 3q8pq20 and non-3q8pq20 subtypes. Two of the candidate loci (HOXA11 and MT1A), selected from analysis of published data, failed to validate in our cohort. Only HOXA9 was found to be differentially methylated in 3q8pq20 compared to non-3q8pq20 tumors, as well as between node positive and node negative cases. Although the differences in methylation level reached statistical significance, they are modest and would probably not result in a robust clinical test for nodal status.
Homeobox (HOX) genes are transcription factors with roles in development, regulating patterning during embryogenesis and maintaining differentiated states. De-regulated expression of HOX genes is reported in cancers [5]. They can be overexpressed and act as oncogenes or they can act as tumor suppressors with expression down regulated via promoter methylation. Our data are consistent with HOXA9 acting as a tumor suppressor in oral cancer. Methylation of HOXA9 has been reported previously in oral cavity cancer [6], and methylation and loss of expression of HOXA9 reported in breast [4,7,8], lung [9], ovarian [10] and bladder cancer [11], whereas HOXA9 is well known to act as an oncogene in leukemia [5]. The tumor suppressor function of HOXA9 has been extensively investigated in breast cancer where it has been shown that HOXA9 directly regulates BRCA1[4] and a number of other genes involved in invasion, growth and metastasis [7]. While we show here that HOXA9 also appears to positively regulate BRCA1 expression in oral cancer cells, further studies will be required to fully understand how HOXA9 functions as a tumor suppressor in oral cancer. Indeed, the oncogenic and tumor suppressive activities of deregulated HOXA9 expression in different tissues highlight the importance of tissue context for the functioning of deregulated developmental genes in cancer.
Conclusions
The HOXA9 promoter is frequently methylated in oral SCC and diminished expression of HOXA9 confers a growth advantage to oral SCC cells. Higher levels of HOXA9 methylation are present in tumors with greater risk of metastasis.
Abbreviations
SCC: Squamous cell carcinoma; NPV: Negative predictive value.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KU, RV and BH carried out the molecular assays and cell culture experiments. AB, BLS and DGA assembled the patient cohort and associated clinical and molecular information. KU and DGA drafted the manuscript. DGA conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Comparison of methylation levels in 3q8pq20 cancers vs. non-3q8pq20 cancers + normals (data from NCBI GEO, GSE20939 and GSE20742). Table S2. Methylation status of HOXA9 in oral cancers.
Differentially methylated loci. Copy number and methylation data from Poage et al. [1] were obtained from NCBI GEO (Accession numbers GSE20742 and GSE20939, respectively). Copy number data were segmented and samples were assigned to the 3q8pq20 or non-3q8pq20 subtype based on the presence of copy number changes, +3q, -8p, +8q, and/or +20 as described previously [2]. A nonlinear transformation was applied to the methylation data beta values [s = sqrt(beta) - sqrt(1 - beta)], which increases the Gaussian character of the data and has the effect of reducing the number of false positives. The transformed data were then quantile normalized across samples. Probes were tested for differential methylation between 3q8pq20 and non-3q8pq20 subtypes plus normal cases using the limma package. The probes for each comparison were filtered on absolute mean difference in methylation level (> 0.05) and adjusted p-value (< 0.05, FDR) [3]. This analysis yielded 15 probes differentially methylated between 3q8pq20 and non-3q8pq20 samples. Figure S2. Colony formation by SCC4 cells expressing HOXA9 and control SCC4 cells. Shown are images of six-well plates following staining with crystal violet. SCC4-HOXA9 and SCC4-empty vector cells were cultured in the presence of 1 μg/mL doxycycline. Smaller colonies are present in the plates of SCC4-HOXA9 cells in which HOXA9 was re-expressed (top two panels) compared to SCC4 cells with an empty vector (SCC4-empty vector, bottom two panels).
Contributor Information
Kenichiro Uchida, Email: k.uchida@yamaguchi-u.ac.jp.
Ratna Veeramachaneni, Email: VeeramachaneniR@cc.ucsf.edu.
Bing Huey, Email: bhuey@cc.ucsf.edu.
Aditi Bhattacharya, Email: draditibhattacharya@gmail.com.
Brian L Schmidt, Email: bls322@nyu.edu.
Donna G Albertson, Email: da66@nyu.edu.
Acknowledgements
We thank members of the Helen Diller Family Comprehensive Cancer Center Genome Analysis shared resource for their help with quantitative RT-PCR assays and Teruyuki Muraguchi and Mauting Lin for assistance with preparation of HOXA9 expressing SCC4 cells. Some of the pyrosequencing work was performed at the Protein and Nucleic Acid facility, Stanford University.
Funding
This work was supported by NIH grants CA118323 and CA131286 to DGA, and CA113833 to BLS. AB was the recipient of a pre-doctoral fellowship from the California Tobacco-Related Disease Research Program (18DT-0011).
