Abstract
In addition to the two large clostridial cytotoxins (TcdA and TcdB), some strains of Clostridium difficile also produce an actin-specific ADP-ribosyltransferase, called binary toxin CDT. We used a PCR method and Southern blotting for the detection of genes encoding the enzymatic (CDTa) and binding (CDTb) components of the binary toxin in 369 strains isolated from patients with suspected C. difficile-associated diarrhea or colitis. Twenty-two strains (a prevalence of 6%) harbored both genes. When binary toxin production was assessed by Western blotting, 19 of the 22 strains reacted with antisera against the iota toxin of C. perfringens (anti-Ia and anti-Ib). Additionally, binary toxin activity, detected by the ADP-ribosyltransferase assay, was present in only 17 of the 22 strains. Subsequently, all 22 binary toxin-positive strains were tested for the production of toxins TcdA and TcdB, toxinotyped, and characterized by serogrouping, PCR ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis. All binary toxin-positive strains also produced TcdB and/or TcdA. However, they had significant changes in the tcdA and tcdB genes and belonged to variant toxinotypes III, IV, V, VII, IX, and XIII. We could differentiate 16 profiles by using typing methods, indicating that most of the binary toxin-positive strains were unrelated.
Clostridium difficile is a spore-forming, anaerobic, gram-positive bacillus that was first recognized in the late 1970s to be the main cause of pseudomembranous colitis (PMC) (8, 29). C. difficile is responsible for almost all cases of PMC and for 15 to 25% of cases of antibiotic-associated diarrhea (AAD) (9). In the last 20 years, C. difficile has also emerged as a major cause of nosocomial diarrhea in adult patients and has been responsible for large outbreaks in hospital settings (15, 30). For hospitalized patients, the overall incidence of C. difficile-associated diseases has been found to vary widely, from 0.1 to 2 per 100 patient admissions (13, 15, 30, 31). In many hospitals, C. difficile is the most frequently isolated enteropathogen.
Two large toxin proteins (TcdA [or toxin A] and TcdB [or toxin B]) are thought to be the primary virulence factors of C. difficile (3). These toxins are encoded by two separate genes, named tcdA and tcdB. Together with three additional genes they form a 19.6-kb pathogenicity locus called PaLoc (14, 17). TcdA and TcdB both disrupt the actin cytoskeleton of intestinal epithelial cells by the UDP-glucose-dependent glucosylation of proteins from the Rho and Ras subfamilies (26, 27).
Some strains of C. difficile also produce an actin-specific ADP-ribosyltransferase called CDT (22, 36, 42-45) which was first described by Popoff et al. in 1988 (36). The binary toxin CDT is unrelated to the well-characterized toxins TcdA and TcdB. It belongs to the group of clostridial binary toxins, which include the iota toxin of Clostridium perfringens type E, the toxin of Clostridium spiroforme, and the C2 toxin of Clostridium botulinum C and D (25, 34, 36, 37). Binary toxins consist of two independent unlinked protein chains, designated CDTa (enzymatic component) and CDTb (binding component) in C. difficile (18, 33). The binding component recognizes a cell surface receptor, resulting in the internalization of the enzymatic component into the cytosol, which catalyzes the ADP-ribosylation of monomeric actin and leads to disorganization of the cytoskeleton (1). The binary toxin locus from strain CD196 has been cloned and sequenced (33). It contains two genes (cdtA and cdtB), with an organization and sequences similar to the genes of the iota toxin of C. perfringens: the protein sequences of CDTa and CDTb are 81 and 84% similar, respectively, to the corresponding iota toxin proteins. CDT is cytotoxic to Vero cells in culture (23, 33, 36). The changes observed in Vero cells include the rounding and depolymerization of actin filaments, as shown by fluorescein isothiocyanate-phalloidin staining, and are similar to those induced by the C. spiroforme toxin. The cytotoxic effect of the CDT is neutralized by anti-Ib antibodies directed against the iota toxin of C. perfringens.
Binary toxins produced by some other clostridia have been implicated in human and animal digestive diseases. For example, C. perfringens type E isolates produce iota toxin and have been implicated in animal enterotoxemia (11). The binary toxin is the only virulence factor identified to date for C. spiroforme, which has been implicated in rabbit enteritis and in one case of colitis in a human (2, 35). The toxigenic C. botulinum C2 toxin induces necrotic and hemorrhagic lesions of the intestine and lungs (25). In addition, C. difficile CD196, the strain in which CDT was first identified, was isolated from a patient with severe PMC (36).
Since the majority of strains isolated from symptomatic patients produce only TcdA and TcdB, one can conclude that CDT is not required for the virulence of C. difficile, but it may serve as an additional virulence factor and may function in synergy with the large clostridial cytotoxins. The aims of this study were first to determine the prevalence of C. difficile containing the binary toxin among strains isolated in Paris and the surrounding region and second to characterize these binary toxin-positive strains.
MATERIALS AND METHODS
Bacterial strains.
We studied 369 C. difficile strains recovered from 288 adult patients with suspected AAD or antibiotic-associated colitis (AAC). The patients were hospitalized at 17 care facilities in Paris or the surrounding area in the year 2000. The strains were isolated from stools cultured on selective medium (taurocholate, cefoxitin, cycloserine brain heart infusion agar supplemented with 5% defibrinated horse blood, 0.1% sodium taurocholate, 10 μg of cefoxitin ml−1, and 250 μg of cycloserine ml−1) in an anaerobic atmosphere for 48 h.
