Abstract
Viral escape from HIV-1-specific CD8+ T cells has been demonstrated in numerous studies previously. However, the qualitative features driving the emergence of mutations within epitopes are still unclear. In this study, we aimed to distinguish whether specific functional characteristics of HLA-B*5701-restricted CD8+ T cells influence the emergence of mutations in high-risk progressors (HRPs) versus low-risk progressors (LRPs). Single genome sequencing was performed to detect viral mutations (variants) within seven HLA-B*5701-restricted epitopes in Gag (n = 4) and Nef (n = 3) in six untreated HLA-B*5701 subjects followed from early infection up to seven years. Several well-characterized effector markers (IFN-γ, IL-2, MIP-1β, TNF, CD107a and perforin) were identified by flow cytometry following autologous (initial and emerging variant/s) epitope stimulations. This study demonstrates that specific functional attributes may facilitate the outgrowth of mutations within HLA-B*5701-restricted epitopes. A significantly lower fraction of IL-2 producing cells and a decrease in functional avidity and polyfunctional sensitivity were evident in emerging epitope variants compared to the initial autologous epitopes. Interestingly, the HRPs mainly drove these differences, while the LRPs maintained a directed and maintained functional response against emerging epitope variants. In addition, LRPs induced improved cell cycle progression and perforin up-regulation after autologous and emerging epitope variant stimulations in contrast to HRPs. The maintained quantitative and qualitative features of the CD8+ T cell responses in LRPs toward emerging epitope variants provide insights into why HLA-B*5701 subjects have different risks of HIV-1 disease progression.
Keywords: HIV-1, Immunity, cellular, CD8-Positive T-Lymphocytes, HLA-B57 Antigen, Disease progression
Introduction
CD8+ T cells are critical in the immune control of HIV-1 infection. Epitope mutations that evade these immune responses can become fixed in the viral population as a result of the selective pressure from HIV-specific CD8+ T cells (1–5). Viral mutations can have an impact on peptide-MHC class I–T cell receptor (TCR) interactions (6, 7), binding of the peptide to the MHC class I molecules (5, 8–10), and the intracellular processing of the viral peptides (11–13). Several mutations completely abrogate an epitope-specific CD8+ T cell response while others are cross-recognized by the TCR of available T cell clones or induce recruitment of newly developed T cell clonotypes (14). However, the exact mechanisms driving the fixation of mutations within epitopes to which CD8+ T cell responses are directed remain elusive.
The factors that constitute an effective HIV-specific CD8+ T cell response are still debated, but differences in the functional T cell characteristics and specificities are likely to influence the efficacy. It has previously been described that both cytolytic and non-cytolytic antiviral effects are associated with the rate of HIV-1 disease progression (15–19). Other studies have shown associations between polyfunctionality and viral control (20, 21). Qualitative features surely represent an important part of an effective immune response, but most of the data presented so far have been generated in cross-sectional studies that do not include responses to individual epitope variants presented by a single HLA-allele.
The rate of progression in untreated HIV-1 infected subjects varies substantially between individuals. Factors that have been linked to the predicted course of the infection include clinical, virological and immunological parameters. In HIV infection, HLA-B*57 is the most consistent host factor that has an impact on the viral load set-point and associated with a better prognosis in HIV-1 infection (22–28). Nevertheless, not all subjects carrying protective HLA-B*57 alleles have a slow progression rate. The underlying mechanisms defining the rate of disease progression is not fully understood but likely involves both virological and immunological characteristics (29). As different HLA-B*57 alleles have similar peptide-binding motifs they are frequently grouped together when studying qualitative differences of the CD8+ T cell responses and/or their association to clinical outcome. However, small genetic differences between the HLA-B*57 alleles and the closely related HLA-B*5801 allele has been proven to have an impact on the immunogenicity, ability to select for viral mutations, and control viral replication in an HIV subtype C infected cohort in South Africa (10). To avoid the impact of genetic differences in the analysis of functional differences between initial and emerging epitope variants, this study was restricted to HIV subtype B infected individuals carrying the protective HLA-B*5701 allele (27).
We have previously demonstrated that subjects with HLA-B*5701 had a more robust polyfunctional Gag-specific CD8+ T cell response, coupled with higher IL-2 production in early infection if their CD4+ T cell count was >750 cells/mm3 at baseline (29). The immunological profile in these subjects was coupled with a lower genetic diversity and more constrained mutational profile in the gag p24 region compared to subjects with a lower CD4+ T cell count at baseline (median 13 estimated weeks post infection). In this study, we investigated whether the initial autologous versus corresponding major and minor viral variants of HLA-B*5701-restricted epitopes in the Gag and Nef regions revealed any functional differences prior to the emergence of mutations. The analysis was restricted to epitope variants recognized by the HIV-specific CD8+ T cells. By employing this design, functional features of the HLA-B*5701-restricted CD8+ T cell responses were traced to the emergence of mutations and coupled to the risk of disease progression.
Materials and Methods
Study subjects
Six HLA-B*5701 male patients infected with HIV-1 subtype B were recruited from the OPTIONS cohort, at the University of California, San Francisco (30), and followed from early infection (10–18 weeks) up to seven years. Based on baseline CD4 T+ cell count, three subjects (P1–P3) were classified as high-risk progressors (HRPs, <750 cells/mm3) and three (P4–P6) as low-risk progressors (LRPs, >750 cells/mm3) (29, 31). The University of California, San Francisco (UCSF) Committee on Human Research and the Regional Ethical Council in Stockholm, Sweden (2008/1099-31) approved this study and all patients provided written informed consent.
RNA extraction, cDNA synthesis and PCR amplification
RNA extraction and HIV-1 gag p24 single genome sequencing of longitudinal plasma samples (29) were performed as previously described. The nef sequences were obtained by performing cDNA synthesis using the ThermoScript RT-PCR System (Invitrogen) with gene-specific primer 5′-CCAGTACAGGCRAAAAGC-3′ (HXB2 nt position 9523-9540) (0.1 uM). Designed subtype B-specific primers were selected to amplify the HIV-1 region of nef, using a nested PCR with Platinum Taq DNA Polymerase (Invitrogen). First round PCRs used forward primer 5′-CATACCTASAAGAATAAGACARGG-3′ (HXB2 nt position 8797-8814) and reverse primer (described above) and the nested PCRs used forward primer 5′-ATGGGTGGCAARTGGTC-3′ (HXB2 nt position 1171–1189) and reverse primer 5′-AGTACAGGCARAAAGCRGC-3′ (HXB2 nt position 9520-9538). Purification and sequencing was performed as previously described (32). Sequences were imported and manually edited using Sequencher software and aligned in BioEdit. The HIV gag p24 and nef sequences included in this study were deposited at GenBank (http://www.ncbi.nlm.nih.gov/genbank/). GenBank accession numbers for the gag p24 sequences are: JX234575-234615, JX234645-234745, JX234801-234826, JX234855-234911, JX234994-235091, JX235120-235147, JX235167-235192, JX235220-235242, JX235266-235286, and JX235310-235332. GenBank accession numbers for the nef sequences are: KJ493407-KJ493601.
PBMC stimulation and flow cytometric analysis
Optimal peptides (9–11 mers) corresponding to autologous and variant HLA-B*5701-restricted epitopes in the HIV-1 Gag p24 (n=4) and Nef (n=3) region were used to measure the immunogenicity by CD8+ T cells. The protocols for PBMC stimulation and flow cytometry stainings have previously been described in detail (29, 33). Briefly, PBMCs were thawed, rested in media containing DNase (Sigma Aldrich) and supplemented with 2ug/mL of optimal peptides. For the peptide dilution experiments, additional peptide concentrations ranging from 10−4–10−8 ug/mL was used. The cells were incubated with peptides for 6–10 hrs in the short-term cultures together with Brefeldin A (Sigma Aldrich). When degranulation was measured, anti-CD107a PE-CF594 (clone: H4A3, BD Bioscience) was added already during the stimulation period together with monensin (BD Bioscience). For the long-term culture experiments, cells were incubated with 2ug/mL of peptides for 3 days and then re-stimulated with the same peptide concentration together with Brefeldin A, anti-CD107a and monensin.
The PBMCs were washed and stained with the following extracellular markers for different panels: anti-CD14 V500 (Clone M5E2), anti-CD19 V500 (Clone B43), anti-HLA-DR BV605 (Clone G46) (BD Bioscience); anti-PD-1 BV421 (clone EH12.2H7), (Biolegend); and LIVE/DEAD Fixable Aqua or Violet dyes (Life Technologies) to discriminate dead cells.
