Abstract
Quantification of hepatitis C virus (HCV) core antigen and RNA in serum samples leads to a highly variable ratio of both. It is not clear whether this is due to the inaccuracy of RNA quantification or whether both are independent parameters in a certain range. We established a real-time reverse transcription (RT)-PCR for HCV RNA that combines very high sensitivity with a large dynamic range and minimal standard deviations. The assay was calibrated with the first international standard, 96/790, and the international genotype panel for HCV from the National Institute of Biological Standardisation and Control. A linear readout was obtained between 200 and 5 × 107 IU/ml. The detection limit was 80 IU/ml, the reproducibility was <0.05 log, and the standard error within one run was <0.01. Comparison of the method with the Roche Monitor competitive RT-PCR revealed its high accuracy. The core protein concentration was determined within a range from 1.5 to 400 pg/ml by using the preliminary trak-C assay from Ortho Clinical Diagnostics. Correlating the HCV RNA levels with core antigen concentrations in 197 serum samples from 23 interferon-treated patients, a average ratio of 7,900 IU of HCV RNA per pg of core antigen was estimated, but the variability of this ratio exceeded largely the variability of the two assays, ranging from 50 to 20,000 IU/pg. Theoretically, HCV should contain ca. 43,000 IU of RNA/pg core. In conclusion, the core antigen assay seems to detect, in addition to complete virions, RNA-free core protein structures, which enhances its sensitivity (98% in this group). The variable ratio of RNA and core protein is not mainly due to standard deviations of quantification but could be an additional parameter for treatment follow-up and state of viral replication.
The accurate quantification of virus particles in the plasma of persons infected with hepatitis C virus (HCV), the so-called viral load, is essential for decisions on the therapy with interferon and ribavirin and the monitoring of this therapy (1, 5, 13). For this purpose, several tests for the detection of HCV RNA genomes are commercially available, which lead to sometimes conflicting results for HCV RNA copies per milliliter or international units per milliliter. It appears that some divergences are caused by saturation phenomena in competitive nucleic acid amplification tests (4). Different reactivities of the assays with various HCV genotypes other than genotype 1 may also contribute to the problem (3, 11). With the real-time technology for in situ signal generation during DNA amplification (e.g., using the TaqMan or LightCycler system), saturation phenomena can be overcome.
Some efforts have been made to establish real-time protocols for HCV quantification by using the SYBR Green format on LightCycler (17) or the TaqMan technology (8, 10). These published real-time-based techniques are two-step procedures with separate reverse transcription (RT), or they use only the nonspecific SYBR Green for generation of the signal.
One aim of our study was to establish a convenient protocol by using all the options of the LightCycler for improving specificity, such as hybridization probes and a touch-down annealing program for temperature, and to increase the sensitivity by optimized one-tube RT-PCR conditions. After this optimization, the assay was validated with the international standard for HCV RNA and a World Health Organization (WHO) reference panel for HCV genotypes, and results were compared with those of the Roche (Mannheim, Germany) Monitor HCV assay.
HCV particles contain a core protein which most likely encapsidates the viral RNA. Recently, a sensitive enzyme immunoassay (EIA) for detection of the HCV core protein has been developed (2, 18) which detects 1.5 pg of core protein even in the presence of HCV antibodies. It is not clear whether there exists a defined ratio of the number of HCV RNA genomes to the number HCV core protein molecules, but it has been suggested that such an assay of HCV core antigen may be suitable for monitoring of interferon and ribavirin therapy (2, 20). Because conventional RNA quantification methods show a high standard deviation, it is not clear whether the variable ratio of HCV RNA and core antigen is due to the inaccuracy of the assays or whether both have no simple stoichiometric relation. Since our in-house real-time PCR seemed to be very accurate, it appeared suitable to assess the ratio of RNA to core protein more precisely than before.
MATERIALS AND METHODS
Patient sera.
Serial serum samples (256) came from 23 patients involved in a clinical study on the kinetics of HCV RNA under interferon and ribavirin therapy conducted by C. G. Schüttler, W. H. Gerlich, and J. Lohmeyer at the university hospital of the Justus Liebig University. The study was approved by the ethics committee of the hospital.
Twenty-one sera with HCV RNA from the diagnostic laboratory of our institute were tested in parallel by the competitive RT-PCR Amplicor Monitor test kit (Roche Diagnostics) and in duplicate (except for two sera) by the LightCycler assay.
All sera were immediately stored at −20°C, aliquoted, and thawed only once before HCV RNA testing and the HCV core antigen assay.