For all experiments, three strains of C. difficile were used as reference strains: C. difficile CD196 was used as a control for the detection of the cdtA and cdtB genes (36), C. perfringens 10748, which produces the iota toxin, was used as a control in Western blot experiments (36), and C. difficile ATCC 43596 (serogroup C) was used as a reference strain for the detection of the tcdB gene and for the cytotoxicity assay used to detect the toxin TcdB (5).
DNA preparation.
For PCR assays, the template nucleic acid was extracted by use of an Instagene Matrix kit (Bio-Rad, Ivry, France). For Southern blot analysis, genomic DNAs were prepared by a standard phenol-chloroform procedure, as described by Wren and Tabaqchali (46).
Materials and conditions used for amplifications.
Primer sequences (Genome Express, Meylan, France) and PCR fragment lengths are given in Table 1. All PCR experiments used deoxynucleoside triphosphates (dNTPs) and Taq DNA polymerase from Amersham Biosciences (Orsay Cedex, France) and were carried out in a GeneAmp 9700 PCR system (PE Applied Biosystems, Foster City, Calif.). With the exception of PCR ribotyping, the amplified DNA products were analyzed by 0.8% agarose gel electrophoresis and comparison to a UV table after ethidium bromide staining.
TABLE 1.
Primers used for amplification
| PCR product | Primer | Sequence (5′-3′) | Fragment length (kb) |
|---|---|---|---|
| 16S rDNA | PG48 | CTCTTGAAACTGGGAGACTTGA | 0.27 |
| B | CCGTCAATTCATTTAAGTTT | ||
| cdtA | Cdta pos | TGAACCTGGAAAAGGTGATG | 0.353 |
| Cdta rev | AGGATTATTTACTGGACCATTTG | ||
| cdtB | Cdtb pos | CTTATTGCAAGTAAATACTGAG | 0.49 |
| Cdtb rev | ACCGGATCTCTTGCTTCAGTC | ||
| tcdB | NK104 | GTGTAGCAATGAAAGTCCAAGTTTACGC | 0.2 |
| NK105 | CACTTAGCTCTTTGATTGCTGCACCT | ||
| A3a | A3C | TATTGATAGCACCTGATTTATATACAAG | 3.1 |
| A4N | TTATCAAACATATATTTTAGCCATATATC | ||
| B1a | B1C | AGAAAATTTTATGAGTTTAGTTAATAGAAA | 3.1 |
| B2N | CAGATAATGTAGGAAGTAAGTCTATAG | ||
| AP-PCR | AP4 | TCACGCTGCA | |
| PCR ribotyping | 16S | GTGCGGCTGGATCACCTCCT | |
| 23S | CCCTGCACCCTTAATAACTTGACC |
A3 and B1 are PCR fragments that are amplified for toxinotyping.
Amplification of genes for 16S rRNA.
Primers PG48 and B, derived from the C. difficile 16S rRNA gene, were used to confirm the identification of strains by PCR (24) and to control the quality of crude DNA extracts (positive PCR controls). 16S rRNA gene amplification confirmed that all 369 strains had been correctly identified as C. difficile. For all experiments, all three reference strains gave the expected results.
Amplification of cdtA and cdtB genes.
PCR primers designed to amplify regions of the binary toxin genes cdtA and cdtB, previously described by Stubbs et al. (45), were used. PCRs were performed in 50-μl mixtures (50 mM KCl, 10 mM Tris-HCl [pH 9], 0.1% Triton X-100, 1.5 mM MgCl2, a 200 μM concentration of each dNTP, a 0.15 μM concentration of each primer, 1 U of Taq polymerase, and 5 μl of template DNA). PCR amplifications were performed with 30 cycles of 94°C for 45 s, 52°C for 1 min, and 75°C for 1 min 20 s.
Detection of cdtA and cdtB by Southern blot analysis.
Genomic DNAs were digested with HindIII (Ozyme, Saint Quentin en Yvelines, France), and the restriction fragments were separated by electrophoresis in a 0.8% agarose gel. They were then transferred to a nylon membrane (Hybond N; PharmaciaBiotech) and probed with internal fragments of the cdtA and cdtB genes (353 and 490 bp, respectively) from strain CD196. The probe fragments were obtained by PCR as described above and were labeled with [32P]dCTP (Boehringer, Mannheim, Germany) according to the manufacturer's instructions.
Detection of binary toxin CDT by Western blotting and ADP-ribosyltransferase assay.
Western blotting was performed as described by Popoff et al. (36). All strains that were positive for cdtA and cdtB were cultured in Wilkins-Chalgren broth for 48 h. Proteins were then precipitated from the culture supernatant with 70% ammonium sulfate (32, 36). These supernatant proteins (30 μg) were dissolved in 200 μl of distilled water, dialyzed overnight against phosphate-buffered saline, and concentrated with Aquacide II (Calbiochem Biosciences, Inc., La Jolla, Calif.). They were then subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis in 10% acrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked by incubation for 1 h in 5% skim milk powder in phosphate-buffered saline and were incubated overnight at room temperature with rabbit polyclonal antisera (1:5,000) specific for the enzymatic (Ia) or binding (Ib) component of the C. perfringens iota toxin. Bound antibodies were detected with peroxidase-labeled protein A and a Signal Plus kit (Pierce Chemical Co., Rockford, Ill.).