Cells were permeabilized and fixed with the cytofix/perm kit (BD Bioscience) for assessment of functional characteristics while the FOXP3 staining kit (eBioscience) was used to detect intra-nuclear proteins (Ki-67). The intracellular markers that were used for different flow panels included: anti-CD3 APC-H7 (Clone SK7), anti-CD4 V500 (clone RPA-T4), anti-CD8 PerCP (clone SK1), anti-IFN-γ AF700 and FITC (Clone B27), anti-IL-2 APC (Clone MQ1-17H12), anti-TNF PE-Cy7 and FITC (Clone MAb11), anti-MIP-Iβ PE-Cy7 (clone D21-1351), anti-Ki-67 FITC (Clone b56) (BD Bioscience); anti-Perforin PE (clone D48) (Biolegend and Tepnel); anti-TNF eFluor450 (Clone MAb11) (eBioscience); and anti-CD8 Qd565 (Clone 3B5), anti-CD4 PE-Cy5.5 (Clone S3.5) (Life Technologies).
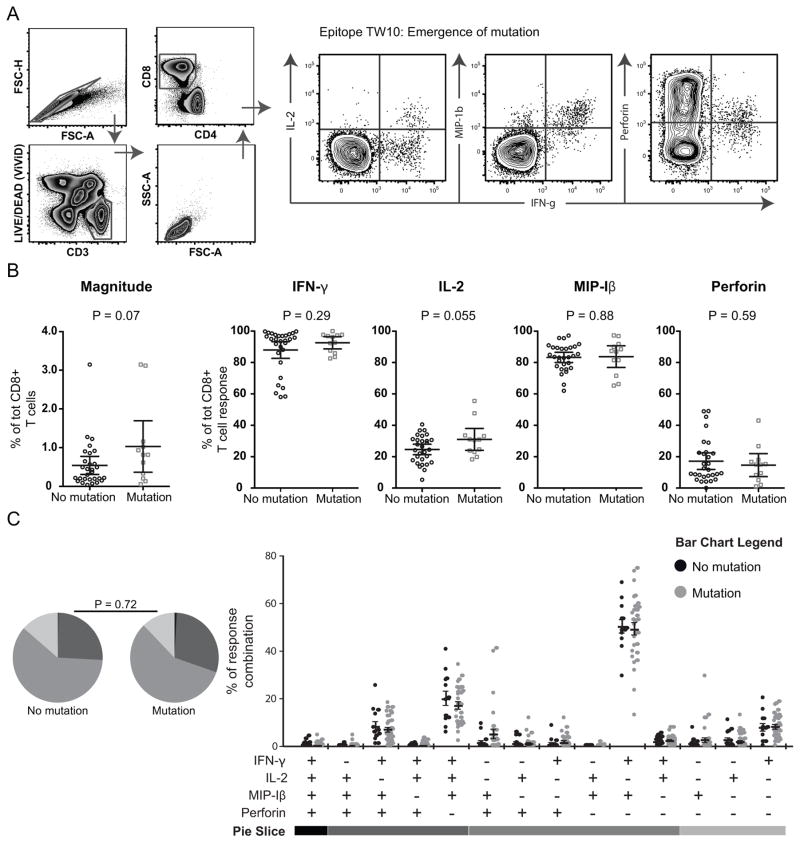
Cells were then washed, fixated and run on a 4 laser LSR Fortessa or Canto II (BD Bioscience). Antibody capture beads (BD Biosciences) were used for compensation and FlowJo 8.8.7 (Treestar) for gating analyses. Most manual gatings were based on fluorescence minus one (FMO) gating strategies. A typical T cell gating strategy to distinguish CD8+ T cell polyfunctionality is shown in Fig. 1A, where a response was considered positive if the frequency of IFN-γ producing cells were >0.05% of total CD8+ T cells after background reduction and twice the negative background.
Fig. 1. Functional characterization of HLA-B*5701-restricted epitopes developing mutations or not.
A) FACS plots illustrating the gating scheme to distinguish viable CD8+ T cells and a typical IFN-γ response together with the other functional markers. B) Scatter plots illustrating the magnitude and average production of IFN-γ, IL-2, MIP-1β and perforin (all time points) for all autologous epitopes developing mutations (grey) or not (black). Significant differences between the groups were analyzed using un-paired t-tests, where upper and lower whiskers show SEM. C) Pie charts demonstrating the functional diversity (for all time points) of all autologous epitope-specific responses between epitopes where mutations emerged or not. Permutation tests were conducted to compare significant differences between the pie charts. In the lower row, the functional combinations (for all time points) of all mutating (grey dots) or non-mutating (black dots) autologous epitope-specific responses are depicted. Mean and SEM are provided and student’s t-test in SPICE was used to assess significant differences between the groups.
Statistical analysis
All statistical tests are described in corresponding figure legends. Statistical comparisons between two groups of individuals were performed using Graphpad Prism 5.0 software and pie charts were analyzed using SPICE version 5.21 (34).
Results
Six HLA-B*5701 subjects, three HRPs and three low-risk progressors LRPs, based on CD4 T+ cell count at baseline, were followed longitudinally from early infection up to seven years (29). Between 18 to 34 gag p24 epitope sequences (details given in (29)) and 1 to 29 nef epitope sequences were obtained by single genome sequencing from each time point (Table I). CD8+ T cell responses against HLA-B*5701-restricted epitopes, i.e. peptides matching both the autologous founder virus sequence and emerging sequence variants within the epitope regions, were measured at three different time points for each subject (Table I).
Table I.
Sequence variation for all tested HLA-B*57-restricted epitopes and variants in the HIV-1 Gag p24- and Nef-region for all subjects.

|
HXB2 is used as the reference sequence for the HLA-B*57-restricted epitopes in Gag p24 (ISW9, KF11, TW10 and QW9) and Nef (KL10, HQ10 and YT9).
Patient identity.
Weeks post infection; sequences and immunological data were obtained from plasma and PBMC samples respectively, from the same time point in the majority of the patients. Wpi in parenthesis are PBMC samples taken at a different time point compared to the plasma samples.
The number of sequenced single nef genomes are indicated after each sequence
HRPs (P1–P3): high-risk progressors; LRPs (P4–P6): low-risk progressors. The epitopes corresponding to the major viral population at each time point are marked in boldface. Two epitopes marked in boldface at the same time point correspond to a 50–50 proportion of the respective variants. The epitopes with a positive CD8+ T cell response are filled in gray.
Sequence data for the Nef-region was not obtained for all time points.
Assessment of polyfunctionality and magnitude against autologous HLA-B*5701-restricted epitopes
We first sought to determine whether several functional parameters (IFN-γ, IL-2, MIP-1β and perforin) of CD8+ T cells were linked to HLA-B*5701-restricted epitope escape. The CD8+ T cell functionality against conserved epitopes, for which no mutations occurred between two time intervals (n = 29), were therefore compared to responses towards epitopes for which mutations emerged (n = 12) between the same time intervals. We observed trends towards elevated magnitude (p = 0.07) and greater IL-2 (p = 0.055) expression in the CD8+ T cell responses against mutating (n = 12) epitopes compared to the conserved (n = 29) epitopes (Fig. 1B). Overall however, neither mono- nor polyfunctional features of epitope-specific CD8+ T cells were significantly associated with protection against emergence of mutations in HLA-B*5701-restricted epitopes (P = 0.72, Fig. 1C).
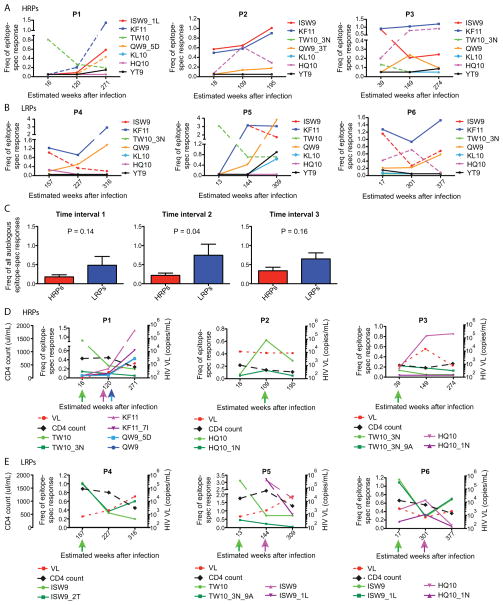
We further plotted the magnitude of the responses against all epitopes corresponding to the initial (first time-point) autologous sequence in all subjects over time. The subjects were divided into HRPs (Fig. 2A) and LRPs (Fig. 2B). The LRPs had in general an average higher magnitude of the responses against the immunodominant HLA-B*5701-restricted epitopes (> 1 % of the CD8+ T cells) at all the time points tested (Fig. 2C). As previously described, the depicted early immunodominant TW10 response in P1 and P5 was associated with the development of the TW10-3N and TW10-3N-9A escape mutations (Fig. 2D and E, respectively) (6, 35, 36). Surprisingly, epitopes where no mutations emerged during the entire study period showed the highest magnitudes at all tested time-points for most individuals. For instance, KF11 was measurable for all patients and had the highest magnitude at numerous time-points for one HRP (P3) and all three LRPs (P4, P5 and P6). A response towards KF11 has previously been shown to provide virological control (37). In this study, the KF11 epitope variant (KF11_7I) was only detected in two individuals (P1 and P6) of which one (P1) had a low response against the autologous sequence. These results indicate that the magnitude of epitope-specific responses by themself are not a complete predictive factor for emergence of viral mutations within HLA-B*5701-restricted epitopes.