Primers and probes.
The primers used were NCR-s (5′-TGC GGA ACC GGT GAG TAC A; positions −193 to −175) and NCR-as (5′-CTT AAG GTT TAG GAT TCG TGC TCA T; positions 24 to 1) (nucleotide positions are relative to the sequence of GenBank accession no. M58335, first codon). The hybridization probes were HCV-NCR-LR (5′-LC Red640-TGC CTG ATA GGG TGC TTG CGA GT-p; positions −52 to −30) and HCV-NCR-FL (5′-GGT CGC GAA AGG CCT TGT GGT A-FL; positions −75 to −54). Primer and probe sequences were selected for optimal conservation between HCV isolates.
RNA extraction and preparation.
Viral RNA was extracted from 140 μl of serum or plasma by using the QiaAmp viral RNA kit (catalog no. 52904; Qiagen, Hilden, Germany) according to the manufacturer's instructions. To avoid the addition of salt to the RT-PCR mixture, one step was altered in the standard protocol: instead of elution with 60 μl of the proposed elution buffer, we used 60 μl of RNase-free water. Because the target sequence in the end of the 5′ noncoding region (NCR) region of the HCV genome contains stable secondary structures of the internal ribosomal entry site (7, 12), 10 μl of RNA was mixed with 1 μl (10 pmol) of antisense primer, incubated for 1 min at 94°C for breaking secondary structures and 1 min at 56°C for annealing of the primer, and then stored on ice. From this freshly prepared RNA extract, 9 μl was used for the one-tube RT-PCR.
One-tube RT and amplification.
The RT-PCR was performed by using the LightCycler RNA amplification kit for tests with hybridization probes (catalog no. 2015145; Roche Diagnostics) with 6.5 mM MgCl2. The hybridization probes were added in unequal molarity with 8 pmol of the detection probe (HCV-NCR-LR) and 4 pmol of the anchor probe (HCV-NCR-FL). Adding 10 pmol each of primer (NCR-s and NCR-as) also results in an asymmetric concentration, since the same amount of primer NCR-as was already added for preincubation of the RNA extract. This results in an increased efficiency of amplification.
The RT-PCR conditions were 12 min at 55°C for RT, 30 s at 95°C for denaturation of the reverse transcriptase, followed by 45 cycles of 3 s at 95°C (slope 20°C/s) and 7 s at 64°C (first annealing temperature) to 54°C (touchdown second annealing temperature), with a decreasing step size of 1°C per cycle, and extension for 14 s at 72°C (slope 2°C/s). After the last cycle, a melting curve analysis was performed from 50 to 90°C. The fit point mode for the LightCycler was used for evaluation.
HCV core antigen.
For the measurement of the HCV core antigen, we used a preliminary second-generation HCV core EIA (trak-C; Ortho Clinical Diagnostics, Raritan, N.J.) with an optical density cutoff of 1.5 pg of core antigen/ml. Samples with core antigen values above 150 pg/ml were diluted and measured again.
HCV RNA standard.
To calibrate the quantification, a highly HCV-positive plasma sample from a blood donor was tested in parallel to the first international standard, 96/790, for HCV (500,000 IU/ml) (15) and then diluted with HCV-negative plasma to a concentration of 106 IU/ml. The calibration of this secondary standard was validated in an international trial for quantifying the WHO standard 96/790 (15).