For the ADP-ribosyltransferase assay (45), concentrated supernatant proteins (10 μg) prepared as described above were incubated at 37°C for 1 h with 50 μl of a mixture containing 50 mM triethanolamine-HCl (pH 7.5), 5 mM MgCl2, 10 mM dithiothreitol, 10 mM thymidine, and [32P]NAD (106 cpm per reaction). The reaction mix also contained brain extract as a source of actin (10 μg). The proteins were precipitated by the addition of 20 μl of a solution containing 1 mg of bovine serum albumin ml−1, 10% (wt/vol) SDS, and 0.5 ml of trichloroacetic acid (10% [wt/vol]); the reaction mixture was then incubated on ice for 1 h. The precipitate was dissolved in distilled water and subjected to SDS-polyacrylamide gel electrophoresis in 10% acrylamide gels. After radiography, qualitative differences in ADP-ribosyltransferase activity were assessed visually.
Production of TcdA and TcdB in vitro.
Strains were incubated anaerobically in brain heart infusion broth for 5 to 7 days as described by Barbut et al. (4). TcdA production was detected by use of the Premier C. difficile toxin A assay (Meridian Diagnostics, Cincinnati, Ohio) according to the manufacturer's instructions. TcdB was detected by a cytotoxicity assay using MRC-5 monolayers (bioMérieux, Marcy l'Etoile, France) as follows. Supernatants were filtered through a 0.45-μm-pore-size filter and 10× dilutions were applied to cells grown in 96-well plates. The characteristic cytotoxic effect (cell rounding) was observed after 18 h at 37°C (4).
Amplification of gene for TcdB.
We used the primer set NK104 and NK105 from the nonrepetitive part of the C. difficile tcdB gene for its amplification (7, 28). PCRs were performed in 100-μl mixtures (50 mM KCl, 10 mM Tris-HCl [pH 8.8], 1.5 mM MgCl2, a 200 μM concentration of each dNTP, 92 ng of each primer for tcdB, 2.5 U of Taq polymerase, and 10 μl of template DNA) (7, 28). PCR amplifications were performed with 40 cycles of 95°C for 15 s, 62°C for 2 min, and 72°C for 40 s.
Toxinotyping.
Toxinotyping was used to search for genetic variations in parts of the tcdA and tcdB genes. The oligonucleotide primers used were based on the published sequence of the VPI 10463 pathogenicity region (EMBL database accession no. X928982) and were described previously by Rupnik et al. (38, 39, 40). The first 3.1 kb of tcdB (PCR fragment B1) and a 3.1-kb fragment spanning the repetitive region of tcdA (PCR fragment A3), obtained with primer sets A3C-A4N and B1C-B2N, respectively, were screened for variations. Amplification was done for 30 cycles (fragment B1) or 35 cycles (fragment A3). PCR programs were as follows: initial denaturation for 3 min at 93°C; annealing and extension for 8 min at 47°C, followed by 4 s of denaturation at 93°C; and then a final extension cycle of 10 min at 47°C. The resulting amplicons were separated by electrophoresis in 0.8% gel agarose and were visualized under UV light by ethidium bromide staining. The products were also analyzed after restriction enzyme gel electrophoresis using EcoRI (for PCR fragment A3) or AccI and HincII (for PCR fragment B1). The toxinotype was then determined according to the combination of restriction patterns for PCR fragments A1 and B3 (38, 39; http://www.uni-lj.si/∼bfbcdiff).
Serogrouping.
Serogrouping of the C. difficile strains was performed by the method of Delmée et al. (19, 20). Eleven antisera specific for serogroups A1, A5, A8, A9, A10, C, D, F, G, H, and K were used for enzyme-linked immunosorbent assays, as described previously (20).
AP-PCR.
For arbitrarily primed PCRs (AP-PCRs), the 10-mer primer AP4 was used and PCRs were performed in 100-μl volumes (50 mM KCl, 10 mM Tris-HCl [pH 8.8], 4 mM MgCl2, a 200 μM concentration of each dNTP, 25 pmol of primer, 2.5 U of Taq polymerase, and 10 μl of template DNA). PCR amplifications were performed for 45 cycles of 1 min at 94°C, 1 min at 36°C, and 2 min at 75°C. Strains with patterns differing by at least one high-intensity band were assigned to different types (10).
PCR ribotyping.
Specific primers complementary to the 3′ end of the 16S rRNA gene and the 5′ end of the 23S rRNA gene were used to amplify the variable-length intergenic spacer region (10). The sequence of the 16S primer corresponded to bases 1482 to 1501 of the 16S rRNA gene and that of the 23S primer corresponded to bases 1 to 24 of the 23S rRNA gene of C. difficile (10). PCRs were performed in 100-μl volumes (50 mM KCl, 10 mM Tris-HCl [pH 8.8], 1.5 mM MgCl2, a 200 μM concentration of each dNTP, 25 pmol of each primer, 2.5 U of Taq polymerase, and 10 μl of template DNA). PCR amplifications were performed for 35 cycles of 1 min at 94°C, 1 min at 57°C, and 1 min at 75°C. PCR products were analyzed in a 3% Resophor agarose gel (Bio-Rad, Ivry, France). As described above, strains with patterns differing by at least one high-intensity band were assigned to different types.