Fig. 2. Frequencies of autologous and emerging epitope variant HLA-B*5701-restricted responses over time.
The frequencies of all Gag (n=4) and Nef (n=3) autologous HLA-B*5701-restricted responses are depicted over time in A) HRPs and B) LRPs. The Y-axis illustrates the epitope-specific frequencies of total CD8+ T cells, where the colors represent the magnitude of each initial autologous HLA-B*5701-restricted responses. The dashed lines depict the responses against those epitopes that eventually developed mutations. The X-axis represents the estimated weeks after infection as the responses were measured. C) Un-paired comparisons between the magnitude of all autologous epitope-specific responses between HRPs (red) and LRPs (blue) at time interval 1–3. P-values from un-paired t-tests are provided and mean and SEM are depicted for the bars. The frequency of all autologous and emerging epitope variant HLA-B*5701-restricted responses are depicted over time in D) HRPs and E) LRPs. The dashed lines represent the HIV-1 viral load (red) and CD4 count (black) over time, where the CD4 count is depicted on the far left Y-axis and HIV-1 viral load on each subjects’ right Y-axis. The left Y-axis for each subject represents the epitope-specific frequencies of total CD8+ T cells, where the colors demonstrates the magnitude of each autologous and corresponding emerging epitope variant-restricted response. The colored arrows under the X-axis clarifies the time-points for when and which autologous and emerging epitope variants that where tested for immunogenicity.
Functional diversity of the CD8+ T cell response against autologous and emerging HLA-B*5701-restricted epitope variants
We next investigated in depth the functional patterns of the CD8+ T cell responses against the epitopes where mutations emerged (n = 12) during the infection. In these subsequent analyses, the epitope variants corresponding to the autologous founder virus sequence for the tested time point were defined as the “autologous epitopes” (n = 12). Epitope variants that predominated (greater than 50% of viral variants) at the subsequent time point were entitled “emerging epitope variants” (n = 10), while minority variants (less than 50% of viral variants) at the subsequent time interval were entitled “minor epitope variants” (n = 7). All of these epitopes were tested and compared directly before the viral mutations emerged, and the magnitudes and time-points for when all epitope-specific responses were measured are depicted in Fig. 2D–E. Most of the HRPs developed mutations within a diverse set of B*5701-restricted epitopes, while in all LRPs mutations emerged within the ISW9 epitope. In general, the LRPs were able to preserve the response against the emerging epitope variants over time (Fig. 2E).
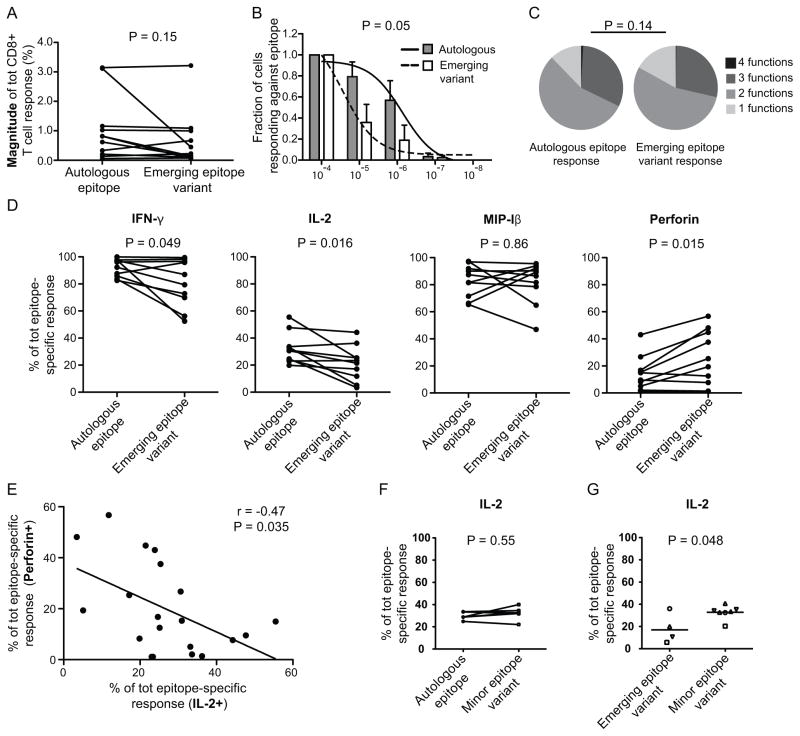
We further determined whether the magnitude of the CD8+ T cell response against the autologous and emerging epitope variants differed, but surprisingly found that the response was similar before mutations occurred (P = 0.15, Fig. 3A). However, these experiments were performed under saturated peptide concentrations (2 ug/ml) and we therefore conducted peptide-MHC class I (pMHC) avidity experiments. In these analyses, the autologous epitopes displayed higher ex vivo pMHC avidity than the emerging epitope variants (P = 0.05, Fig. 3B), which indicate that these emerging mutations truly represent epitope escape variants from CD8+ T cell responses. Furthermore, the combined functional characteristics (IFN-γ, IL-2, MIP-1β and perforin) of autologous and emerging epitope variant-specific CD8+ T cell responses were compared, but no significant differences in polyfunctionality were detected (P = 0.14, Fig. 3C and Supplemental Fig. 1A). However, the frequency of cells with three (including IL-2) to four functions was surprisingly greater for the autologous compared to the emerging epitope variants (Table II). By deciphering the frequencies of specific functional characteristics, we found that higher fractions of IL-2 (P = 0.016), but lower direct ex vivo perforin (P = 0.015) production by CD8+ T cells were present against the autologous epitopes (Fig. 3D). A statistically significant difference was also observed for IFN-γ production (P = 0.049), but not for MIP-1β expression (P = 0.86; Fig. 3D). In conjunction to these results, an inverse correlation was found between IL-2 and perforin production when assessing all autologous and emerging epitope-specific responses (r = −0.47, P = 0.035; Fig. 3E).
Fig. 3. The outgrowth of emerging epitope variants is associated with decreased pMHC avidity and IL-2 production in HLA-B*5701 subjects.
A) Paired comparisons of the magnitudes between autologous and emerging variant epitope-specific responses. P-value from a paired t-test is provided. B) pMHC avidity comparison between autologous and emerging epitope variant-specific responses at 5 different peptide concentrations. The Y-axis depicts the fraction of the response from the first peptide concentration (1 ug/mL) and subsequent peptide concentrations. The bars illustrate the mean (and SEM) fraction of the response for autologous (grey bars) and emerging epitope variants (white bars), while the sigmoidal curves are overlaid for autologous (solid line) and emerging epitope variants (dashed line). The p-values were calculated based on the area-under-curve for each epitope-specific response and then compared using paired t-tests. C) Pie charts illustrating the functional diversity of ten autologous and emerging variant epitope-specific responses. Permutation tests were conducted to compare significant differences between the pie charts. D) Paired comparisons between autologous and emerging variant epitope-specific responses showing the average production of IFN-γ, IL-2, MIP-1β and perforin. P-values from paired t-tests are provided. E) Correlation between IL-2 and perforin production for all autologous and emerging variant epitope-specific responses using the Spearman non-parametric test. G) Paired comparisons between autologous and minor variant epitope-specific responses for the average production of IL-2. P-values from paired t-tests are provided. H) Scatter plots demonstrating the average (mean) production of IL-2 in response to emerging and minor epitope variants. Each symbol of the dots represents different epitopes and corresponding variants. P-values were obtained from un-paired t-tests.
Table II.
