Abstract
Faithful transmission of genetic material is essential for cell viability and organism health. The occurrence of DNA damage, due to either spontaneous events or environmental agents, threatens the integrity of the genome. The consequences of these insults, if allowed to perpetuate and accumulate over time, are mutations that can lead to the development of diseases such as cancer. Alkylation is a relevant DNA lesion produced endogenously as well as by exogenous agents including certain chemotherapeutics. We sought to better understand the cellular response to this form of DNA damage using mass spectrometry-based proteomics. For this purpose, we performed sub-cellular fractionation to monitor the effect of MMS treatment on protein localization to chromatin. The levels of over 500 proteins were increased in the chromatin-enriched nuclear lysate including histone chaperones. Levels of ubiquitin and subunits of the proteasome were also increased within this fraction, suggesting that ubiquitin-mediated degradation by the proteasome has an important role in the chromatin response to MMS treatment. Finally, the levels of some proteins were decreased within the chromatin-enriched lysate including components of the nuclear pore complex. Our spatial proteomics data demonstrate that many proteins that influence chromatin organization are regulated in response to MMS treatment, presumably to open the DNA to allow access by other DNA damage response proteins. To gain further insight into the cellular response to MMS-induced DNA damage, we also performed phosphorylation enrichment on total cell lysates to identify proteins regulated via post-translational modification. Phosphoproteomic analysis demonstrated that many nuclear phosphorylation events were decreased in response to MMS treatment. This reflected changes in protein kinase and/or phosphatase activity in response to DNA damage rather than changes in total protein abundance. Using these two mass spectrometry-based approaches, we have identified a novel set of MMS-responsive proteins that will expand our understanding of DNA damage signaling.
Keywords: mass spectrometry, proteomics, MMS, chromatin, phosphorylation
1. Introduction
Maintaining genome integrity over the course of each cell cycle requires fidelity in DNA replication and segregation as well as a robust DNA damage response (DDR) [1, 2]. The lattermost is particularly challenging because of the number and scope of potential DNA lesions that may be encountered by the cell. Sources of DNA damage can be endogenous and exogenous. It has been estimated that each cell experiences approximately 20,000 spontaneous DNA damage events each day. If not repaired properly, DNA damage can be mutagenic, possibly leading to diseases such as cancer. Therefore, the DDR coordinates cellular proteins and pathways to regulate cell cycle progression, DNA repair and apoptosis. Many cancer therapeutic agents utilize the DDR to induce cytotoxicity [3].
Alkylating agents represent an important source of DNA damage. For example, S-adenosyl methionine (SAM), which serves as a methyl donor in various metabolic reactions, can also target DNA in a potentially mutagenic reaction. N-nitrosamine, a carcinogen found in tobacco, is an exogenous source of DNA alkylation. Finally, cancer chemotherapeutics such as nitrogen mustards including cyclophosphamide and bis-chloroethylnitrosourea (BCNU) are alkylating agents. In the laboratory, methyl methanesulfonate (MMS) and methylnitronitrosoguanidine (MNNG) have been used frequently to study the consequences of DNA alkylation [4]. The major adduct formed by treatment with the above agents is 7-methylguanine (N7-MeG). Methylation at this site frequently leads to generation of an abasic site via hydrolysis of the modified base. The base excision repair (BER) pathway is frequently called upon to remedy lesions caused by alkylation [5, 6].
The effect of MMS treatment on cellular signaling pathways has been primarily explored in yeast. Initially, microarray analysis was used to determine the transcriptional response to MMS [7]. A library of GFP-tagged proteins was screened by flow cytometry and led to the identification of 157 MMS-induced gene products [8]. Screens performed on haploid deletion libraries have identified many non-essential genes whose deletion renders cells MMS sensitive [9-11]. More recently, epistatic miniarray profiles (E-MAPs) have been assembled to identify genetic interactions among 418 yeast genes that display differential interactions upon MMS treatment [12]. DNA damage signaling events have also been examined by comparing phosphorylation in wild-type and checkpoint kinase null cells, leading to the identification of 62 DDR-regulated phosphorylation sites [13]. Systematic examination of the proteins involved in response to MMS treatment has not been extensively performed in higher eukaryotes, although an RNAi screen has been carried out in Drosophila cells [14].
Mass spectrometry-based proteomics is a powerful tool for identifying and quantifying protein expression, protein modifications and protein interactions that is widely used in biological investigation of cellular processes [15]. Multi-dimensional protein identification technology (MudPIT), which utilizes orthogonal liquid chromatography (LC) separations of peptides prior to tandem MS analysis, is routinely used to interrogate the protein constituents of complex biological samples [16]. Stable isotope labeling with amino acids in cell culture (SILAC) can be used to perform relative quantification of protein and protein modifications in combination with mass spectrometry [17]. The use of mass spectrometry-based proteomics in studies of the DDR has led to a significant leap forward in our understanding of the cellular signaling pathways engaged by human cells in response to DNA damage. In particular, spatial proteomics and phosphoproteomics have been performed in several studies.
DNA damage occurs in the context of chromatin and must be repaired in this environment as well [18, 19]. This means that nucleosome positioning, histone modifications and variants as well as other DNA binding proteins are all features of chromatin that must be regulated during DNA repair. Numerous specialized structures are present within the nucleus and it is likely that they also influence how DNA repair proceeds. Recently, several efforts have been made to understand the chromatin landscape in response to DNA damage. Biochemical sub-cellular fractionation into nuclear or chromatin-enriched fractions that were subsequently probed using mass spectrometry has been used to identify DNA-binding proteins sensitive to treatments such as etoposide, ultraviolet (UV) light or ionizing radiation (IR) [20-22].
Reversible protein post-translational modification (PTM) is a dynamic regulatory mechanism widely used in cellular signaling pathways including the DDR. PTMs may influence protein stability, protein activity, protein localization and protein interactions. Phosphorylation, ubiquitylation and sumoylation are PTMs frequently utilized as part of the DDR [23]. In particular, checkpoint kinases such as ATM and ATR are activated in response to multiple forms of DNA damage to phosphorylate and regulate their substrates. Several studies have used “substrate” antibodies that recognize phosphorylated S/T-Q sites to identify hundreds of targets of these kinases in the context of hydroxyurea, UV light or IR [24-26]. However, the activities of other protein kinases are also affected by DNA damage [27-29]. Unbiased phosphoproteomics studies performed using etoposide, neocarzinostatin (NCS) or IR have identified additional phosphorylation sites regulated by the DDR [30-32].
Although many of the proteins that comprise the initial response to MMS have been identified, the complete repertoire of downstream DDR events induced by MMS remains poorly understood. Mass spectrometry-based proteomics is well-suited to discover additional MMS responsive proteins. This approach was applied to two themes of the DNA damage response, protein recruitment to chromatin and protein post-translational modification, focusing on phosphorylation.