PFGE analysis.
For pulsed-field gel electrophoresis (PFGE), strains were cultured in prereduced Wilkins-Chalgren broth for 17 h at 37°C and pelleted by centrifugation at 5,000 × g. Bacterial cells were embedded in agarose plugs and lysed by incubation for 18 h at 37°C with lysozyme-lysostaphin and overnight at 50°C with proteinase K, using a GenePath Group A kit (Bio-Rad) according to the manufacturer's recommendations (6). Plugs were digested with 25 U of SmaI overnight at 25°C. Electrophoresis was performed in a CHEF DRII instrument (Bio-Rad) with running conditions of 50 to 500 V for 19 h (program 14).
RESULTS
Detection of cdtA and cdtB.
Of the 369 strains studied (isolated from 288 patients with suspected AAD or AAC), 22 (6%) strains, isolated from 20 patients, gave amplicons of the expected sizes. These results were confirmed by Southern blotting (data not shown): a hybridization fragment was obtained with the cdtA probe and two hybridization fragments were observed with the cdtB probe, consistent with the HindIII restriction site in the cdtB probe. For each probe, all strains harbored the same hybridization profile.
The 22 positive strains corresponded to patients who were hospitalized in 11 of the 17 institutions. In the five hospitals in which >25 C. difficile strains were isolated, the prevalence of binary toxin-positive strains ranged from 0 to 13%.
Production and activity of the binary toxin.
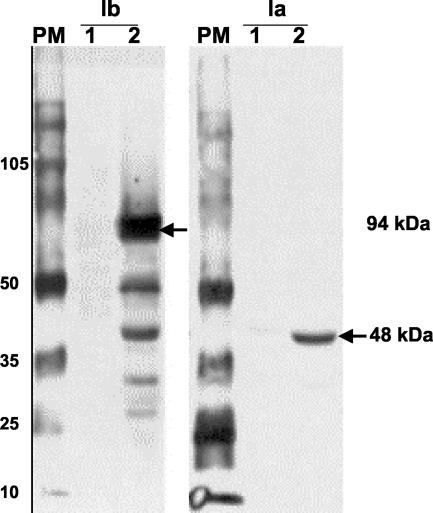
Strains with cdt genes were tested for production of the binary toxin (Table 2). On Western blots (Fig. 1), 19 of the 22 strains reacted with antisera against the iota toxin of C. perfringens (anti-Ia and anti-Ib). Strains 22270, 37405, and 48961 did not react with the anti-Ia and anti-Ib antisera.
TABLE 2.
Toxin production and phenotypic and genotypic characterization of strains that are positive for cdtA and cdtB
| Strain(s)a | Binary toxin production and activityb
|
Production of TcdB | Phenotypic or genotypic markerd
|
|||||
|---|---|---|---|---|---|---|---|---|
| Western blot analysis | ADP-ribosyltransferase activity | Toxinotype | Serogroup | PCR ribotype | AP-PCR type | PFGE type | ||
| 58850*, 55312, 54953* | + | + | + | V | NT | A | a | P1 |
| 6583 | + | − | + | V | NT | A | a | P1 |
| 50079 | + | + | + | V | NT | A | a | P3 |
| 54402, 20222, 55832 | + | + | + | V | NT | A | a | P2 |
| 9459 | + | + | + | V | NT | A | b | nt |
| 22270 | − | − | + | V | NT | A | c | nt |
| 37405 | − | − | + | V | NT | A | e | nt |
| 38780 | + | − | + | V | NT | A | f | nt |
| 52582 | + | + | + | V | NT | A | g | nt |
| 13376 | + | + | + | VII | NT | B | h | nt |
| 17153 | + | ++ | + | IX | NT | C | i | nt |
| 37078 | + | ++ | +c | IX | C | G | d | nt |
| 48961** | − | − | + | IV | NT | E | j | nt |
| 20183 | + | + | + | IV | A1 | E | j | nt |
| 50942** | + | + | + | IV | A1 | E | f | nt |
| 11419, 17852 | + | ++ | + | III | G | D | d | nt |
| 50232 | + | + | + | XIII | A5 | F | k | nt |
*, strains from patient A; **, strains from patient B.
−, negative; +, low levels of activity; ++, high levels of activity.
Atypical cytotoxic effect.
NT, not typeable; nt, not tested.
FIG. 1.
Detection of CDTa (94 kDa) and CDTb (48 kDa) components by Western blotting using antisera against Ia and Ib from C. perfringens. Lanes: PM, Rainbow molecular marker RPN 800 (Amersham Biosciences); 1, strain 22270; 2, strain 37078.
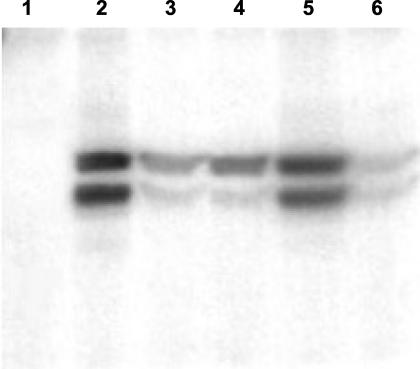
ADP-ribosyltransferase activity (Fig. 2) was detected in 17 of the 22 strains tested. Five strains (including the three giving negative results on Western blots) displayed no ADP-ribosyltransferase activity (strains 6583, 22270, 37405, 38780, and 48961).