Frequency of CD8+ T cells with specific functional characteristics targeting autologous versus major epitope variants.
| No. of functions | Functional combination | Autologous epitope variantsa | Major epitope variantsa |
|---|---|---|---|
| 4 | IFN-γ+, IL-2+, MIP-1β+, Perforin+ | 1.00 (−0.04–2.04) | 0.46 (0.34–0.89) |
| 3 | IFN-γ+, IL-2+, MIP-1β+, Perforin− | 20.63 (12.44–28.81) | 15.72 (6.93–24.51) |
| 3 | IFN-γ+, IL-2+, MIP-1β−, Perforin+ | 0.30 (−0.06–0.66) | 0.12 (−0.09–0.34) |
| 3 | IFN-γ+, IL-2−, MIP-1β+, Perforin+ | 8.36 (2.85–13.86) | 12.45 (0.16–24.73) |
| 3 | IFN-γ−, IL-2+, MIP-1β+, Perforin+ | 0.14 (−0.17–0.44) | 0.11 (−0.14–0.37) |
| 2 | IFN-γ+, IL-2+, MIP-1β−, Perforin− | 2.31 (0.98–3.64) | 1.88 (0.79–2.97) |
| 2 | IFN-γ+, IL-2−, MIP-1β+, Perforin− | 49.93 (42.2–57.65) | 40.11 (29.24–50.97) |
| 2 | IFN-γ+, IL-2−, MIP-1β−, Perforin+ | 1.36 (−0.56–3.29) | 2.27 (−0.37–4.91) |
| 2 | IFN-γ−, IL-2+, MIP-1β+, Perforin− | 0 | 0.11 (−0.14–0.35) |
| 2 | IFN-γ−, IL-2+, MIP-1β−, Perforin+ | 1.73 (0.04–3.42) | 1.8 (−0.26–3.86) |
| 2 | IFN-γ−, IL-2−, MIP-1β+, Perforin+ | 1.40 (−0.75–3.55) | 8.24 (1.58–14.89) |
| 1 | IFN-γ+, IL-2−, MIP-1β−, Perforin− | 8.18 (3.56–12.81) | 7.78 (4.808–10.76) |
| 1 | IFN-γ−, IL-2+, MIP-1β−, Perforin− | 3.14 (0.25–6.04) | 4.14 (−3.42–11.71) |
| 1 | IFN-γ−, IL-2−, MIP-1β+, Perforin− | 1.53 (−0.18–3.24) | 4.80 (−2.97–12.58) |
mean (95% CI)
We next evaluated whether fluctuating IL-2 production could be detected between autologous and corresponding minor epitope variants. No significant differences for IL-2 production (P = 0.55; Fig. 3G) or any other marker was found (Supplemental Fig. 1B). However, by comparing epitope-specific emerging and minor variant responses, significantly lower IL-2 (P = 0.048) production was found for the emerging variants, despite the small number of available data points (Fig. 3H). No other markers showed these differences (Supplemental Fig. 1C).
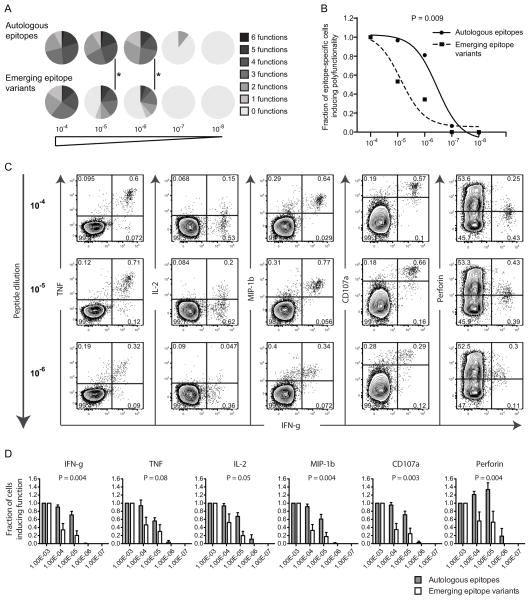
Additional peptide dilution experiments were conducted to verify whether differences could be distinguished in terms of polyfunctional sensitivity using different peptide concentrations. In addition to IFN-γ, IL-2, MIP-1β and perforin, also TNF and CD107a expression were measured in these analyses. The autologous epitope-specific CD8+ T cell responses revealed increased polyfunctional characteristics (Fig. 4A) and sensitivity (P = 0.009, Fig. 4B) at lower peptide concentrations. As previously described (38), most functional markers showed a decrease in median fluorescence intensity (MFI) and percentage as a consequence of lower peptide concentrations (Fig. 4C). Nevertheless, all functional markers except TNF declined more rapidly at lower peptide concentrations in response to the emerging epitope variants (P < 0.05) compared to the response against autologous epitopes (Fig. 4D). Interestingly, the fraction of perforin producing cells increased as a consequence of lower peptide concentrations (Fig. 4C–D), particularly against autologous epitopes, and was potentially due to the down-regulation of CD8 molecules at higher peptide concentrations. We also assessed the MFI of PD-1 and HLA-DR in the peptide dilution analyses to determine whether the level of exhaustion and activation of the CD8+ T cell repertoire against autologous and emerging epitope variants differed. However, the intensity of neither marker changed significantly (P > 0.05) after peptide dilutions or differed between the groups (data not shown). Despite that polyfunctionality declined, these functional results were primarily driven by the lack of response against the emerging epitope variants at lower peptide concentrations in specific subjects.
Fig. 4. Different polyfunctional sensitivity between autologous and emerging epitope variant-specific responses.
A) The number of functions (0–6) where calculated based on combined IFN-γ, TNF, IL-2, MIP-1β CD107a and perforin production and illustrated using pie charts for the different peptide concentrations (10−4–10−8). P-values < 0.05 are depicted with * (permutation test). B) The polyfunctional sensitivity was compared between autologous and emerging epitope variant-specific responses at 5 different peptide concentrations. The Y-axis depicts the fraction of polyfunctionality (number of functions) from the first peptide concentration (1 ug/mL) and subsequent peptide concentrations. The sigmoidal curves are illustrated for autologous (solid line) and emerging epitope variants (dashed line) where the symbols illustrate the average number of functions in each group at different peptide concentrations. The p-values were calculated based on the area-under-curve for each epitope-specific response and then compared between the groups using paired t-tests. C) FACS plots of a representable example of an autologous epitope-specific response. IFN-γ production in combination with the other functional markers is depicted for those different peptide concentrations generating a positive response. D) Bar plots showing the fraction of all functions from baseline (1 ug/mL) and subsequent peptide concentrations. The bars illustrate the mean (and SEM) fraction of the response for autologous (grey bars) and emerging epitope variants (white bars), where the p-values were obtained using area-under-curve calculations for each epitope-specific response and then compared using paired t-tests.
Qualitative and quantitative differences in HLA-B*5701-restricted responses between HRPs and LRPs
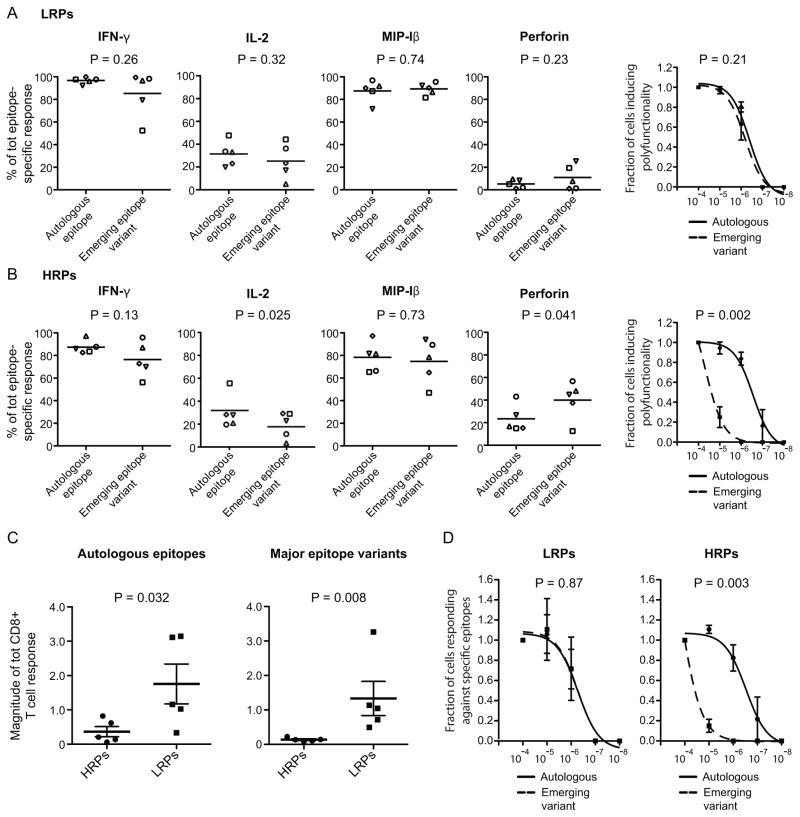
Next, we sought to assess whether the fluctuating IL-2, perforin and IFN-γ expression between autologous epitopes and emerging epitope variants (depicted in Fig. 3D) was linked to disease progression. There was no statistically significant difference in the production of these markers for LRPs (Fig. 5A). However, among HRPs there was significantly higher IL-2 production (P = 0.025, Fig. 5A) and lower perforin production (P = 0.041, Fig. 5A) for autologous epitopes. Both the LRPs and HRPs showed non-significant higher IFN-γ expression for autologous epitopes (P = 0.26 and 0.13, respectively, Fig. 5A and B). It was also confirmed that LRPs maintained a similar polyfunctional response between autologous and emerging epitope variants even at lower peptide concentrations (P = 0.21, Fig. 5A), while the HRPs lost their functional response against emerging variants (P = 0.002, Fig. 5B).