2. Materials and Methods
2.1 Cell culture
HeLa cells were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. SILAC was performed for more than 6 passages to differentially label proteins. HeLa cells were cultured in either “light” SILAC media (unlabeled lysine and arginine) or “heavy” SILAC media (13C6-lysine and 13C6,15N4-arginine) [Invitrogen]. For DNA damage treatment, separate populations of HeLa cells grown in either “light” or “heavy” SILAC media were seeded in equal cell numbers onto 150 mm dishes. The following day the cells were treated with thymidine [Sigma-Aldrich] at a concentration of 2 mM for approximately 16 hours. HeLa cells grown in “light” SILAC media were simultaneously treated with MMS [Sigma-Aldrich] diluted to a final concentration of 0.05% during the final hour of thymidine treatment. Cells were then harvested for the acute time point. For the S-phase time point, both cell populations were washed twice with phosphate buffered saline (PBS) and then replaced with fresh SILAC media. Cells were grown for an additional four hours prior to harvest.
2.2 Cell lysis
Sub-cellular fractionation was performed as previously described with minor modifications [33]. Cells were lysed in Buffer A (1 mM Hepes pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT and protease inhibitors) supplemented with 0.1% TritonX-100 for 10 minutes at 4 °C. Cells were centrifuged at 4000 rpm for 5 minutes to pellet the nuclei. The supernatant was clarified by re-centrifuging at full speed and this lysate was used as the cytosol-enriched fraction. The nuclei were washed in Buffer A for 10 minutes at 4 °C followed by centrifugation at 4000 rpm for 5 minutes; the supernatant was then discarded. The nuclei were lysed in Buffer B (3 mM EDTA, 0.2 M EGTA, 1 mM DTT and protease inhibitors) for 30 minutes at 4 °C and then centrifuged at 4000 rpm for 5 minutes. The supernatant was removed and the pellet was re-suspended in Buffer C (50 mM Tris-Cl pH 8, 250 mM NaCl, 1% NP40, 1 mM DTT and protease inhibitors) for 20 minutes at 4 °C. Following centrifugation at full speed for 5 minutes, the resulting supernatant was used as the chromatin-enriched nuclear fraction.
Whole cell protein lysates were prepared by incubating cells in 50 mM Hepes pH 7.0, 250 mM NaCl, 0.1% NP-40, PhosSTOP phosphatase inhibitor cocktail [Roche Applied Science] and protease inhibitors for 30 minutes at 4 °C. Cells were centrifuged at full speed for 10 minutes and the resulting supernatant was saved for either direct analysis or phosphorylation enrichment.
2.3 Sample preparation for mass spectrometry
Samples were first denatured in 8 M urea and then reduced and alkylated with 10 mM Tris(2-carboxyethyl)phosphine hydrochloride [Roche Applied Science] and 55 mM iodoacetamide [Sigma-Aldrich] respectively. The urea concentration was diluted to 2 M with 100 mM Tris pH 8.5 and then the samples were incubated overnight at 37 °C with trypsin [Promega] at a final protease to protein ratio of 1:50.
For phosphorylation analysis, phosphopeptide enrichment was performed using titanium dioxide magnetic beads [Thermo Scientific] according to the manufacturer's specifications with only the following modification. Enrichments were carried out in eppendorf tubes rather than a 96-well plate and magnetic separations were carried out on a magnetic rack [Invitrogen]. One milligram of digested whole cell protein lysate was enriched for each sample. Briefly, the magnetic beads were washed four times with Binding Buffer and then incubated with the protein digests that had been adjusted to a final solution of 80% acetonitrile/2% formic acid. Afterward, the magnetic beads were washed four more times with Binding Buffer and once with Washing Buffer prior to incubation with Elution Buffer. The final eluates were lyophilized prior to mass spectrometry analysis.
Protein digests or enriched phosphopeptides were pressure-loaded onto 250 micron i.d. fused silica capillary [Polymicro Technologies] columns with a Kasil frit packed with 3 cm of 5 micron Partisphere strong cation exchange (SCX) resin [Whatman] and 3 cm of 5 micron C18 resin [Phenomenex]. After desalting, each bi-phasic column was connected to a 100 micron i.d. fused silica capillary [Polymicro Technologies] analytical column with a 5 micron pulled-tip, packed with 10 cm of 5 micron C18 resin [Phenomenex].
2.4 Mass spectrometry analysis
Each MudPIT column was placed inline with an 1100 quaternary HPLC pump [Agilent Technologies] and the eluted peptides were electrosprayed directly into the mass spectrometer. The buffer solutions used were 5% acetonitrile/0.1% formic acid (buffer A), 80% acetonitrile/0.1% formic acid (buffer B) and 500 mM ammonium acetate/5% acetonitrile/0.1% formic acid (buffer C). For analysis of protein lysates, an 11-step MudPIT was run with salt pulses of 0%, 10%, 20%, 30%, 40%, 50%, 60%, 70% and 100% buffer C and 90% buffer C/10% buffer B (twice). For phosphopeptide analysis, a 6-step MudPIT was run with salt pulses of 0%, 10%, 20%, 40%, and 100% buffer C and 90 buffer C/10% buffer B. In each MudPIT step, the 120 minute elution gradient had the following profile: 10% buffer B beginning at 15 minutes to 40% buffer B at 105 minutes.
Protein analyses of sub-cellular fractions were carried out on an LTQ Orbitrap XL [Thermo Scientific]. A cycle consisted of one full scan mass spectrum (400-2000 m/z) in the Orbitrap at 60,000 resolution followed by five data-dependent collision induced dissociation (CID) MS/MS spectra in the LTQ. Dynamic exclusion was enabled with a repeat count of 1, a repeat duration of 30 seconds, an exclusion list size of 150 and an exclusion duration of 180 seconds. Protein and phosphopeptide analyses of whole cell lysate were carried out on a LTQ Orbitrap Velos [Thermo Scientific]. A cycle consisted of one full scan mass spectrum (300-1600 m/z) in the Orbitrap at 60,000 resolution followed by 12 data-dependent collision induced dissociation (CID) MS/MS spectra in the LTQ Velos. Charge state screening was enabled and unassigned charge states and charge state 1 were rejected. Dynamic exclusion was enabled with a repeat count of 1, a repeat duration of 30 seconds, an exclusion list size of 500 and an exclusion duration of 120 seconds. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system [Thermo Scientific].