RESULTS AND DISCUSSION
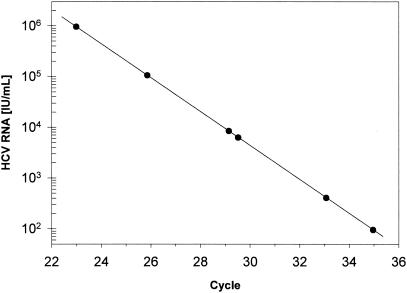
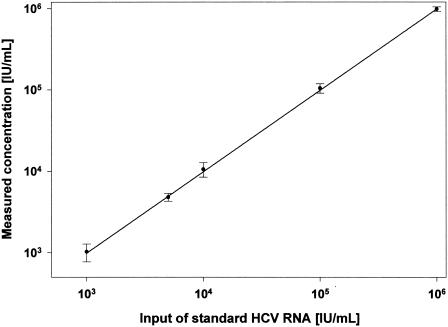
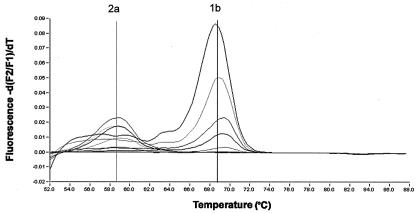
The standard LightCycler protocol includes the hybridization of two hybridization probes to one of the amplified strands. One probe is labeled with a fluorescent dye emitting a shorter wavelength, e.g., fluorescein, and the other probe is labeled with a dye emitting a longer wavelength, e.g., LightCycler red. The fluorescence resonance energy transfer between both probes depends on close proximity, and this occurs only if they are both bound to their target sequence. Thus, the long-wave fluorescence can be used as quantitative signal for the amount of amplified target DNA. The signal strength, which is significantly higher than the nonspecific signal, defines the crossing point, i.e., the number of amplification cycles necessary to reach that signal. With the described RT-PCR protocol, an accurate linear relationship between cycle number at the crossing point and the amount of reference HCV RNA from 102 to 106 IU/ml was obtained (Fig. 1). The intratest precision, calculated as the standard error of the calibration curve, was <0.01 (r2 > 0,9998), and the intertest precision was <0.05 log (Fig. 2). The LightCycler system offers the option to measure the temperature at which binding of one or both of the probes to the target DNA is opened. The decrease of the fluorescence signal with temperature is recorded as the first derivative, df1/dt, for better definition of the melting point (Fig. 3). The melting point analysis showed a melting point of 68.5 to 69.8°C for the hybridization probes with all HCV genotypes except for two (see below).
FIG. 1.
Calibration curve obtained with six dilutions of the secondary standard serum plotted as cycle number at the crossing point versus log international units per milliliter.
FIG. 2.
Reproducibility and linearity of 24 real-time PCR runs by using the dilution series made from 24 independent extractions of the reference serum. Standard deviations are indicated by error bars.
FIG. 3.
Melting curve analysis of the fluorescence resonance energy transfer signal generated by the binding of the two hybridization probes to the plus strands of the amplicon. Data for five dilutions of genotype 1b standard serum (melting point [Tm] = 68.8°C) and five sera of genotype 2a (Tm = 58.7°C) are shown as the first derivative [d(f2/f1)] versus temperature.
The average detection limit was 80 IU/ml, or 5 IU/run. The upper confidence interval of the detection limit was 200 IU/ml, which correlates to 11 IU/run according to the extraction volume and the sample volume added to the reaction mixture. In samples containing 80 to 120 IU/ml, the fluorescence signal was in some cases insufficient to generate an amplification curve which is accepted by the LightCycler evaluation program for calculation of the crossing point. However, the melting curve usually showed a detectable peak even for 100 IU/ml. These samples, therefore, were positive without a quantitative result.
The use of specific primers, specific hybridization probes, and touchdown annealing temperature combined with a one-tube system is for the quantification of HCV RNA first described in this study. This combination ascertains a high overall specificity of the procedure. The touch-down protocol also reduces the amplification of undesirable side products. The small number of pipetting steps due to the one-tube RT-PCR and the combined amplification and signal generation within a closed system prevented contaminations. All negative controls remained negative during melting curve analysis as well as during agarose gel electrophoresis (data not shown).
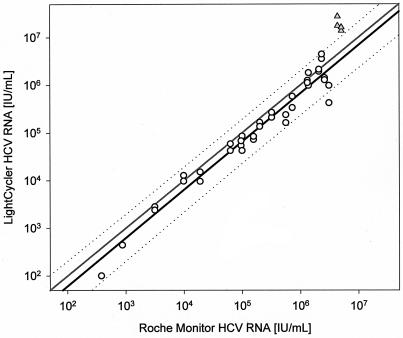
Testing 21 samples in parallel with the routinely used Roche HCV Monitor test confirmed the sensitivity of our in-house RT-PCR (Fig. 4). All 21 samples positive by the Monitor test were also positive by the in-house test. The results shown in Fig. 4 suggest that the Monitor test reaches saturation at 5 × 106 IU/ml. Two samples with that result had >107 IU/ml by the in-house test and were outside the 95% prediction interval of correlation (Fig. 4). The two assays were almost perfectly proportional to each other below the saturation level of the Monitor test (correlation coefficient, 0.9578; regression slope, 1.01), particularly at low concentrations. Adding the values >107 IU/ml to the regression, the correlation coefficient is 0.9397 and the slope 1.092. But the real-time PCR generated, on the average, 1.5-times-lower results. Given a 95% prediction interval of around factor 2.5 for the correlation of the two assays (Fig. 4), this difference is comparatively insignificant. Due to the very good reproducibility of the real-time assay, the relatively large variance of correlation may be due to the competitive Monitor assay, which is in general believed to have a 95% confidence interval of ca. a half log10 step (14).