FIG. 2.
ADP-ribosyltransferase assay using a thymidine-containing brain extract as a source of actin. Lanes: 1, strain 8549 (negative control); 2, strain CD196 (positive control); 3, strain 13376; 4, strain 17153; 5, 11419; 6, strain 9459. The two bands visible on the figure represent the two isoforms of actin G.
Detection of tcdB gene and production of TcdA and TcdB in vitro.
We tested for the presence of the toxin gene coding for TcdB in all 369 strains: the nonrepetitive domain of the tcdB gene was detected in 214 strains.
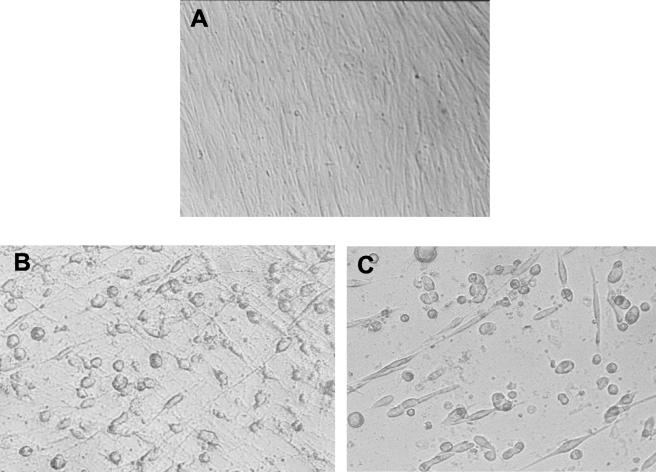
The production of TcdA and TcdB was tested only for the 22 binary toxin-positive isolates. The Premier C. difficile Toxin A assay was positive for all strains except strain 13376. For this strain, the A3-targeted PCR used for toxinotyping indicated a deletion of 300 bp in the repetitive domain of the tcdA gene. All 22 strains gave positive results in the cytotoxicity assay for detecting TcdB. One strain (37078) had a cytotoxic effect on MRC-5 cells that differed from that of the reference strain, ATCC 43596 of serogroup C. This strain induced rounding of the cells with no spindle formation and tended to promote the formation of a discrete cluster of cells (Fig. 3).
FIG. 3.
Cytotoxic effect of TcdB on MRC-5 monolayers. (A) Negative control. (B) Reference strain ATCC 43596 (serogroup C). (C) Strain 37078.
Toxinotyping.
All binary toxin-positive strains had changed tcdA and/or tcdB genes, and six different toxinotypes (III, IV, V, VII, IX, and XIII) were found (Table 2).
Phenotypic and genotyping characterization of binary toxin-positive strains.
Only six strains could be typed with the antisera used. They belonged to the following serogroups: A1 (strains 20183 and 50942), G (strains 11419 and 17582), A5 (strain 50232), and C (strain 37078) (Table 2). The remaining 16 strains were nontypeable. With molecular typing methods, seven PCR ribotypes (A, B, C, D, E, F, and G) and 11 AP-PCR profiles (a to k) were identified. PFGE analysis was used to differentiate the eight strains of toxinotype V and both ribotype A and AP-PCR profile a. Three different PFGE profiles were distinguished (P1, P2, and P3). The findings for the different markers related well and showed that 16 of the 22 binary toxin-positive strains were epidemiologically unrelated
The three patients who had strains with identical profiles (toxinotype V, PCR ribotype A, AP-PCR profile a, and PFGE profile P1) were hospitalized in three different institutions. Two pairs of strains were recovered from two patients: in one case (strains 53850 and 54953), the strains had identical profiles consistent with a relapse of infection, whereas the strains isolated from the other patient (48961 and 50942) were different by serogrouping and AP-PCR, indicating a reinfection.
DISCUSSION
Little is known about the clinical relevance and pathogenic role of the ADP-ribosylating toxin CDT in C. difficile infections. C. difficile induces diseases of varying severity, from mild diarrhea to PMC, toxic megacolon, and even fulminant colitis. Differences in the levels of production of toxins TcdA and TcdB alone cannot account for the wide spectrum of clinical presentations. Georges et al. (21) reported that there were no significant differences between clinical presentations according to the titer of the cytotoxin. Similarly, there was no correlation between virulence in a hamster model of AAC and the production of TcdA and TcdB in vitro (12). Therefore, the binary toxin CDT produced by some C. difficile strains could be responsible for these differences. CDT is a potent cytotoxin, and the damage it causes to mucosal barrier functions may prepare the way for the action of typical clostridial cytotoxins. Alternatively, CDT may act in synergy with other toxins, depolymerizing the actin cytoskeleton by a complementary mechanism. C. difficile strain CD196 was investigated because it causes severe PMC and it is possible that the production of this additional toxin exacerbates the symptoms of PMC (36).