2.5 Data analysis
MS/MS spectra were extracted using RawXtract (version 1.9.9) [34]. MS/MS spectra were searched with the ProLuCID algorithm against human Uniprot database concatenated to a decoy database in which the sequence for each entry in the original database was reversed [16]. For protein identifications, the “light” ProLuCID search was performed using no enzyme specificity and static modification of cysteine due to carboxyamidomethylation (57.02146). A second “heavy” ProLuCID search was performed and additionally considered static modification of arginine (10.0124) and lysine (6.0204). For phosphopeptide analysis, the ProLuCID search was performed using full enzyme specificity and a differential modification of serine, threonine and tyrosine due to phosphorylation (79.9663) was considered in addition to the above static modifications.
Both “light” and “heavy” ProLuCID search results were assembled and filtered using the DTASelect (version 2.0) algorithm [35], requiring a minimum of one peptide per protein identification. For protein identifications, peptides were required to be at least partially tryptic and for phosphopeptide identifications, peptides were required to be fully tryptic. All peptide-spectra matches had less than 10 ppm mass deviation. For protein identification, the protein false positive rate was kept below one percent and the average mass deviation was less than 2 ppm. For phosphopeptide identification, only modified peptides were considered and the peptide false positive rate was kept below one percent.
Phosphorylation site localization was performed using AScore [36]. Phosphorylation sites with an AScore of 13 or greater were considered to be confidently localized. For phosphopeptide and protein quantification, DTASelect was re-run to allow subset proteins and to separately display peptide identifications in multiple salt steps. Quantification was performed using the Census algorithm and the proline conversion correction option was selected [37]. A determinant factor of 0.5 was used to filter quantified peptides. SILAC ratios report are the MMS-treated divided by the control. Therefore, numbers greater than one indicate an increased protein level in response to MMS treatment and numbers less than one indicate a decreased protein level in response to MMS treatment. Log2 ratio values are also reported for the purpose of generating volcano plots.
Comparison of replicate analyses and statistics were performed using the Integrated Proteomics Pipeline (Integrated Proteomics Applications). Direct ratio comparison of SILAC labeled proteins utilized individual peptide measurements [38]. Statistical analysis of direct ratio comparisons was performed using a t-test and a Benjamini-Hochberg correction was applied. A corrected p-value of 0.05 or less was considered significant. Ratio/ratio comparisons utilized replicate protein measurements. Statistical analysis of ratio/ratio comparisons was performed using analysis of variance (ANOVA) and a Benjamini-Hochberg (B-H) correction was applied. A corrected p-value of 0.05 or less was considered significant. For the generation of volcano plots, log10 of the B-H corrected p-values are reported. Statistically significant protein measurements were required to have been quantified in at least three of four replicate analyses.
Gene ontology (GO) analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) [39]. MMS-responsive proteins were compared against a background of all human proteins. Statistically significant terms were examined using the functional annotation clustering tool and the functional annotation chart tool. The Biological Database Network (bioDBnet) was used to map Uniprot identifiers to gene ID, gene symbol and Uniprot protein name [40]. Motif-X was used to identify enriched motifs within the phosphoproteomic data [41]. The default parameters were chosen.
2.6 Flow cytometry
Cells were labeled with 10 μM BrdU for 1 hour prior to harvest. Time points were taken at 0, 2, 4, 6, 8 and 10 hours post-release from thymidine block. Cells were harvested by trypsinization and fixed in ethanol overnight at 4 degrees Celsius. Cells were then incubated in 2N HCl with 0.5%Triton X-100 and 1mg/mL pepsin for 20 minutes at room temperature. Next, cells were incubated with 0.1 M sodium borate for 5 minutes. Cells were re-suspended in one percent BSA in PBS and incubated with 20 μl FITC conjugated anti-BrdU (Becton-Dickinson) for 30 minutes in the dark. Cells were re-suspended in PBS containing 5 μg/ml propidium iodide prior to analysis.
2.8 Antibodies
The following antibodies were used for immunoblotting or immunofluorescence staining: phospho-CHK2 Thr68 (Cell Signaling – 2661), DNMT1 (GeneTex – GTX116011), phospho-H2AX Ser 139 (Millipore – 05-636), KAP1 (GeneTex – GTX102226), NAP1L4 (Abcam – ab21631), NUP107 (GeneTex – GTX116664), NUP155 (GeneTex – GTX120945), NUP160 (Bethyl Labs – A301-791A), SET (Bethyl Labs – A302-261A) and ubiquitin (Santa Cruz – sc8017).
3. Results and Discussion
Given the prevalence of DNA damage cause by alkylating agents, a more complete understanding of the cellular proteins and pathways involved in the DNA damage response is needed. We used mass spectrometry-based proteomics to investigate the DDR following MMS treatment. It is worth noting that MMS can also alkylate other macromolecules such as proteins. Protein alkylation was not monitored in this study but could contribute to changes in protein levels and localizations that were observed.
Stable isotope labeling with amino acids in cell culture (SILAC) was utilized for protein quantification by mass spectrometry. For this purpose, one population of HeLa cells was grown in medium containing heavy [13C6,15N4] arginine and [13C6] lysine and the other population of HeLa cells was grown in medium containing light arginine and lysine. HeLa cells were chosen for this study because they are easy to grow and synchronize and because they have been successfully used in siRNA screens to identify proteins involved in DDR signaling [42, 43]. Both HeLa cell populations were treated with 2 mM thymidine overnight to synchronize cells in late G1/S-phase. The “treated” population of cells was concurrently exposed to 0.05% MMS during the final hour of thymidine treatment while the “control” population was not exposed to MMS. Both populations were washed twice with PBS and fresh medium was added to allow cells to resume cell cycle progression. This was monitored by flow cytometry using propidium iodide (PI) and BrdU incorporation to monitor DNA content and DNA replication respectively (Figure S1A). Based on this analysis, both populations entered S-phase at the same rate, peaking at 4 hours post-release. After this time point, the “control” population began to pass out of S-phase while the “treated” population continued to incorporate the same level of BrdU at all time points taken (up to 10 hours post-release). Based on these observations, we initially harvested cells for protein analysis at two time points. An acute time point was taken immediately at the conclusion of thymidine/MMS treatment (no release) and an S-phase time point was taken 4 hours post-release.
3.1 Spatial proteomics
Spatial proteomics was employed to specifically investigate how MMS treatment affected the proteome of sub-cellular compartments. This should allow changes in protein localization to be monitored in response to MMS treatment that may otherwise have been obscured in the analysis of total protein levels. Sub-cellular fractionation was performed on each sample to generate a chromatin-enriched nuclear lysate and a cytosol-enriched lysate for analysis by mass spectrometry. Twenty micrograms from a trypsin digestion of a 1:1 mixture of “control” and “treated” lysates were analyzed on an LTQ Orbitrap XL using MudPIT. Peptide spectra matches were generated using the ProLuCID search algorithm and filtered using DTASelect with a protein false discovery rate of less than one percent. Peptide/protein quantification was performed using Census.