Fig. 5. HRPs elicit an inverse ex vivo IL-2/perforin production, lower magnitude and functional avidity against autologous and emerging epitope variants.
Scatter plots demonstrating the mean production of IFN-γ, IL-2, MIP-1β and perforin in response to autologous epitopes and emerging epitope variants for A) LRPs and B) HRPs. Each symbol of the dots represents different epitopes and corresponding variants. P-values were obtained from paired t-tests. The right plots illustrate the polyfunctional sensitivity between the autologous (solid line) and emerging epitope variants (dashed line) where the symbols illustrate the average number of functions (and SEM) in each group at different peptide concentrations. The p-values were calculated based on the area-under-curve for each epitope-specific response and then compared between the groups using paired t-tests. Magnitude comparisons (mean and SEM) between HRPs and LRPs for C) autologous and emerging epitope variant-specific responses. P-values from un-paired t-tests are provided. D) pMHC avidity analysis in HRPs and LRPs where the autologous (solid line) and emerging epitope variants (dashed line) are depicted in the sigmoidal curves. The symbols illustrate the average number of functions in each group at different peptide concentrations. The p-values were calculated based on the area-under-curve for each epitope-specific response and then compared between the groups using paired t-tests.
Additionally, the magnitude of CD8+ T cell responses was significantly higher against autologous (P = 0.032), as well as emerging epitope variants (P = 0.008), in LRPs compared to HRPs (Fig. 5C). Similarly to the polyfunctional characteristics, LRPs maintained the pMHC avidity against the emerging epitope variants (P = 0.87), while the HRPs lost the response against these variants at lower peptide concentrations (P = 0.003, Fig. 5D).
In conclusion, these data demonstrate that LRPs maintain primarily IL-2 production, but also polyfunctionality at lower peptide concentrations, and have a higher magnitude and pMHC avidity toward emerging epitope variants compared to HRPs.
Relationship between ex vivo IL-2 production and perforin up-regulation after long-term epitope stimulations
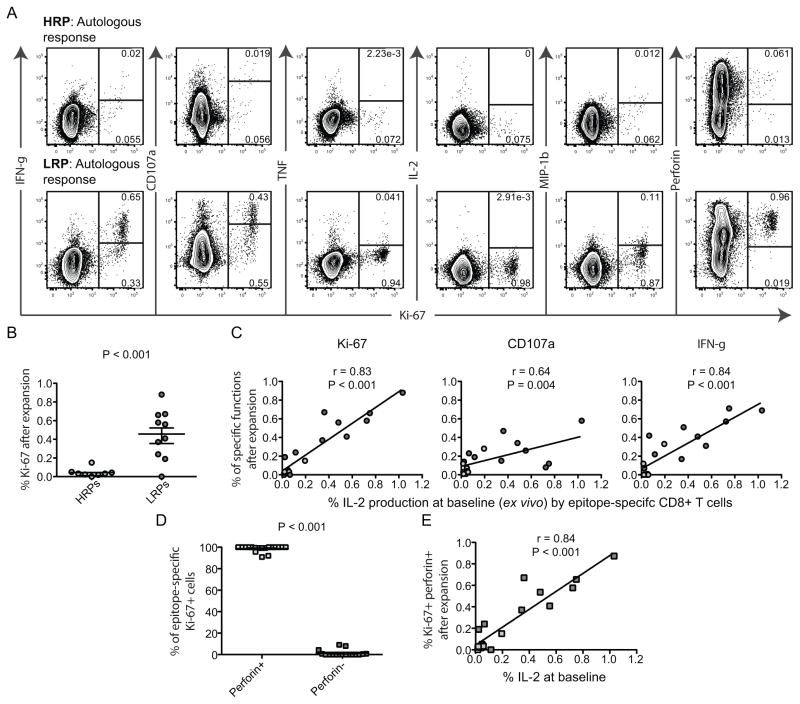
Individuals with protective MHC I alleles after vaccination (39) and those controlling HIV-1 replication (18) have previously been shown to induce supreme CD8+ T cell proliferation and consequently up-regulation of cytolytic functions. We therefore elucidated whether autocrine (ex vivo) IL-2 expression might induce improved cell cycle progression and up-regulation of perforin in long-term cultures. PBMCs were incubated with the autologous and emerging epitope variants for 3 days to assess the expression of Ki-67 together with the other functional markers (Fig 6A). Most of the autologous and emerging epitope variant-specific cells in LRPs were efficient to up-regulate Ki-67, while HRPs showed limited cell cycle progression (P < 0.001, Fig 6A–B). Importantly, the frequency of baseline ex vivo frequencies of IL-2 producing CD8+ T cells was highly correlated with Ki-67 (P < 0.001, r = 0.82), CD107a (P = 0.004, r = 0.64) and IFN-γ (P < 0.001, r = 0.84) expression following long-term incubations with autologous and emerging epitope variants (Fig. 6C). The overall ex vivo magnitude of the epitope-specific responses was also correlated with the Ki-67 up-regulation (P = 0.003, r = 0.64), but IL-2 was a better predictor of cell proliferation. TNF, IL-2 and MIP-Iβ production were poorly expressed in cells after expanding the autologous and emerging epitope-variant-specific cells in cultures (Fig. 6A), indicating that the CD8+ T cell clones exhibited late effector maturity. The majority of the Ki-67+ cells also possessed enriched levels of perforin (P < 0.001, Fig. 6D) and likewise, IL-2 production ex vivo was associated with Ki-67+perforin+ production after 3 day incubations (P < 0.001, r = 0.84, Fig. 6E). These data suggest that LRPs maintain the magnitude and autocrine IL-2 secretion against the autologous and emerging epitope variants, leading to increased T-cell turnover and hence up-regulation of perforin that potentially induce cytolysis of virus-infected cells.
Fig. 6. LRPs show profound up-regulation of Ki-67 and perforin after 3-day stimulations with autologous and emerging epitope variants.
A) FACS plots illustrating the expression of Ki-67 (X-axis) together with the other functional markers (Y-axis) after autologous peptide stimulations for 3 days. The upper row shows a representative autologous-specific response in a HRP, and the lower a LRP response. B) Scatter plot demonstrating the Ki-67 up-regulation of total CD8+ T cells for HRPs and LRPs after autologous and emerging epitope variant stimulations. P-values were obtained from un-paired t-tests and mean (SEM) are depicted in the plot. C) Correlation between the baseline (6 hr stimulation) magnitudes of IL-2 producing CD8+ T cells (X-axis) and Ki-67, CD107a and IFN-γ expression after 3 days in culture (Y-axis) with autologous and emerging epitope variants. The darker grey circles represent LRPs and the lighter grey circles HRPs. All correlations were based on Spearman non-parametric test. D) Graph demonstrating the % of Ki-67+ CD8+ T cells (3 days incubation) that up-regulate perforin or not (paired t-test). E) Spearman correlation analysis of baseline IL-2 production by CD8+ T cells (X-axis) and Ki-67+perforin+ expression (Y-axis) after 3 days in culture with epitopes.