In this study, we found that 22 of 369 strains (6%) isolated from 288 patients harbored binary toxin genes. An interest in binary toxin-producing strains of C. difficile has only developed in recent years (43), and the prevalence of binary toxin genes varies from one country to another. In 2000, a study indicated that 6.4% of the toxigenic isolates of C. difficile referred to the Anaerobe Reference Unit for UK hospitals possessed the cdtA and cdtB genes (45). In 2003, Rupnik et al. (42) found a prevalence of 1.6% for the toxin in 310 isolates from various hospitals in Japan and Korea and from healthy infants in Indonesia. In France, Branger et al. (C. Branger, P. Ferron, L. Noussair, N. Lambert-Zikowsky, Abstr. 21st Interdisciplinary Meet. Anti-Infect. Chemother., abstr. 129/P1, 2001) reported that 9.3% of the toxigenic strains from a group of patients with diarrhea harbored the cdtA and cdtB genes. In our series of 369 strains, 214 were toxigenic (positive by tcdB PCR). The deduced prevalence of binary toxin genes in these 214 toxigenic strains (10.3%) is high, but it is similar to that obtained by Branger et al. (Abstr. 21st Interdisciplinary Meet. Anti-Infect. Chemother.). Binary toxin-producing isolates have also been isolated from patients throughout Europe and were recently reported for strains isolated in Asia, indicating that such strains are widespread (42-45). Discrepancies in the findings of prevalence reports may be partly due to the differences in patient (symptomatic or not, adult or children) or strain selection. In addition, as was shown recently by Rupnik et al., we observed that the distribution of binary toxin-positive strains can vary between hospitals (42).
The locations of the cdtA and cdtB genes in the chromosomes of C. difficile strains have not yet been reported. The identical hybridization fragments observed for all strains with either a cdtA or cdtB probe strongly suggest that our strains have a binary toxin locus inserted at the same site in the chromosome.
Only a few studies have detected the production of the CDT in strains with binary toxin genes (33, 45). The culture supernatants of most PCR-positive strains reacted with anti-Ia and anti-Ib antisera for the detection of CDTa and CDTb, but those of three strains did not. Four of the 13 strains tested by Stubbs et al. also failed to react with anti-Ia and anti-Ib antisera (45). There may be several reasons for the observed lack of reaction, including (i) differences in amino acid sequences, (ii) mutations in the gene promoters, and (iii) little or no expression of the cdtA and cdtB genes. In addition, no ADP-ribosylating activity was observed for five strains, including the three strains that were negative in Western blots. Similarly, C. difficile strains with probable silent genes for binary toxins were reported previously (33). Interestingly, two ADP-ribosyltransferase-negative strains (6583 and 38780) produced the CDTa and CDTb components. Only one similar isolate with an expressed binary toxin but with low activity was reported previously (45). Such strains could be used to identify the amino acids that are required for enzymatic activity.
Strain 37078 (toxinotype IX) had an atypical effect on MRC-5 cells which was very different from that of a reference strain (ATCC 43596). This atypical effect was, however, similar to that of TcdB of serogroup F strains (toxinotype VIII), although strain 37078 does not belong to serogroup F. The atypical effect of serogroup F strains has been shown to be similar to the effect of the lethal toxin of Clostridium sordellii (16). It has been suggested that the TcdB toxin in these strains may be a functional hybrid of the TcdB toxin of C. difficile and the lethal toxin of C. sordellii. An atypical cytotoxic effect has also been described for strains of toxinotype X (41), but not for strains of toxinotype IX. We are currently investigating the tcdB gene of this particular strain.
As reported by others, binary toxin genes were detected only in strains with a tcdA and/or tcdB gene that was significantly different from that of VPI10463, which is the reference strain for toxinotyping (toxinotype 0) (39, 45). The only exception was reported recently by Spigaglia et al., who described one binary toxin-producing strain belonging to toxinotype 0 (44). In addition, our study confirms that toxinotype VIII is the only group of strains with significant changes in the large clostridial toxin genes that do not possess the genes for the binary toxin (45).
The characterization of strains indicated that most strains with CDT genes belonged to the rarer serogroups. Various genotypic typing methods have been employed for epidemiological studies of C. difficile. We used those that were reported to have the most discriminatory power, such as PCR ribotyping, AP-PCR, and PFGE (10). The results of the combined markers were consistent and showed that most binary toxin-producing strains were epidemiologically unrelated and not derived from a common ancestor. Moreover, consecutive strains isolated in the same setting corresponded to different types. The results concerning the two pairs of strains isolated from two patients indicated a relapse and a reinfection. Previous studies have reported that almost half of all clinical recurrences are due to reinfections with a different strain (5).
In conclusion, we showed here that the prevalence of binary toxin genes in strains of C. difficile isolated in the Paris area is 6% and that these genes are only present in variant strains. Clinical studies are now needed to define the exact role of the binary toxin of C. difficile in human pathogenesis.
Acknowledgments
This study was supported by grant no. EA2392 from the Unité Propre de Recherche de l'Enseignement Supérieur (UPRES).
We thank M. R. Popoff (Pasteur Institute, Paris, France) for providing strain CD196 and the antisera against the iota toxin of C. perfringens. We are deeply indebted to Maja Rupnik for her comments and discussions about the manuscript.