3.1.1 Acute time point
We began by confirming that activation of the DDR was detectable immediately following MMS treatment using two commonly used markers. We examined Ser139 phosphorylation of gamma-H2AX by immunofluorescence and observed a strong signal in MMS-treated cells (Figure S1B). We also observed increased Thr68 phosphorylation of CHK2 following MMS treatment (Figure S1C). Based on these observations, DDR signaling appears to be initiated under the conditions of this study. We expected that further analysis of the acute time point by mass spectrometry would identify additional sensors of the DNA damage response. Unfortunately, the levels of very few proteins were observed to be reproducibly altered in either fraction at this time point (Table S1, Table S2). There are several explanations for this occurrence. The level of DNA damage may not have been sufficient to induce changes in protein levels between the two fractions. This is unlikely given the strong effect observed on cell cycle progression. Alternatively, some of the proteins that respond to DNA damage caused by MMS treatment may be of low abundance, and therefore not observed in this analysis. This explanation is most probable, given that many DNA-binding proteins are known to be lowly expressed in cells.
Although we did not observe many large changes in protein levels at the acute time point, there were some noteworthy events. Within the chromatin-enriched nuclear fraction (Table S2), SUMO2/3 levels increased greater than 2-fold in response to MMS treatment. Levels of FEN1, MSH2, XRCC5 and XRCC6 were also increased in this fraction. Components of the nucleosome remodeling and histone deacetylase (NuRD) complex were also observed to be increased within the chromatin-enriched fraction. This included MBD3, MTA2, GATAD2A and CHD4. Previous studies have also identified components of this complex in the response to DNA damage caused by UV light and IR treatment [21, 22, 44]. This indicates that the NuRD complex plays an early and potentially universal role in response to many forms of DNA damage. Since the changes in protein abundance observed at the acute time point were generally small, we did not pursue this time point further. We reasoned that the effects of MMS treatment would be easier to observe in the S-phase time point, when more dramatic changes in protein levels would have had time to develop.
3.1.2 S-phase time point
Four replicate experiments were performed on each sample to demonstrate that changes were due to biological origin rather than variation in procedure. In total, 2088 proteins were quantified in the cytosol-enriched lysate (Table S3). Of these, 1007 proteins were quantified in at least three of four replicate analyses (Figure 1A,B). In total, 2604 proteins were quantified in the chromatin-enriched nuclear lysate (Table S4). Of these 1453 proteins were quantified in at least three of four replicate analyses (Figure 1A,B). Only proteins quantified in at least three of four replicate analyses were considered in downstream analysis, although proteins with fewer measurements may also be of interest.
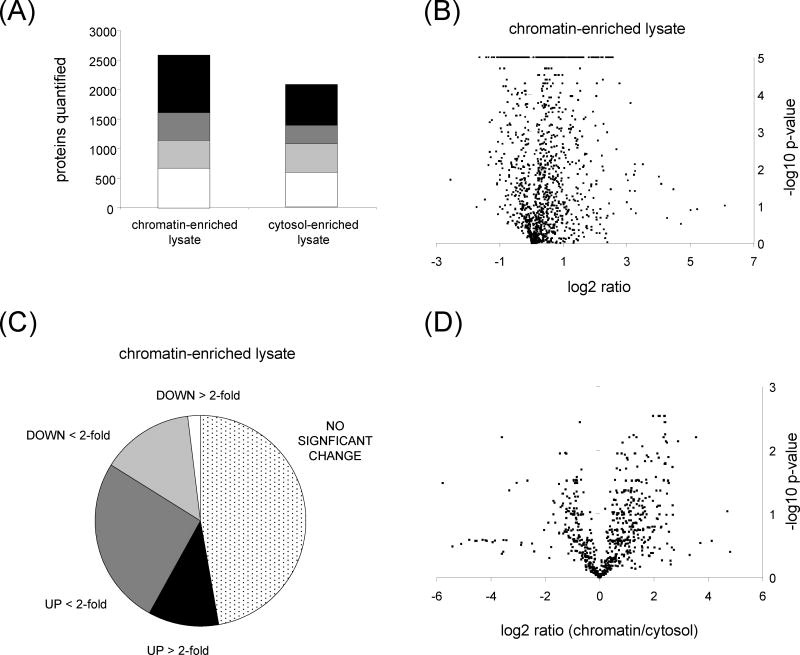
Figure 1.
Analysis of proteins quantified in each sub-cellular fraction following MMS treatment at the S-phase time point. (A) Number of proteins quantified in one (white), two (light gray), three (dark gray) or four (black) replicates in either the chromatin-enriched nuclear lysate or the cytosol-enriched lysate. (B) Volcano plot for proteins from chromatin-enriched nuclear lysate quantified by at least three replicates, showing the log2 ratio (x-axis) and log10 B-H corrected p-value (y-axis). (C) Distribution of protein quantification following MMS treatment in either the chromatin-enriched nuclear lysate, showing the percentage of proteins increased 2-fold or greater (black), proteins increased less than 2-fold (dark gray), proteins that decreased less than 2-fold (light gray), proteins decreased 2-fold or greater (white) and proteins whose levels were not significantly changed (dotted). (D) Volcano plot for proteins in both chromatin-enriched nuclear lysate and cytosol-enriched lysate, showing the log2 ratio (x-axis) and log10 B-H corrected p-value (y-axis).
Gene ontology enrichment was used to examine the cellular component of the two sub-cellular fractions (Table S5). This was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID). Proteins quantified in the cytosol-enriched lysate showed a clear enrichment for the gene ontology term “cytosol.” However, proteins from other sub-cellular compartments including the nucleus were also present. These contaminants probably represent either proteins that are capable of shuttling between multiple locations or proteins that leaked into the cytosol during the fractionation procedure. This observation is consistent with other studies employing biochemical fractionation of sub-cellular compartments subsequently analyzed by mass spectrometry [20]. This also underscores the fact that these procedures represent enrichments rather than purifications and that the sensitivity of mass spectrometers permits the analysis of low abundance constituents within complex mixtures. Despite the presence of nuclear proteins within the cytosol-enriched fraction, we found a stronger enrichment for the gene ontology term “chromatin” in the chromatin-enriched nuclear fraction. This demonstrates that DNA bound proteins were successfully partitioned by our fractionation procedure. Surprisingly, the gene ontology terms “mitochondria” and “endoplasmic reticulum” were also enriched in the chromatin fraction. This likely reflects the fact that membranes were pelleted along with chromatin prior to release of associated proteins. Therefore, this fraction may also yield information about proteins from these two other organelles.