Discussion
HIV-1-specific CD8+ T cell responses represents a major factor predicting the outcome of HIV-1 disease progression. Although neither the STEP trial or the RV144 trial showed evidence of CD8+ T cell responses affecting set-point viremia or protection in the vaccine, recent studies have demonstrated that vector induced T cell responses can limit HIV-1 RNA level in subjects carrying protective HLA alleles (B-27, 57 and 5801) (40) as well as limit SIV replication and possibly clear the infection (41). Thus, it still remains important to identify correlates of effective CD8+ T cell responses establishing pressure on founder viral sequences of HIV-1. In the present study, HLA-B*5701-restricted CD8+ T cell responses were closely examined from early infection in subjects with different risk of disease progression. This allowed us to characterize the functional features of the CD8+ T cell response generating pressure on the autologous founder virus as identified by emerging mutations within the epitopes.
Polyfunctional characteristics have been associated with viral control in the chronic phase of HIV-1 infection (20, 21). However, in cross-sectional settings the “true characteristics” of efficient CD8+ T cell responses might be misleading due to other factors exerting pressure on the autologous virus (42). Similar to previous results (43, 44), no statistically associations were found between CD8+ T cell polyfunctionality and the outgrowth of HLA-B*5701-restricted epitope mutants. However, by assessing the polyfunctional sensitivity under conditions of lower peptide concentrations, clear differences were found between response against autologous and emerging variants of the virus. These differences were driven by the HRPs that completely lost the response and functional characteristics against emerging epitope variants under lower peptide concentrations. These results are in agreement with the data from Almeida et al in HLA-B27 subjects (21), showing that polyfunctionality is determined by antigen sensitivity and suggest that HLA-B*5701 individuals possessing a maintained functional response against emerging escape variants of HIV-1 might have a lower risk of disease progression (45). Furthermore, our findings are in line with recent data from Pohlmeyer et al (46) illustrating that HLA-B*57 elite suppressors are able to control the replication of engineered viral escape variants. Whether the determining factor of these diverse features is driven by the TCR repertoire remains to be proven, but public clonotypes have been linked to development of MHC I-restricted escape (14, 47) and elite control in former studies (48).
MHC I-restricted epitope escape has been shown to dramatically reduce the magnitude of the CD8+ T cell response (49). Previous studies have also found associations between higher magnitude of T cell responses and rapid escape (43, 50). We identified a similar trend, with higher magnitude responses against mutating epitopes compared to conserved epitopes. In agreement with earlier studies however, it was demonstrated that HLA-B*57-restricted epitope variants do not necessarily impact the CD8+ T cell magnitude at higher peptide concentrations (51). Nevertheless, by conducting peptide dilution experiments it was verified that particularly HRPs had poor pMHC avidity against emerging epitope variants and the magnitude of CD8+ T cell responses was significantly higher against autologous and emerging epitope variants in LRPs compared to HRPs. It was recently revealed that the development of high avidity cross-reactive KK10-specfic CD8+ T cell clonotypes contributes to the viral control in HLA-B*2705 study subjects (14), suggesting that the plasticity of the TCR recognizing viral epitope variants may explain the different rates of disease progression in subjects carrying protective HLA alleles.
By comparing the functional profiles we found that IL-2 production declined while perforin expression increased in response to the mutated epitope variants compared to the autologous epitopes for HRPs, but not for LRPs. This indicates that the magnitude of CD8+ T cell responses, as well as maintaining a IL-2 production towards both autologous and emerging epitope variants, may be linked with lower risk to progress towards AIDS. An inverse correlation between ex vivo IL-2 and perforin production for virus-specific CD8+ T cells has previously been demonstrated (52), but not in the context of viral escape. Neither has it been shown that functional differences exist between emerging and minor epitope variants that may have an impact on the selection of mutations. Non-cytolytic CD8+ T cell responses have been associated with viral escape (53) and may be an important influence on set-point viremia (54, 55). In agreement with these studies, IL-2 producing CD8+ T cells might thus represent a non-cytolytic mechanism that drives fixation of epitope mutations. Another potential explanation could be that autocrine IL-2 production is linked to increased proliferation and cytolytic gene expression (16, 56). In this study, we verified these characteristics and showed that LRPs were able to go through more extensive cell cycle progression and perforin up-regulation after both autologous and emerging epitope variant stimulations. Therefore, the data suggest that IL-2 and perforin are linked together although different memory CD8+ T cell subsets usually express these functions. Thus, non-cytolytic and cytolytic features most probably cooperate to induce the pressure on the founder virus.
An interesting observation was that CD8+ T cell responses against minor epitope variant displayed a significantly higher IL-2 production than emerging epitope variants. This indicates that some minor viral populations might not grow in size due to the pressure from IL-2 producing cells. However, these data were generated with a small number of available data points. The small study cohort is a general limitation, reflecting the restricted possibilities of obtaining unique HLA-B*5701 patient samples longitudinally from early infection. Also, the study includes no samples from acute phase (Fiebig stage I/II) of infection and only HLA-B*5701-restricted epitopes were studied. Nevertheless, significant differences were still observed between diverse variables and the two groups of patients.
In summary, these results indicate that HLA-B*5701 subjects that have a lower risk of HIV-1 disease progression maintain the functional avidity and possess higher percentage of IL-2 producing CD8+ T cells towards emerging epitope variants, compared to subjects with higher risk of progression. These findings suggest that the magnitude and cooperation between non- and cytolytic CD8+ T cell responses exert pressure on autologous HLA-B*5701-restricted epitopes, which might be of importance in the future design of anti-HIV-1 therapeutic antigens.
Supplementary Material
Acknowledgments
Grant support
This work was supported by grants from the Swedish Research Council (K2010-56X-20345-04-3 and K2014-57X-22451-01-5), Swedish Agency for International Development Cooperation-SIDA (2005-001756), Erik and Edith Fernströms foundation, Karolinska Institutet, Åke Wibergs Foundation (40418186), Magnus Bergvalls Foundation, the Swedish Physicians Against AIDS Research Foundation (FOa2011-0021), and the Swedish Society of Medicine (SLS-101021). This work was also supported by US NIH Program Project Grant P01 AI071713 from the National Institute of Allergy and Infectious Diseases.