References
- Bhattacharya A, Roy R, Snijders AM, Hamilton G, Paquette J, Tokuyasu T, Bengtsson H, Jordan RC, Olshen AB, Pinkel D, Schmidt BL, Albertson DG. Two distinct routes to oral cancer differing in genome instability and risk for cervical node metastasis. Clin Cancer Res. 2011;17(22):7024–7034. doi: 10.1158/1078-0432.CCR-11-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poage GM, Christensen BC, Houseman EA, McClean MD, Wiencke JK, Posner MR, Clark JR, Nelson HH, Marsit CJ, Kelsey KT. Genetic and epigenetic somatic alterations in head and neck squamous cell carcinomas are globally coordinated but not locally targeted. PLoS One. 2010;5(3):e9651. doi: 10.1371/journal.pone.0009651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Hall GL, Lowe D, Bowers NL, Liloglou T, Field JK, Woolgar JA, Risk JM. CpG island methylation phenotype (CIMP) in oral cancer: associated with a marked inflammatory response and less aggressive tumour biology. Oral Oncol. 2007;43(9):878–886. doi: 10.1016/j.oraloncology.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Gilbert PM, Mouw JK, Unger MA, Lakins JN, Gbegnon MK, Clemmer VB, Benezra M, Licht JD, Boudreau NJ, Tsai KK, Welm AL, Feldman MD, Weber BL, Weaver VM. HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. J Clin Invest. 2010;120(5):1535–1550. doi: 10.1172/JCI39534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- Guerrero-Preston R, Soudry E, Acero J, Orera M, Moreno-Lopez L, Macia-Colon G, Jaffe A, Berdasco M, Ili-Gangas C, Brebi-Mieville P, Fu Y, Engstrom C, Irizarry RA, Esteller M, Westra W, Koch W, Califano J, Sidransky D. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev Res (Phila) 2011;4(7):1061–1072. doi: 10.1158/1940-6207.CAPR-11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Song CX, Huang H, Frankenberger CA, Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C, Rosner MR. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci U S A. 2013;110(24):9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds PA, Sigaroudinia M, Zardo G, Wilson MB, Benton GM, Miller CJ, Hong C, Fridlyand J, Costello JF, Tlsty TD. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J Biol Chem. 2006;281(34):24790–24802. doi: 10.1074/jbc.M604175200. [DOI] [PubMed] [Google Scholar]
- Son JW, Jeong KJ, Jean WS, Park SY, Jheon S, Cho HM, Park CG, Lee HY, Kang J. Genome-wide combination profiling of DNA copy number and methylation for deciphering biomarkers in non-small cell lung cancer patients. Cancer Lett. 2011;311(1):29–37. doi: 10.1016/j.canlet.2011.06.021. [DOI] [PubMed] [Google Scholar]
- Wu Q, Lothe RA, Ahlquist T, Silins I, Trope CG, Micci F, Nesland JM, Suo Z, Lind GE. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol Cancer. 2007;6:45. doi: 10.1186/1476-4598-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert T, Borre M, Christiansen A, Hermann GG, Orntoft TF, Dyrskjot L. Diagnosis of bladder cancer recurrence based on urinary levels of EOMES, HOXA9, POU4F2, TWIST1, VIM, and ZNF154 hypermethylation. PLoS One. 2012;7(10):e46297. doi: 10.1371/journal.pone.0046297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of methylation levels in 3q8pq20 cancers vs. non-3q8pq20 cancers + normals (data from NCBI GEO, GSE20939 and GSE20742). Table S2. Methylation status of HOXA9 in oral cancers.
Differentially methylated loci. Copy number and methylation data from Poage et al. [1] were obtained from NCBI GEO (Accession numbers GSE20742 and GSE20939, respectively). Copy number data were segmented and samples were assigned to the 3q8pq20 or non-3q8pq20 subtype based on the presence of copy number changes, +3q, -8p, +8q, and/or +20 as described previously [2]. A nonlinear transformation was applied to the methylation data beta values [s = sqrt(beta) - sqrt(1 - beta)], which increases the Gaussian character of the data and has the effect of reducing the number of false positives. The transformed data were then quantile normalized across samples. Probes were tested for differential methylation between 3q8pq20 and non-3q8pq20 subtypes plus normal cases using the limma package. The probes for each comparison were filtered on absolute mean difference in methylation level (> 0.05) and adjusted p-value (< 0.05, FDR) [3]. This analysis yielded 15 probes differentially methylated between 3q8pq20 and non-3q8pq20 samples. Figure S2. Colony formation by SCC4 cells expressing HOXA9 and control SCC4 cells. Shown are images of six-well plates following staining with crystal violet. SCC4-HOXA9 and SCC4-empty vector cells were cultured in the presence of 1 μg/mL doxycycline. Smaller colonies are present in the plates of SCC4-HOXA9 cells in which HOXA9 was re-expressed (top two panels) compared to SCC4 cells with an empty vector (SCC4-empty vector, bottom two panels).