3.1.2.1 Proteins increased within the chromatin-enriched nuclear lysate
In the chromatin-enriched lysate, 159 proteins were increased 2-fold or greater in response to MMS treatment (Figure 1C). These represent proteins recruited to the chromatin in response to DNA damage. We used gene ontology to examine prevalent biological processes among the increased proteins. The term “cell cycle” was enriched. It was expected that MMS-induced DNA damage should affect cell cycle-related processes via checkpoint activation. The term “regulation of protein ubiquitylation” was also enriched. Examination of the proteins belonging to this term showed that they were primarily proteasome subunits. In fact, 37 individual subunits of the proteasome were quantified in the chromatin-enriched lysate, all of which trended toward increased chromatin association in response to MMS treatment (Table 1). Previous studies have indicated that the proteasome is recruited to chromatin in response to certain forms of DNA damage and protein post-translational modification by ubiquitin is regulated in response to DNA damage [18]. We also noted that ubiquitin was quantified in the chromatin-enriched lysate and that the levels of ubiquitin increased in response to MMS treatment. We confirmed by immunoblotting that there was an increase in ubiquitin conjugates within the chromatin-enriched lysate from MMS treated cells (Figure 2A). These data suggest that ubiquitin-mediated protein degradation by the proteasome is a key mechanism that is part of the DDR.
Table 1. Proteasome subunits quantified in chromatin-enriched lysate.
| gene symbol | average ratio | p-value |
|---|---|---|
| PSMB5 | 5.63 | 3.57E-02 |
| PSMA2 | 4.13 | 2.09E-02 |
| PSME3 | 3.82 | 0.00E+00 |
| PSMD5 | 3.54 | 4.01E-01 |
| PSMC2 | 3.01 | 6.63E-02 |
| PSMC3 | 2.87 | 0.00E+00 |
| PSMA1 | 2.83 | 1.00E-05 |
| PSMA4 | 2.76 | 1.40E-04 |
| PSMD2 | 2.65 | 0.00E+00 |
| PSMD8 | 2.61 | 0.00E+00 |
| PSMA5 | 2.55 | 1.00E-05 |
| PSMD7 | 2.51 | 0.00E+00 |
| PSMD1 | 2.43 | 1.10E-04 |
| PSMA6 | 2.42 | 3.80E-04 |
| PSMD13 | 2.42 | 1.09E-01 |
| PSMD14 | 2.26 | 3.06E-03 |
| PSMC4 | 2.13 | 1.23E-01 |
| PSMB1 | 2.1 | 1.00E-05 |
| PSME4 | 2.05 | X |
| PSMC6 | 1.98 | 2.52E-01 |
| PSMD11 | 1.97 | 2.66E-02 |
| PSMC5 | 1.91 | 3.43E-03 |
| PSMC1 | 1.84 | 1.17E-01 |
| PSMD4 | 1.78 | 1.13E-01 |
| PSMB3 | 1.75 | 7.80E-04 |
| PSMA7 | 1.73 | 1.00E-05 |
| PSMA8 | 1.73 | 1.00E-05 |
| PSMB4 | 1.72 | 6.81E-02 |
| PSMB6 | 1.66 | 3.00E-05 |
| PSME2 | 1.64 | 8.70E-04 |
| PSME1 | 1.44 | 3.61E-01 |
| PSMD3 | 1.41 | 1.21E-01 |
| PSMB7 | 1.39 | 5.72E-03 |
| PSMD6 | 1.24 | 0.00E+00 |
| PSMA3 | 1.18 | 2.40E-01 |
| PSMB2 | 0.96 | 6.77E-01 |
| PSMB9 | 0.7 | X |
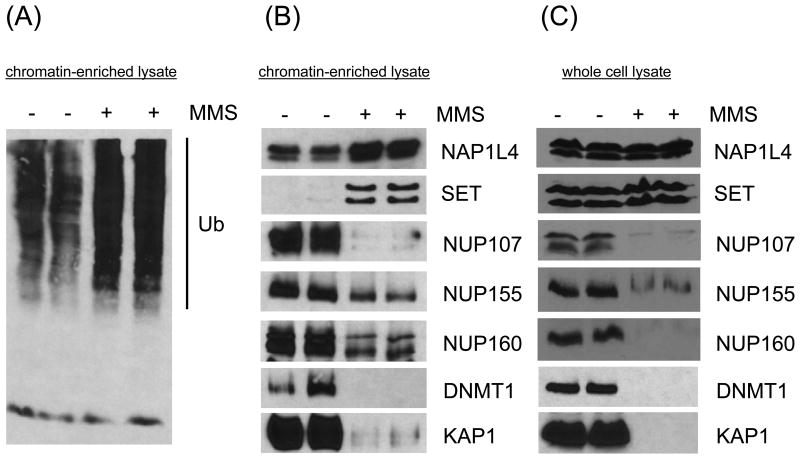
Figure 2.
Immunoblotting of proteins from the chromatin-enriched nuclear lysate in response to MMS treatment at the S-phase time point is in agreement with quantification measurements by mass spectrometry. Two independently generated samples were analyzed for each condition. (A) Immunoblot showing that high molecular weight ubiquitin conjugates are elevated in the chromatin-enriched nuclear lysate following MMS treatment. (B) Immunoblot showing validation of selected proteins from the chromatin-enriched nuclear lysate following MMS treatment. (C) Immunoblot showing the same proteins from the whole cell lysate following MMS treatment.
We looked for other proteins with shared function among those increased in the chromatin-enriched lysate in response to MMS treatment and discovered three histone chaperones, SET, NAP1L1 and NAP1L4. We confirmed SET and NAP1L4 by immunoblotting (Figure 2B). Histone chaperones play an important role in the addition and removal of histones from DNA [45]. Several studies have demonstrated a role for the histone chaperones ASF1 and CAF-1 in the DNA damage response [46]. These histone chaperones specifically bind H3/H4, while the histone chaperones identified in this study associate with H2A/H2B. SUPT16H, which is a component of the facilitates chromatin transcription (FACT) complex, has been previously shown to regulate H2AX exchange on chromatin, and this activity is coordinated with H2AX phosphorylation [47]. The placement of histones within chromatin can influence the accessibility of other DNA-binding proteins to their relevant binding sites. This is critical for components of the DDR to access site of DNA damage. Additionally, post-translational modification of histone tails serves to recruit chromatin-binding proteins, which is another critical component of DDR signaling. Exchange of histones by chaperones represents a means of quickly resetting histone post-translational modifications. This could be performed at the initiation of the DDR and/or during recovery.