Abbreviations
- HRPs
high-risk progressors
- LRPs
low-risk progressors
References
- 1.Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, Ogunlesi AO, Elvin JG, Rothbard JA, Bangham CR, Rizza CR, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 2.Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 3.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, McNevin J, Cao J, Zhao H, Genowati I, Wong K, McLaughlin S, McSweyn MD, Diem K, Stevens CE, Maenza J, He H, Nickle DC, Shriner D, Holte SE, Collier AC, Corey L, McElrath MJ, Mullins JI. Selection on the human immunodeficiency virus type 1 proteome following primary infection. Journal of virology. 2006;80:9519–9529. doi: 10.1128/JVI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlsson AC, Iversen AK, Chapman JM, de Oliviera T, Spotts G, McMichael AJ, Davenport MP, Hecht FM, Nixon DF. Sequential broadening of CTL responses in early HIV-1 infection is associated with viral escape. PloS one. 2007;2:e225. doi: 10.1371/journal.pone.0000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, John AS, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 7.Hoof I, Perez CL, Buggert M, Gustafsson RK, Nielsen M, Lund O, Karlsson AC. Interdisciplinary analysis of HIV-specific CD8+ T cell responses against variant epitopes reveals restricted TCR promiscuity. J Immunol. 2010;184:5383–5391. doi: 10.4049/jimmunol.0903516. [DOI] [PubMed] [Google Scholar]
- 8.Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, Brander C, Ogg G, Sullivan JS, Dyer W, Jones I, McMichael AJ, Rowland-Jones S, Phillips RE. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson AC, Deeks SG, Barbour JD, Heiken BD, Younger SR, Hoh R, Lane M, Sallberg M, Ortiz GM, Demarest JF, Liegler T, Grant RM, Martin JN, Nixon DF. Dual pressure from antiretroviral therapy and cell-mediated immune response on the human immunodeficiency virus type 1 protease gene. Journal of virology. 2003;77:6743–6752. doi: 10.1128/JVI.77.12.6743-6752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kloverpris HN, Stryhn A, Harndahl M, van der Stok M, Payne RP, Matthews PC, Chen F, Riddell L, Walker BD, Ndung’u T, Buus S, Goulder P. HLA-B*57 Micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure, and HIV immune control. Journal of virology. 2012;86:919–929. doi: 10.1128/JVI.06150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen TM, Altfeld M, Yu XG, O’Sullivan KM, Lichterfeld M, Le Gall S, John M, Mothe BR, Lee PK, Kalife ET, Cohen DE, Freedberg KA, Strick DA, Johnston MN, Sette A, Rosenberg ES, Mallal SA, Goulder PJ, Brander C, Walker BD. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. Journal of virology. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draenert R, Le Gall S, Pfafferott KJ, Leslie AJ, Chetty P, Brander C, Holmes EC, Chang SC, Feeney ME, Addo MM, Ruiz L, Ramduth D, Jeena P, Altfeld M, Thomas S, Tang Y, Verrill CL, Dixon C, Prado JG, Kiepiela P, Martinez-Picado J, Walker BD, Goulder PJ. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J Exp Med. 2004;199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokomaku Y, Miura H, Tomiyama H, Kawana-Tachikawa A, Takiguchi M, Kojima A, Nagai Y, Iwamoto A, Matsuda Z, Ariyoshi K. Impaired processing and presentation of cytotoxic-T-lymphocyte (CTL) epitopes are major escape mechanisms from CTL immune pressure in human immunodeficiency virus type 1 infection. Journal of virology. 2004;78:1324–1332. doi: 10.1128/JVI.78.3.1324-1332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ladell K, Hashimoto M, Iglesias MC, Wilmann PG, McLaren JE, Gras S, Chikata T, Kuse N, Fastenackels S, Gostick E, Bridgeman JS, Venturi V, Arkoub ZA, Agut H, van Bockel DJ, Almeida JR, Douek DC, Meyer L, Venet A, Takiguchi M, Rossjohn J, Price DA, Appay V. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity. 2013;38:425–436. doi: 10.1016/j.immuni.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 16.Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M, Ehler L, Metcalf J, Liu S, Connors M. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nature immunology. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 17.Pantaleo G, Koup RA. Correlates of immune protection in HIV-1 infection: what we know, what we don’t know, what we should know. Nature medicine. 2004;10:806–810. doi: 10.1038/nm0804-806. [DOI] [PubMed] [Google Scholar]
- 18.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, Lloyd M, Roby G, Kwan R, McLaughlin M, Stallings S, Rehm C, O’Shea MA, Mican J, Packard BZ, Komoriya A, Palmer S, Wiegand AP, Maldarelli F, Coffin JM, Mellors JW, Hallahan CW, Follman DA, Connors M. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, Kovacs CM, Rodriguez B, Sieg SF, Teixeira-Johnson L, Gudonis D, Goepfert PA, Lederman MM, Frank I, Makedonas G, Kaul R, Walker BD, Betts MR. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS pathogens. 2010;6:e1000917. doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin AG, Douek D, Autran B, Appay V. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. The Journal of experimental medicine. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 23.Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, Urban TJ, Zhang K, Gumbs CE, Smith JP, Castagna A, Cozzi-Lepri A, De Luca A, Easterbrook P, Gunthard HF, Mallal S, Mussini C, Dalmau J, Martinez-Picado J, Miro JM, Obel N, Wolinsky SM, Martinson JJ, Detels R, Margolick JB, Jacobson LP, Descombes P, Antonarakis SE, Beckmann JS, O’Brien SJ, Letvin NL, McMichael AJ, Haynes BF, Carrington M, Feng S, Telenti A, Goldstein DB. Common genetic variation and the control of HIV-1 in humans. PLoS genetics. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O’Brien SJ, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann DL. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 26.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 27.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O’Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, Altfeld M, Henn MR, Le Gall S, Streeck H, Haas DW, Kuritzkes DR, Robbins GK, Shafer RW, Gulick RM, Shikuma CM, Haubrich R, Riddler S, Sax PE, Daar ES, Ribaudo HJ, Agan B, Agarwal S, Ahern RL, Allen BL, Altidor S, Altschuler EL, Ambardar S, Anastos K, Anderson B, Anderson V, Andrady U, Antoniskis D, Bangsberg D, Barbaro D, Barrie W, Bartczak J, Barton S, Basden P, Basgoz N, Bazner S, Bellos NC, Benson AM, Berger J, Bernard NF, Bernard AM, Birch C, Bodner SJ, Bolan RK, Boudreaux ET, Bradley M, Braun JF, Brndjar JE, Brown SJ, Brown K, Brown ST, Burack J, Bush LM, Cafaro V, Campbell O, Campbell J, Carlson RH, Carmichael JK, Casey KK, Cavacuiti C, Celestin G, Chambers ST, Chez N, Chirch LM, Cimoch PJ, Cohen D, Cohn LE, Conway B, Cooper DA, Cornelson B, Cox DT, Cristofano MV, Cuchural G, Jr, Czartoski JL, Dahman JM, Daly JS, Davis BT, Davis K, Davod SM, DeJesus E, Dietz CA, Dunham E, Dunn ME, Ellerin TB, Eron JJ, Fangman JJ, Farel CE, Ferlazzo H, Fidler S, Fleenor-Ford A, Frankel R, Freedberg KA, French NK, Fuchs JD, Fuller JD, Gaberman J, Gallant JE, Gandhi RT, Garcia E, Garmon D, Gathe JC, Jr, Gaultier CR, Gebre W, Gilman FD, Gilson I, Goepfert PA, Gottlieb MS, Goulston C, Groger RK, Gurley TD, Haber S, Hardwicke R, Hardy WD, Harrigan PR, Hawkins TN, Heath S, Hecht FM, Henry WK, Hladek M, Hoffman RP, Horton JM, Hsu RK, Huhn GD, Hunt P, Hupert MJ, Illeman ML, Jaeger H, Jellinger RM, John M, Johnson JA, Johnson KL, Johnson H, Johnson K, Joly J, Jordan WC, Kauffman CA, Khanlou H, Killian RK, Kim AY, Kim DD, Kinder CA, Kirchner JT, Kogelman L, Kojic EM, Korthuis PT, Kurisu W, Kwon DS, LaMar M, Lampiris H, Lanzafame M, Lederman MM, Lee DM, Lee JM, Lee MJ, Lee ET, Lemoine J, Levy JA, Llibre JM, Liguori MA, Little SJ, Liu AY, Lopez AJ, Loutfy MR, Loy D, Mohammed DY, Man A, Mansour MK, Marconi VC, Markowitz M, Marques R, Martin JN, Martin HL, Jr, Mayer KH, McElrath MJ, McGhee TA, McGovern BH, McGowan K, McIntyre D, McLeod GX, Menezes P, Mesa G, Metroka CE, Meyer-Olson D, Miller AO, Montgomery K, Mounzer KC, Nagami EH, Nagin I, Nahass RG, Nelson MO, Nielsen C, Norene DL, O’Connor DH, Ojikutu BO, Okulicz J, Oladehin OO, Oldfield EC, 3rd, Olender SA, Ostrowski M, Owen WF, Jr, Pae E, Parsonnet J, Pavlatos AM, Perlmutter AM, Pierce MN, Pincus JM, Pisani L, Price LJ, Proia L, Prokesch RC, Pujet HC, Ramgopal M, Rathod A, Rausch M, Ravishankar J, Rhame FS, Richards CS, Richman DD, Rodes B, Rodriguez M, Rose RC, 3rd, Rosenberg ES, Rosenthal D, Ross PE, Rubin DS, Rumbaugh E, Saenz L, Salvaggio MR, Sanchez WC, Sanjana VM, Santiago S, Schmidt W, Schuitemaker H, Sestak PM, Shalit P, Shay W, Shirvani VN, Silebi VI, Sizemore JM, Jr, Skolnik PR, Sokol-Anderson M, Sosman JM, Stabile P, Stapleton JT, Starrett S, Stein F, Stellbrink HJ, Sterman FL, Stone VE, Stone DR, Tambussi G, Taplitz RA, Tedaldi EM, Telenti A, Theisen W, Torres R, Tosiello L, Tremblay C, Tribble MA, Trinh PD, Tsao A, Ueda P, Vaccaro A, Valadas E, Vanig TJ, Vecino I, Vega VM, Veikley W, Wade BH, Walworth C, Wanidworanun C, Ward DJ, Warner DA, Weber RD, Webster D, Weis S, Wheeler DA, White DJ, Wilkins E, Winston A, Wlodaver CG, van’t Wout A, Wright DP, Yang OO, Yurdin DL, Zabukovic BW, Zachary KC, Zeeman B, Zhao M. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norstrom MM, Buggert M, Tauriainen J, Hartogensis W, Prosperi MC, Wallet MA, Hecht FM, Salemi M, Karlsson AC. Combination of immune and viral factors distinguishes low-risk versus high-risk HIV-1 disease progression in HLA-B*5701 subjects. Journal of virology. 2012;86:9802–9816. doi: 10.1128/JVI.01165-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hecht FM, Busch MP, Rawal B, Webb M, Rosenberg E, Swanson M, Chesney M, Anderson J, Levy J, Kahn JO. Use of laboratory tests and clinical symptoms for identification of primary HIV infection. AIDS. 2002;16:1119–1129. doi: 10.1097/00002030-200205240-00005. [DOI] [PubMed] [Google Scholar]