FIG. 4.
Correlation between the LightCycler real-time and Roche Monitor competitive RT-PCR for 21 serum samples. Linear regression was calculated with real-time HCV RNA values of <107 IU/ml (○) (r2 = 0.9578) and indicated together with the 95% prediction interval (black and dotted lines, respectively). Theoretically, both assays should give identical values, as shown by the grey line. ▵, values of >107 IU/ml.
An international serum panel for HCV genotypes 1 to 6 (16) was also used for validation of the real-time RT-PCR. All HCV genotypes reacted reproducibly within the expected range of quantification. The efficiency of amplification was identical for all genotypes, as could be derived from the similar slopes of the amplification curve within the exponential phase (data not shown).
In the melting curve analysis, all specimens of genotype 2 showed a 10°C lower melting temperature of 58.5 to 60.2°C (Fig. 3) of the hybridization probes due to a conserved C-T exchange at position −72, which is found at the fourth position in the probe HCV-NCR-FL. This lower melting point of the hybridization probe HCV-NCR-FL did not affect the quantification and efficiency of amplification.
The LightCycler method and the HCV core antigen EIA were applied in parallel to 256 serial serum samples from 23 patients undergoing an interferon-ribavirin combination therapy. The sera were from early time points before or within the first 3 days of treatment. One hundred ninety-seven samples were positive by both assays, 4 were only HCV RNA positive, 2 were only HCV core antigen positive, and 51 were negative by both assays. Thus, assuming all results to be truly positive, the sensitivity of the HCV RNA assay was 99.0%, and that of the core assay was 98.0%. The four core-negative samples had between 80 and 940 IU of RNA/ml. The two RNA-negative samples had 2.7 and 3.7 pg of core antigen/ml. Tanaka et al. (18) also reported a sensitivity of 98% for their version of the core assay, but Zanetti et al. (20) found only 82% sensitivity.
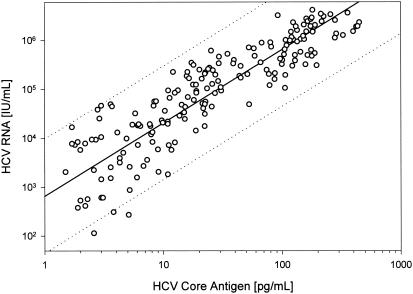
The quantitative relation of RNA and core antigen is shown in Fig. 5. From the mean of the RNA/core ratios of the samples positive by both tests, we calculated that 1 pg of total HCV core antigen corresponds on the average to 7,890 (±410 [standard error]) IU of HCV RNA. Using the same HCV core EIA and the Monitor assay, an almost identical ratio of 8,000 IU/pg was found by Bouvier-Alias et al. (2), but the high standard deviation of the Monitor assay for HCV RNA disabled the interpretation of the high variability of the RNA/core ratio. In our study, the correlation between both parameters was rather low, with r2 = 0.744. In contrast, the correlation (r2) of the measured and the true concentrations in the calibration curves was for both assays always >0.99 and mostly >0.999. Zanetti et al. (20) also found a relatively low correlation (r2 = 0.83) between HCV RNA with the Roche Monitor assay and the preliminary version of the trak-C assay.
FIG. 5.
Correlation between HCV RNA quantification by the LightCycler and measurement of HCV core antigen with EIA (trak-C) for 197 serum samples from follow-up with 23 interferon-treated patients. The linear regression (black line) and 95% prediction interval (dotted line) are indicated.
The ratio between the amount of HCV core antigen and the number of HCV RNA molecules was highly variable in the different samples, ranging from 50 IU/pg to 20,000 IU/pg. This range exceeds by far the expected variations caused by the standard deviations of the real-time PCR (8.3%) or core EIA (21.7%). The 95% confidence interval of the correlation was a factor of 15.5 for the RNA (Fig. 5), whereas for the inaccuracy generated by the RNA assays it would be 1.16 and by the core assay it would be 1.43. An accurate prediction of the ratio of RNA to core is not possible because the structure of HCV particles is not exactly known. Furthermore, there is no generally acknowledged true number of HCV RNA molecules per 1 IU. However, an estimate is possible with some plausible assumptions.