3.1.2.2 Proteins decreased within the chromatin-enriched nuclear lysate
Only 30 proteins detected in the chromatin-enriched lysate were decreased 2-fold or greater in response to MMS treatment (Figure 1C). These represent proteins that are removed from chromatin as part of the DNA damage response. We used gene ontology to see if there were any common components among these proteins and identified the term “nuclear pore complex” among the enriched terms. Indeed, 18 out of 22 members of the nuclear pore complex quantified in the chromatin-enriched lysate were decreased in response to MMS treatment (Table 2). We confirmed this decrease in protein levels by immunoblotting for three nucleoporins, NUP107, NUP155 and NUP160 (Figure 2B). Previous studies have demonstrated an association between nuclear pore complexes and chromatin. Additionally, some individual nucleoporins have been shown to bind sites of chromatin independent from intact nuclear pore complexes [48]. The association of nucleoporins with chromatin is thought to regulate the transcriptional activity of these regions. Individual nucleoporins have also been shown to influence the DNA damage response [49]. Depletion of specific nucleoporins by siRNA can both positively and negatively effect DNA damage signaling [43, 50], potentially be affecting nuclear import and/or nuclear export of proteins including 53BP1.
Table 2. Nucleoporins quantified in chromatin-enriched lysate.
| gene symbol | average ratio | p-value |
|---|---|---|
| NUP153 | 0.25 | 3.90E-02 |
| NUP107 | 0.36 | 3.55E-03 |
| NUP85 | 0.36 | 1.80E-04 |
| NUP160 | 0.37 | 0.00E+00 |
| NUP133 | 0.47 | 0.00E+00 |
| NUP98 | 0.48 | 1.00E-05 |
| NUP205 | 0.51 | 6.23E-03 |
| NUP188 | 0.52 | 5.07E-03 |
| NUP93 | 0.52 | 1.00E-05 |
| NUP155 | 0.62 | 0.00E+00 |
| NUP50 | 0.65 | 1.00E-04 |
| NUP37 | 0.67 | 1.73E-01 |
| NUP62 | 0.7 | 3.52E-01 |
| RAE1 | 0.73 | 2.44E-02 |
| NUP35 | 0.85 | 3.70E-01 |
| NDC1 | 0.88 | 2.17E-01 |
| NUP358 | 0.91 | 3.34E-01 |
| NUP214 | 0.95 | 5.64E-01 |
| TPR | 1.06 | 5.68E-01 |
| NUP88 | 1.65 | 4.93E-01 |
| SEH1 | 1.65 | 2.32E-03 |
| NUP54 | 2.67 | 7.84E-01 |
Proteins can be observed to decrease in relative abundance within one sub-cellular fraction following a perturbation for several reasons. One possibility is that these proteins have changed localization and are present in a different sub-cellular location. In this case, total protein levels would be expected to remain unchanged. Alternatively, these proteins could be degraded. In this case, a decrease in total protein levels should also be observed. We examined the levels of NUP107, NUP155 and NUP160 in whole cell extract by immunoblotting and observed a similarly decrease following MMS treatment as we had seen in the chromatin-enriched lysate (Figure 2C). To determine whether this was a more general phenomenon, we examined two additional proteins, DNMT1 and KAP1. We first confirmed that DNMT1 and KAP1 were decreased in the chromatin-enriched lysate in response to MMS treatment by immunoblotting (Figure 2B). We then examined the expression of these proteins in whole cell lysate. We observed that DNMT1 and KAP1 showed a similar decrease in their levels in whole cell lysate in response to MMS treatment by immunoblotting (Figure 2C). This indicates that they are most likely degraded. There is precedent for the involvement of ubiquitin-mediated proteasome degradation of proteins as part of the DDR [51]. Our data also supports a role for the regulation of chromatin-associated proteins by the proteasome (Table 1). Proteins that are targeted for degradation rather than simply moved to a different sub-cellular address are likely to be key signal transducers that must be silenced in order to properly initiate the DDR.
DNMT1 is responsible for copying methylation patterns onto newly synthesized DNA during S-phase. DNMT1 has been previously reported to be depleted by methylating agents including MMS [17]. Degradation of DNMT1 may be a DDR mechanism to prevent signaling through chromatin modifying proteins that would otherwise respond to the aberrant methylation patterns induced by MMS treatment. KAP1 is an E3 SUMO protein ligase and nuclear co-repressor that can be recruited to chromatin to regulate various cellular processes including transcription [52]. KAP1 has been previously shown to be regulated by the DDR. KAP1 is phosphorylated by the checkpoint kinase ATM on Ser824 in response to several forms of DNA damage. This phosphorylation event leads to the inhibition of its SUMO protein ligase activity. Degradation of KAP1 may represent another mechanism through which the DDR can inhibit its activity. Additionally, degradation of KAP1 would affect target genes that it regulates in its role as a transcriptional co-repressor. These enzymes modify either the protein or DNA component of chromatin. The fact that they are negatively regulated through degradation following MMS treatment demonstrates that changes to chromatin structure and organization represent a critical arm of the DDR.
3.1.2.3 Proteins within the cytosol-enriched lysate
In the cytosol-enriched lysate, only 51 proteins were increased while 8 proteins were decreased greater than 2-fold following MMS treatment. To further explore the regulation of protein localization in response to MMS treatment, we identified proteins present in both the chromatin-enriched nuclear lysate as well as the cytosol-enriched lysate (Table S6). There were 719 such proteins. We then compared the relative changes in protein abundance within each fraction (Figure 1D). Most of these proteins showed a relatively larger increase in abundance within the chromatin-enriched lysate demonstrating a net movement of proteins in this direction. This demonstrates the utility of spatial proteomics to identify changes in protein localization between sub-cellular compartments in response to a perturbation such as DNA damage.
3.2 Post-translational modifications
Regulation of protein post-translational modification impacts all cellular processes. In the DNA damage response, protein phosphorylation, ubiquitylation and/or sumoylation are common themes. These PTMs permit rapid recruitment of substrates to arms of the DNA damage response and the reversible nature of these modifications permits the response to be terminated equally efficiently. The identification of DNA damage regulated substrates for these PTMs represents an important goal for understanding the cellular processes under the control of the DNA damage response.
3.2.1 Protein ubiquitylation and sumoylation
Our protein level analysis of the S-phase time point clearly supports a role for ubiquitin, and potentially SUMO, as part of the DNA damage response within the chromatin-enriched nuclear fraction. Quantification of ubiquitin demonstrated that the levels the protein is increased. SUMO1 was quantified in only one replicate, but showed a 2.64-fold increase. We confirmed the presence of elevated levels of high molecular weight ubiquitin conjugates by immunoblotting using independently generated samples (Figure 2A). Other components of the ubiquitin conjugation pathway were also increased, including the E1 ubiquitin-activating enzyme UBA1 and the E2 ubiquitin-conjugating enzyme L3 (UBE2L3). UBA1 has been previously shown to be required for downstream signaling involving 53BP1 and BRCA1 in response to IR [50]. UBE2L3 is known to be cell cycle regulated and has been shown to function in the intra-S phase checkpoint [53]. These proteins may regulate similar targets in response to MMS treatment. To date, only a few ubiquitylation substrates have been identified in human cells in response to MMS treatment. However, the recent development and optimization of ubiquitin remnant (di-glycine) antibodies should improve this gap in our knowledge [54].