- 31.Mellors JW, Munoz A, Giorgi JV, Margolick JB, Tassoni CJ, Gupta P, Kingsley LA, Todd JA, Saah AJ, Detels R, Phair JP, Rinaldo CR., Jr Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 32.Lindkvist A, Eden A, Norstrom MM, Gonzalez VD, Nilsson S, Svennerholm B, Karlsson AC, Sandberg JK, Sonnerborg A, Gisslen M. Reduction of the HIV-1 reservoir in resting CD4+ T-lymphocytes by high dosage intravenous immunoglobulin treatment: a proof-of-concept study. AIDS research and therapy. 2009;6:15. doi: 10.1186/1742-6405-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buggert M, Norstrom MM, Czarnecki C, Tupin E, Luo M, Gyllensten K, Sonnerborg A, Lundegaard C, Lund O, Nielsen M, Karlsson AC. Characterization of HIV-specific CD4+ T cell responses against peptides selected with broad population and pathogen coverage. PloS one. 2012;7:e39874. doi: 10.1371/journal.pone.0039874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roederer M, Nozzi JL, Nason MX. SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytometry. Part A: the journal of the International Society for Analytical Cytology. 2011 doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feeney ME, Tang Y, Pfafferott K, Roosevelt KA, Draenert R, Trocha A, Yu XG, Verrill C, Allen T, Moore C, Mallal S, Burchett S, McIntosh K, Pelton SI, St John MA, Hazra R, Klenerman P, Altfeld M, Walker BD, Goulder PJ. HIV-1 viral escape in infancy followed by emergence of a variant-specific CTL response. J Immunol. 2005;174:7524–7530. doi: 10.4049/jimmunol.174.12.7524. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, Pillay T, Rousseau C, Mullins JI, Brander C, Walker BD, Stuart DI, Kiepiela P, Goulder P. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. Journal of virology. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 38.Almeida JR, Sauce D, Price DA, Papagno L, Shin SY, Moris A, Larsen M, Pancino G, Douek DC, Autran B, Saez-Cirion A, Appay V. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113:6351–6360. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Migueles SA, Rood JE, Berkley AM, Guo T, Mendoza D, Patamawenu A, Hallahan CW, Cogliano NA, Frahm N, Duerr A, McElrath MJ, Connors M. Trivalent adenovirus type 5 HIV recombinant vaccine primes for modest cytotoxic capacity that is greatest in humans with protective HLA class I alleles. PLoS pathogens. 2011;7:e1002002. doi: 10.1371/journal.ppat.1002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fitzgerald DW, Janes H, Robertson M, Coombs R, Frank I, Gilbert P, Loufty M, Mehrotra D, Duerr A. An Ad5-vectored HIV-1 vaccine elicits cell-mediated immunity but does not affect disease progression in HIV-1-infected male subjects: results from a randomized placebo-controlled trial (the Step study) The Journal of infectious diseases. 2011;203:765–772. doi: 10.1093/infdis/jiq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M, Jr, Lifson JD, Picker LJ. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makedonas G, Betts MR. Living in a house of cards: reevaluating CD8+ T-cell immune correlates against HIV. Immunological reviews. 2011;239:109–124. doi: 10.1111/j.1600-065X.2010.00968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrari G, Korber B, Goonetilleke N, Liu MK, Turnbull EL, Salazar-Gonzalez JF, Hawkins N, Self S, Watson S, Betts MR, Gay C, McGhee K, Pellegrino P, Williams I, Tomaras GD, Haynes BF, Gray CM, Borrow P, Roederer M, McMichael AJ, Weinhold KJ. Relationship between functional profile of HIV-1 specific CD8 T cells and epitope variability with the selection of escape mutants in acute HIV-1 infection. PLoS pathogens. 2011;7:e1001273. doi: 10.1371/journal.ppat.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Streeck H, Brumme ZL, Anastario M, Cohen KW, Jolin JS, Meier A, Brumme CJ, Rosenberg ES, Alter G, Allen TM, Walker BD, Altfeld M. Antigen load and viral sequence diversification determine the functional profile of HIV-1-specific CD8+ T cells. PLoS medicine. 2008;5:e100. doi: 10.1371/journal.pmed.0050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miura T, Brockman MA, Schneidewind A, Lobritz M, Pereyra F, Rathod A, Block BL, Brumme ZL, Brumme CJ, Baker B, Rothchild AC, Li B, Trocha A, Cutrell E, Frahm N, Brander C, Toth I, Arts EJ, Allen TM, Walker BD. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphotye recognition. Journal of virology. 2009;83:2743–2755. doi: 10.1128/JVI.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pohlmeyer CW, Buckheit RW, 3rd, Siliciano RF, Blankson JN. CD8+ T cells from HLA-B*57 elite suppressors effectively suppress replication of HIV-1 escape mutants. Retrovirology. 2013;10:152. doi: 10.1186/1742-4690-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iglesias MC, Almeida JR, Fastenackels S, van Bockel DJ, Hashimoto M, Venturi V, Gostick E, Urrutia A, Wooldridge L, Clement M, Gras S, Wilmann PG, Autran B, Moris A, Rossjohn J, Davenport MP, Takiguchi M, Brander C, Douek DC, Kelleher AD, Price DA, Appay V. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood. 2011;118:2138–2149. doi: 10.1182/blood-2011-01-328781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, Piechocka-Trocha A, Cesa KT, Sela J, Cung TD, Toth I, Pereyra F, Yu XG, Douek DC, Kaufmann DE, Allen TM, Walker BD. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nature immunology. 2012;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoof I, Perez CL, Buggert M, Gustafsson RK, Nielsen M, Lund O, Karlsson AC. Interdisciplinary analysis of HIV-specific CD8+ T cell responses against variant epitopes reveals restricted TCR promiscuity. Journal of immunology. 2010;184:5383–5391. doi: 10.4049/jimmunol.0903516. [DOI] [PubMed] [Google Scholar]
- 50.Liu MK, Hawkins N, Ritchie AJ, Ganusov VV, Whale V, Brackenridge S, Li H, Pavlicek JW, Cai F, Rose-Abrahams M, Treurnicht F, Hraber P, Riou C, Gray C, Ferrari G, Tanner R, Ping LH, Anderson JA, Swanstrom R, Cohen M, Karim SS, Haynes B, Borrow P, Perelson AS, Shaw GM, Hahn BH, Williamson C, Korber BT, Gao F, Self S, McMichael A, Goonetilleke N. Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. The Journal of clinical investigation. 2013;123:380–393. doi: 10.1172/JCI65330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang Y, Huang S, Dunkley-Thompson J, Steel-Duncan JC, Ryland EG, St John MA, Hazra R, Christie CD, Feeney ME. Correlates of spontaneous viral control among long-term survivors of perinatal HIV-1 infection expressing human leukocyte antigen-B57. AIDS. 2010;24:1425–1435. doi: 10.1097/QAD.0b013e32833a2b5b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cellerai C, Perreau M, Rozot V, Enders FB, Pantaleo G, Harari A. Proliferation capacity and cytotoxic activity are mediated by functionally and phenotypically distinct virus-specific CD8 T cells defined by interleukin-7R{alpha} (CD127) and perforin expression. Journal of virology. 2010;84:3868–3878. doi: 10.1128/JVI.02565-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balamurali M, Petravic J, Loh L, Alcantara S, Kent SJ, Davenport MP. Does cytolysis by CD8+ T cells drive immune escape in HIV infection? J Immunol. 2010;185:5093–5101. doi: 10.4049/jimmunol.1002204. [DOI] [PubMed] [Google Scholar]
- 54.Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, Miller MD, Else J, Pandrea I, Estes JD, Apetrei C, Schmitz JE, Ribeiro RM, Perelson AS, Silvestri G. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS pathogens. 2010;6:e1000747. doi: 10.1371/journal.ppat.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong JK, Strain MC, Porrata R, Reay E, Sankaran-Walters S, Ignacio CC, Russell T, Pillai SK, Looney DJ, Dandekar S. In vivo CD8+ T-cell suppression of siv viremia is not mediated by CTL clearance of productively infected cells. PLoS pathogens. 2010;6:e1000748. doi: 10.1371/journal.ppat.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.