Theoretically, HCV, as a member of the Flaviviridae virus family, may contain an icosahedral capsid of T3 or T4 symmetry with 180 or 240 core protein subunits and one RNA genome (6). Assuming 240 subunits, a molecular mass of 19,000 Da for core protein and 1 IU of HCV RNA corresponding to 3 RNA molecules, 1 pg of virus cores ought to contain 130,000 HCV genome molecules. However, the measured ratio never exceeded 20,000 IU or 60,000 genomes per pg of core protein in our study and was on the average ca. 24,000 genomes/pg. Thus, on the average, only 18% of the core protein would be associated with HCV particles containing RNA and 240 core subunits, whereas the majority of the protein would be associated with structures devoid of HCV RNA. Such structures have been proposed by Maillard et al. (9).
Alternatively, particles would contain one genome and ca. 1,000 core subunits on the average. This is very unlikely because the particles are believed to be 50 nm in diameter (6) or less (19), and this size would not accommodate so many subunits.
The excess of core antigen over HCV RNA enhances the clinical sensitivity of the core antigen EIA. Theoretically, at an analytical sensitivity of 1.5 pg/ml, no sample containing <65,000 IU/ml should be detectable by the core EIA. However, 81 core-positive sera from interferon-treated patients contained less RNA, and 12 sera contained <1,000 IU/ml.
The question of whether subviral particles are present in HCV infection is mostly relevant for structural and clinical questions. Our data support the possible existence of RNA-free but core antigen-containing structures, either secreted by the infected cell or generated by in vivo degradation. As in the case of hepatitis B (HBsAg and HBeAg), the excess of viral antigen of HCV could function as a decoy for the immune system. Therefore HCV RNA and HCV core antigen levels in serum are not equivalent or simply stoichiometrically related parameters but could reveal new information about infection status upon treatment follow-up.
In conclusion, we have established a real time quantitative RT-PCR for HCV which is conveniently performed in one tube and uses hybridization probes for increased specificity. The primers used are well conserved in HCV genotypes 1 to 6, thus enabling a quantitative PCR with identical efficiency of all genotypes contained in the WHO reference panel. The comparison with the competitive PCR shows that the larger dynamic range of real-time PCR is required for accurate quantitation without testing expensive dilution series. The comparison with the HCV core EIA shows that the EIA is surprisingly sensitive (98%) in this special interferon-treated group, missing only very few HCV RNA positive samples. However, it appears that levels of HCV core antigen do not strictly correlate to HCV RNA. The factors influencing the ratio of HCV core protein to HCV RNA are the subjects of further studies.
Acknowledgments
The study was supported by Ortho Clinical Diagnostics with test kits for HCV core antigen.
We thank Uli Birk for assistance with the testing of diagnostic sera and W. R. Willems for sera pretested with the Amplicor Monitor.
C.G.S. was supported by a grant of the ENHCV consortium of the European Union.
REFERENCES
- 1.Anonymous. 1999. EASL International Consensus Conference on hepatitis C. Paris, 26 to 27 February 1999. Consensus statement. J. Hepatol. 31(Suppl. 1):3-8. [PubMed] [Google Scholar]
- 2.Bouvier-Alias, M., K. Patel, H. Dahari, S. Beaucourt, P. Larderie, L. Blatt, C. Hezode, G. Picchio, D. Dhumeaux, A. U. Neumann, J. G. McHutchison, and J. M. Pawlotzky. 2002. Clinical utility of total HCV core antigen quantification: a new indirect marker of HCV replication. Hepatology 36:211-218. [DOI] [PubMed] [Google Scholar]
- 3.Damen, M., H. T. Cuypers, H. L. Zaaijer, H. W. Reesink, W. P. Schaasberg, W. H. Gerlich, H. G. Niesters, and P. N. Lelie. 1996. International collaborative study on the second EUROHEP HCV-RNA reference panel. J. Virol. Methods 58:175-185. [DOI] [PubMed] [Google Scholar]