3.2.2 Protein phosphorylation
Protein phosphorylation is a well-known regulator of DNA damage response signaling. Checkpoint kinases such as ATM/CHK2 and ATR/CHK1 are critical transducers whose substrates are effector proteins in the DNA damage signaling cascade. While many studies have focused on these proteins, other kinases have also been shown to be involved in the DNA damage response. Therefore, we took an unbiased approach to identify MMS responsive phosphorylation events. SILAC labeled HeLa cells were grown and treated as described for the spatial proteomics experiments. Cells were harvested at the S-phase time point and whole cell protein lysates prepared from these cells were subject to phosphorylation enrichment using titanium dioxide. AScore was used for phosphorylation site localization.
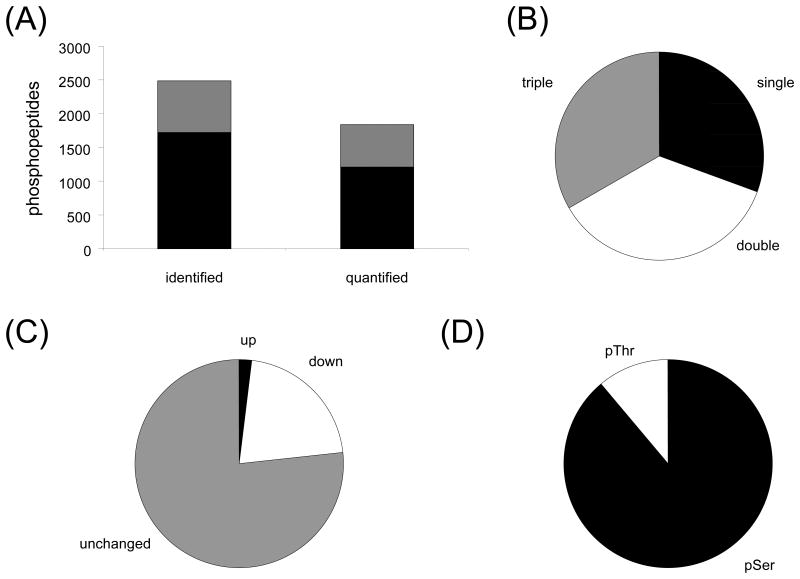
Using this approach, we identified 2487 phosphopeptides corresponding to 1032 unique sequences and 592 proteins (Table S7). We analyzed the functional annotation of these proteins using DAVID and found that Swiss-Prot/Protein Information Resource keywords “phosphoprotein” (566 proteins) and “nucleus” (309 proteins) were highly represented. We also examined gene ontology cellular components and found that many terms associated with the nucleus were enriched. Since we did not perform any sub-cellular fractionation prior to phosphopeptide enrichment, this was unexpected. However, nuclear events are central to the DNA damage response, and so the phosphopeptides in this data set should be very informative. Of the identified phosphopeptides, 1723 were identified in a single experiment while 764 were identified in multiple experiments (Figure 3A). The poor overlap between experiments likely reflects the fact that only a subset of the total phosphoproteome has been identified in this study. More exhaustive phosphoproteomic studies have routinely identified over 10,000 phosphopeptides using much greater amounts of starting material. Our data set consisted of 758 singly phosphorylated peptides, 902 doubly phosphorylated peptides and 827 triply phosphorylated peptides (Figure 3B). This indicates that the enrichment procedure is not biased by the degree of phosphorylation on peptides.
Figure 3.
Analysis of phosphopeptides quantified following MMS treatment at the S-phase time point. (A) Number of phosphopeptides identified or quantified by a single measurement (black) and by two or more measurements (gray) in total cell lysate. (B) Phosphorylation state distribution of identified phosphopeptides following MMS treatment in total cell lysate, showing percentage of singly phosphorylated peptides (black), doubly phosphorylated peptides (gray) and triply phosphorylated peptides (white). (C) Distribution of phosphopeptides quantification following MMS treatment in total cell lysate, showing percentage of proteins increased 2-fold or greater (black), proteins decreased 2-fold or greater (white) and proteins whose levels were unchanged (gray). (D) Phosphoamino acid distribution of quantified phosphopeptides with confident site localization (AScore greater or equal to 13) following MMS treatment in total cell lysate, showing percentage of phosphoserine (black) and phosphothreonine (white).
We quantified 1842 phosphopeptides corresponding to 863 unique sequences and 498 proteins (Table S8). The characteristics of the quantified phosphopeptides and the proteins from which they originated matched those observed for all identified phosphopeptides. We proceeded to examine the changes in phosphorylation. Among all quantified phosphopeptides, there were 560 phosphopeptides corresponding to 235 proteins decreased 2-fold or greater and 69 phosphopeptides corresponding to 51 proteins increased 2-fold or greater. Among the 628 phosphopeptides quantified in multiple experiments, there were 135 phosphopeptides corresponding to 98 proteins decreased 2-fold or greater and 12 phosphopeptides corresponding to 10 proteins increased 2-fold or greater (Figure 3C). Previous studies have mainly focused on phosphorylation events that are induced during the DDR [24-26, 30]. However, these data clearly indicate that decreased phosphorylation among a subset of targets is an important component of the response to MMS treatment. These phosphorylation events likely regulate processes that must be inhibited as part of the DNA damage response. For example, cellular processes such as DNA replication and cell cycle progression, which are regulated in part by protein phosphorylation, are candidates for inhibition by the DDR.
Changes in phosphopeptide abundance can be due to alterations in kinase/phosphatase activity or due to changes in underlying protein abundance. The later, while still of biological interest, is not directly related to phosphorylation. Understanding the origin of the changes in phosphopeptide abundance is helpful to understand the significance of the phosphoproteomic data and the cellular processes responsible for these changes. We began to address this issue by quantifying changes in total protein abundance from the same lysates used to prepare the phosphopeptides. A total of 3638 proteins were quantified and 2152 of these proteins were quantified in at least three replicates (Table S9). We mapped these proteins onto the phosphoproteomic data set. A total of 582 phosphopeptides had corresponding protein quantification data available (Table S10). In most cases, total protein abundance was not dramatically altered, indicating that the change in phosphopeptide abundance was due to a DDR-induced change in the activity of a protein kinase and/or phosphatase.