REFERENCES
- 1.Aktories, K., and A. Wegner. 1992. Mechanisms of the cytopathic action of actin-ADP-ribosylating toxins. Mol. Microbiol. 6:2905-2908. [DOI] [PubMed] [Google Scholar]
- 2.Babudieri, S., S. P. Borrielo, A. Pantosti, I. Luzzi, G. P. Testore, and G. Panichi. 1986. Diarrhoea associated with toxigenic Clostridium spiroforme. J. Infect. 12:278-279. [DOI] [PubMed] [Google Scholar]
- 3.Banno, Y., T. Kobayashi, H. Kono, K. Watanabe, K. Ueno, and Y. Nozawa. 1984. Biochemical characterization and biologic actions of two toxins (D-1 and D-2) from Clostridium difficile. Rev. Infect. Dis. 6:S11-S20. [DOI] [PubMed] [Google Scholar]
- 4.Barbut, F., C. Kajzer, N. Planas, and J. C. Petit. 1993. Comparison of three enzyme immunoassays, a cytotoxicity assay, and toxigenic culture for diagnosis of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 31:963-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbut, F., A. Richard, K. Hamadi, V. Chomette, B. Burghoffer, and J. C. Petit. 2000. Epidemiology of recurrences or reinfections of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2386-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbut, F., V. Lalande, B. Burghoffer, H. V. Thien, E. Grimprel, and J. C. Petit. 2002. Prevalence and genetic characterization of toxin A variant strains of Clostridium difficile among adults and children with diarrhea in France. J. Clin. Microbiol. 40:2079-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barroso, L. A., S. Z. Wang, C. J. Phelps, J. L. Johnson, and T. D. Wilkins. 1990. Nucleotide sequence of Clostridium difficile toxin B gene. Nucleic Acids Res. 18:4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartlett, J. G. 1994. Clostridium difficile: history of its role as an enteric pathogen and the current state of knowledge about the organism. Clin. Infect. Dis. 18:S265-S272. [DOI] [PubMed] [Google Scholar]
- 9.Bartlett, J. G. 2002. Antibiotic-associated diarrhea. N. Engl. J. Med. 346:334-339. [DOI] [PubMed] [Google Scholar]
- 10.Bidet, P., F. Barbut, V. Lalande, B. Burghoffer, and J. C. Petit. 1999. Development of the new PCR-ribotyping method for Clostridium difficile. FEMS Microbiol. Lett. 175:261-266. [DOI] [PubMed] [Google Scholar]
- 11.Borriello, S. P., and R. J. Carman. 1985. Clostridial diseases in the gastrointestinal tract in animals, p. 191-221. In S. P. Borriello (ed.), Clostridia in gastrointestinal disease. CRC Press, Boca Raton, Fla.
- 12.Borriello, S. P., J. M. Ketley, T. J. Mitchell, F. E. Barclay, A. R. Welch, A. B. Price, and J. Stephen. 1987. Clostridium difficile—a spectrum of virulence and analysis of putative virulence determinants in the hamster model of antibiotic-associated colitis. J. Med. Microbiol. 24:53-64. [DOI] [PubMed] [Google Scholar]
- 13.Bowen, K. E., L. V. McFarland, R. N. Greenberg, M. M. Ramsey, K. E. Record, and J. Svenson. 1995. Isolation of Clostridium difficile at a university hospital: a two-year study. Clin. Infect. Dis. 20:S261-S262. [DOI] [PubMed] [Google Scholar]
- 14.Braun, V., T. Hundsberger, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29-38. [DOI] [PubMed] [Google Scholar]
- 15.Cartmill, T. D. I., K. Orr, R. Freeman, P. R. Sisson, and N. F. Lightfoot. 1992. Nosocomial infection with Clostridium difficile investigated by pyrolysis mass spectrometry. J. Med. Microbiol. 37:352-356. [DOI] [PubMed] [Google Scholar]
- 16.Chaves-Olarte, E., P. Low, E. Freer, T. Norlin, M. Weidmann, C. von Eichel-Streiber, and M. Thelestam. 1999. A novel cytotoxin from Clostridium difficile serogroup F is a functional hybrid between two other large clostridial cytotoxins. J. Biol. Chem. 274:11046-11052. [DOI] [PubMed] [Google Scholar]
- 17.Cohen, S. H., Y. J. Tang, and J. Silva. 2000. Analysis of the pathogenicity locus in Clostridium difficile strains. J. Infect. Dis. 181:959-963. [DOI] [PubMed] [Google Scholar]
- 18.Considine, R. V., and L. L. Simpson. 1991. Cellular and molecular actions of binary toxins possessing ADP-ribosyltransferase activity. Toxicon 29:913-936. [DOI] [PubMed] [Google Scholar]
- 19.Delmée, M., Y. Laroche, V. Avesani, and G. Cornelis. 1986. Comparison of serogrouping and polyacrylamide gel electrophoresis for typing of Clostridium difficile. J. Clin. Microbiol. 24:991-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delmée, M., C. Depitre, G. Corthier, A. Ahoyo, and V. Avesani. 1993. Use of an enzyme-linked immunoassay for Clostridium difficile serogrouping. J. Clin. Microbiol. 31:2526-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georges, W. L., R. D. Rolfe, and S. M. Finegold. 1982. Clostridium difficile and its cytotoxin in feces of patients with antimicrobial agent-associated diarrhea and miscellaneous conditions. J. Clin. Microbiol. 15:1049-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geric, B., S. Johnson, D. N. Gerding, M. Grabnar, and M. Rupnik. 2003. Frequency of binary toxin genes among Clostridium difficile strains that do not produce large clostridial toxin. J. Clin. Microbiol. 41:5227-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gülke, I., G. Pfeifer, J. Liese, M. Fritz, F. Hofmann, K. Aktories, and H. Barth. 2001. Characterization of the enzymatic component of the ADP-ribosyltransferase toxin CDTa from Clostridium difficile. Infect. Immun. 69:6004-6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gumerlock, P. H., Y. J. Tang, F. J. Meyers, and J. Sylva, Jr. 1991. Use of the polymerase chain reaction for the specific and direct detection of Clostridium difficile in human feces. Rev. Infect. Dis. 13:1053-1060. [DOI] [PubMed] [Google Scholar]