- 4.Germer, J. J., P. J. Heimgartner, D. M. Ilstrup, W. S. Harmsen, G. D. Jenkins, and R. Patel. 2002. Comparative evaluation of the VERSANT HCV RNA 3.0, QUANTIPLEX HCV RNA 2.0, and COBAS AMPLICOR HCV MONITOR version 2.0 assays for quantification of hepatitis C virus RNA in serum. J. Clin. Microbiol. 40:495-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gervais, A., M. Martinot, N. Boyer, A. Auperin, W. Le Breton, C. Degott, D. Valla, and P. Marcellin. 2001. Quantitation of hepatic hepatitis C virus RNA in patients with chronic hepatitis C. Relationship with severity of disease, viral genotype and response to treatment. J. Hepatol. 35:399-405. [DOI] [PubMed] [Google Scholar]
- 6.Heinz, F. X., M. S. Collett, R. H. Purcell, E. A. Gould, C. R. Howard, M. Houghton, R. J. M. Moormann, C. M. Rice, and H.-J. Thiel. 2000. Genus Hepacivirus, p. 872-878. In M. H. V. van Regenmortel, C. M. Fauquet, and D. H. L. Bishop (ed.), Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 7.Kieft, J. S., K. Zhou, R. Jubin, M. G. Murray, J. Y. Lau, and J. A. Doudna. 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 292:513-529. [DOI] [PubMed] [Google Scholar]
- 8.Komurian-Pradel, F., G. Paranhos-Baccala, M. Sodoyer, P. Chevallier, B. Mandrand, V. Lotteau, and P. Andre. 2001. Quantitation of HCV RNA using real-time PCR and fluorimetry. J. Virol. Methods 95:111-119. [DOI] [PubMed] [Google Scholar]
- 9.Maillard, P., K. Krawczynski, J. Nitkiewicz, C. Bronnert, M. Sidorkiewicz, P. Gounnon, J. Dubuisson, G. Faure, R. Crainic, and A. Budkowska. 2001. Nonenveloped nucleocapsids of hepatitis C virus in the serum of infected patients. J. Virol. 75:8241-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martell, M., J. Gomez, J. I. Esteban, S. Sauleda, J. Quer, B. Cabot, R. Esteban, and J. Guardia. 1999. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J. Clin. Microbiol. 37:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellor, J., A. Hawkins, and P. Simmonds. 1999. Genotype dependence of hepatitis C virus load measurement in commercially available quantitative assays. J. Clin. Microbiol. 37:2525-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odreman-Macchioli, F. E., S. G. Tisminetzky, M. Zotti, F. E. Baralle, and E. Buratti. 2000. Influence of correct secondary and tertiary RNA folding on the binding of cellular factors to the HCV IRES. Nucleic Acids Res. 28:875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosen, H. R., R. R. Ribeiro, L. Weinberger, S. Wolf, M. Chung, D. R. Gretch, and A. S. Perelson. 2002. Early hepatitis C viral kinetics correlate with long-term outcome in patients receiving high dose induction followed by combination interferon and ribavirin therapy. J. Hepatol. 37:124-130. [DOI] [PubMed] [Google Scholar]
- 14.Ross, R. S., S. Viazov, S. Sarr, S. Hofmann, A. Kramer, and M. Roggendorf. 2002. Quantitation of hepatitis C virus RNA by third generation branched DNA-based signal amplification assay. J. Virol. Methods 101:159-168. [DOI] [PubMed] [Google Scholar]
- 15.Saldanha, J., N. Lelie, and A. Heath. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. W. H. O. Collaborative Study Group. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 16.Saldanha, J., A. Heath, and the Collaborative Study Group. 2003. Collaborative Study to Calibrate HCV Genotypes 2-6 against the HCV International Standard, 96/780 (genotype 1). Vox Sang. 84:20-27. [DOI] [PubMed] [Google Scholar]
- 17.Schröter, M., B. Zöllner, P. Schäfer, R. Laufs, and H. H. Feucht. 2001. Quantitative detection of hepatitis C virus RNA by light cycler PCR and comparison with two different PCR assays. J. Clin. Microbiol. 39:765-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka E., C. Ohue, K. Aoyagi, K. Yamaguchi, S. Yagi, K. Kiyosawa, and H. J. Alter. 2000. Evaluation of a new enzyme immunoassay for hepatitis C virus (HCV) core antigen with clinical sensitivity approximating that of genomic amplification of HCV RNA. Hepatology 32:388-393. [DOI] [PubMed] [Google Scholar]
- 19.Yuasa, T., G. Ishikawa, S. Manabe, S. Sekiguchi, K. Takeuchi, and T. Miyamura. 1991. The particle size of hepatitis C virus estimated by filtration through microporous regenerated cellulose fibre. J. Gen. Virol. 72:2021-2024. [DOI] [PubMed] [Google Scholar]
- 20.Zanetti, A. R., L. Romano, M. Brunetto, M. Colombo, G. Bellati, and C. Tackney. 2003. Total HCV core antigen assay: a new marker of hepatitis C viremia for monitoring the progress of therapy. J. Med. Virol. 70:27-30. [DOI] [PubMed] [Google Scholar]