- 25.Hateway, C. L. 1990. Toxigenic clostridia. Clin. Microbiol. Rev. 3:66-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Just, I., J. Selzer, M. Wilm, C. von Eichel-Streiber, M. Mann, and K. Aktories. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375:500-503. [DOI] [PubMed] [Google Scholar]
- 27.Just, I., M. Wilm, J. Selzer, G. Rex, C. von Eichel-Streiber, and M. Mann. 1995. The enterotoxin from Clostridium difficile (toxA) monoglycosylates the Rho proteins. J. Biol. Chem. 270:13932-13936. [DOI] [PubMed] [Google Scholar]
- 28.Kato, N., C. Y. Ou, H. Kato, S. L. Bartlett, V. K. Brown, Jr., V. R. Dowell, and K. Ueno. 1991. Identification of toxigenic Clostridium difficile by the polymerase chain reaction. J. Clin. Microbiol. 29:33-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly, C. P., C. Pothoulakis, and J. T. LaMont. 1994. Clostridium difficile colitis. N. Engl. J. Med. 330:257-262. [DOI] [PubMed] [Google Scholar]
- 30.McFarland, L. V., M. E. Mulligan, R. Y. Kwok, and W. E. Stamm. 1989. Nosocomial acquisition of Clostridium difficile infection. N. Engl. J. Med. 320:204-210. [DOI] [PubMed] [Google Scholar]
- 31.Olson, M., C. J. Shanholtzer, J. T. Lee, Jr., and D. N. Gerding. 1994. Ten years of prospective Clostridium difficile disease surveillance and treatment at the Minneapolis VA Medical Center, 1982-91. Infect. Control Hosp. Epidemiol. 15:371-381. [DOI] [PubMed] [Google Scholar]
- 32.Perelle, S., M. Gibert, P. Boquet, and M. R. Popoff. 1993. Characterization of Clostridium perfringens Iota toxin genes and expression in Escherichia coli. Infect. Immun. 61:5147-5156. (Author's correction, 63:4967, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perelle, S., M. Gibert, P. Bourlioux, G. Corthier, and M. R. Popoff. 1997. Production of a complete binary toxin (actin-specific ADP-ribosyl transferase) by Clostridium difficile CD 196. Infect. Immun. 65:1402-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petit, L., M. Gibert, and M. R. Popoff. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7:104-110. [DOI] [PubMed] [Google Scholar]
- 35.Popoff, M. R., and P. Boquet. 1988. Clostridium spiroforme toxin is a binary toxin which ADP-ribosylates cellular actin. Biochem. Biophys. Res. Commun. 152:1361-1368. [DOI] [PubMed] [Google Scholar]
- 36.Popoff, M. R., E. J. Rubin, D. M. Gill, and P. Boquet. 1988. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect. Immun. 56:2299-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popoff, M. R., F. W. Milward, B. Bancillon, and P. Boquet. 1989. Purification of the Clostridium spiroforme binary toxin and activity of the toxin on HEp-2 cells. Infect. Immun. 57:2462-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rupnik, M., V. Braun, F. Soehn, M. Janc, M. Hofstetter, R. Laufenberg-Feldmann, and C. von Eichel-Streiber. 1997. Characterization of polymorphisms in the toxin A and B genes of Clostridium difficile. FEMS Microbiol. Lett. 148:197-202. [DOI] [PubMed] [Google Scholar]
- 39.Rupnik, M., V. Avésani, M. Janc, C. von Eichel-Streiber, and M. Delmée. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 36:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rupnik, M., J. S. Brazier, B. I. Duerden, M. Grabnar, and S. L. J. Stubbs. 2001. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology 147:439-447. [DOI] [PubMed] [Google Scholar]
- 41.Rupnik, M. 2001. How to detect Clostridium difficile variant strains in a routine laboratory. Clin. Microbiol. Infect. 7:417-420. [DOI] [PubMed] [Google Scholar]
- 42.Rupnik, M., N. Kato, M. Grabnar, and H. Kato. 2003. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J. Clin. Microbiol. 41:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rupnik, M., M. Grabnar, and B. Geric. 2003. Binary toxin producing Clostridium difficile strains. Anaerobes 9:289-294. [DOI] [PubMed] [Google Scholar]
- 44.Spigaglia, P., and P. Mastrantonio. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 40:3470-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stubbs, S., M. Rupnik, M. Gilbert, J. Brazier, B. Duerden, and M. R. Popoff. 2000. Production of an actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186:307-312. [DOI] [PubMed] [Google Scholar]
- 46.Wren, B. W., and S. Tabaqchali. 1987. Restriction endonuclease DNA analysis of Clostridium difficile. J. Clin. Microbiol. 25:2402-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]