Next, we tried to better understand the regulation of the phosphorylation events in our data set. Substrates of specific kinases typically conform to a consensus motif. Confident localization of phosphorylation sites is a critical aspect of phosphoproteomics for downstream analysis. We considered any phosphorylation sites with an AScore greater or equal to 13 as confidently localized and filtered our data accordingly. This resulted in 569 phosphopeptides corresponding to 462 unique sequences and 294 proteins (Table S11). This represented 887 total phosphorylation sites, of which 789 were phosphoserine and 98 were phosphothreonine (Figure 3D). We searched for specific sequence motifs within our phosphopeptide data. There were 87 pS-P sites and 18 pT-P sites. Also, there were 7 pS-Q sites, which are likely ATM/ATR targets. The low number of ATM/ATR substrates in our data set, compared to those reported using directed enrichment of ATM/ATR substrates, is likely to reflect the binding preferences of the phosphopeptide enrichment strategy as well as the relative abundance of individual phosphopeptides within the original sample. Ser15 phosphorylation of p53 was elevated following MMS treatment (data not shown), suggesting that ATM was in fact activated by the MMS treatment conditions we used. We further analyzed the data for enriched motifs and found that acidic residues were common features of the enriched motifs, including the CK2 consensus sequence (S-X-X-E/D). The CDK consensus sequence (S-P-X-K) was also enriched to a lesser degree. Decreased CDK activity is most likely a result of checkpoint activation.
Among all localized phosphopeptides, there were 119 phosphopeptides corresponding to 182 phosphorylation sites on 95 proteins decreased 2-fold or greater and 17 phosphopeptides corresponding to 20 phosphorylation sites on 17 proteins increased 2-fold or greater. Among phosphopeptides localized in two or more experiments, there were 32 phosphopeptides corresponding to 56 phosphorylation sites on 31 proteins decreased 2-fold or greater and 5 phosphopeptides corresponding to 5 phosphorylation sites on 5 proteins increased 2-fold or greater.
Phosphorylation of Ser123 on RAD23A was increased 3-fold in response to MMS treatment. There was no measured change in total protein abundance of RAD23A. Although, phosphorylation of Ser123 has been described previously in several large scale phosphoproteomics studies, this is the first indication as to how this phosphorylation of this site is regulated. RAD23A is a UV excision repair protein known to function in DNA repair [55]. Phosphorylation of Ser123 may stimulate the DNA repair function of RAD23A. RAD23A is also involved in ubiquitin-dependent protein degradation by the proteasome, providing a potential link between two post-translational modifications and additionally connecting to our observation that proteasome subunits appear to be recruited to chromatin in response to MMS treatment.
Phosphorylation of Thr722 on MCM3 was decreased 2-fold in response to MMS treatment. This is a known cyclin E/CDK2 phosphorylation site [5]. There was no measured change in total protein abundance of MCM3. Minichromosome maintenance (MCM) proteins function as a replicative helicase during DNA synthesis. MCM proteins are also involved in DNA repair however their exact role is unclear. The checkpoint kinase ATM phosphorylates MCM3 on a nearby residue, Ser728 [24], although this phosphorylation event was not detected in our study. Nevertheless, it may be the case that phosphorylation of Thr722 interferes with the ability of ATM to phosphorylate Ser728, necessitating that the former site be dephosphorylated during the DNA damage response.
4. Conclusions
Using mass spectrometry-based proteomics, we have identified cellular proteins and pathways that are responsive to DNA damage caused by the alkylating agent MMS. Our spatial proteomics results indicate that the protein composition of chromatin is significantly altered following MMS treatment. More specifically, these changes to the chromatin proteome should result in profound remodeling of chromatin organization. Our data show that many chromatin modifying enzymes are regulated in response to MMS treatment. The NuRD nucleosome remodeling complex is recruited to chromatin soon after MMS treatment, presumably to open the DNA to other DDR proteins. Other enzymes such as DNMT1 and KAP1 are removed from chromatin, most probably as a result of protein degradation, to prevent aberrant signaling. The recruitment of histone chaperones following MMS treatment further demonstrates that chromatin reorganization is a key component of the DDR, to allow DNA repair proteins access to the site of DNA damage. Finally, the decrease in nucleoporins associated with chromatin is a sign that changes in global nuclear architecture have occurred in response to MMS treatment. Post-translational protein modification is also an important mediator of DDR signaling. The recruitment of the proteasome to chromatin suggests that ubiquitin-mediated protein degradation plays a role in this re-shaping of the chromatin protein landscape by the DDR. Our phosphoproteomics results indicate that many nuclear phosphorylation events are decreased following MMS treatment and that this reflects changes in protein kinase and/or phosphatase activity in response to DNA damage.
Supplementary Material
Spatial proteomics was used to monitor protein localization following MMS treatment.
MMS treatment affected the levels of many proteins in the chromatin-enriched fraction.
Proteins involved in chromatin organization were responsive to MMS treatment.
MMS treatment also affected components of the ubiquitin proteasome system.
Decreased phosphorylation of many nuclear proteins was observed after MMS treatment.
Acknowledgments
This work was funded by grants from the National Institutes of Health (CA80100 and CA82683 to TH, and the Cancer Center Support Grant (CA14195), the National Center for Research Resources (5P41RR011823-17 to JRY), the National Institute of General Medical Sciences (8P41GM103533-17 to JRY) and the American Cancer Society (PF-07-279-01-CCG to AA). TH is a Frank and Else Schilling American Cancer Society Professor, and holds the Renato Dulbecco Chair in Cancer Research. We would also like to thank all lab members for thoughtful suggestions and comments.
Footnotes
Conflict of interest statement: The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Seminars in cancer biology. 2010;20:281–293. doi: 10.1016/j.semcancer.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nature reviews Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 4.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chemical research in toxicology. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim YJ, Wilson DM., 3rd Overview of base excision repair biochemistry. Current molecular pharmacology. 2012;5:3–13. doi: 10.2174/1874467211205010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cellular and molecular life sciences : CMLS. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee MW, Kim BJ, Choi HK, Ryu MJ, Kim SB, Kang KM, Cho EJ, Youn HD, Huh WK, Kim ST. Global protein expression profiling of budding yeast in response to DNA damage. Yeast. 2007;24:145–154. doi: 10.1002/yea.1446. [DOI] [PubMed] [Google Scholar]
- 9.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Molecular cell. 2004;16:117–125. doi: 10.1016/j.molcel.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svensson JP, Pesudo LQ, Fry RC, Adeleye YA, Carmichael P, Samson LD. Genomic phenotyping of the essential and non-essential yeast genome detects novel pathways for alkylation resistance. BMC systems biology. 2011;5:157. doi: 10.1186/1752-0509-5-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, Fiedler D, Dutkowski J, Guenole A, van Attikum H, Shokat KM, Kolodner RD, Huh WK, Aebersold R, Keogh MC, Krogan NJ, Ideker T. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravi D, Wiles AM, Bhavani S, Ruan J, Leder P, Bishop AJ. A network of conserved damage survival pathways revealed by a genomic RNAi screen. PLoS genetics. 2009;5:e1000527. doi: 10.1371/journal.pgen.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han X, Aslanian A, Yates JR., 3rd Mass spectrometry for proteomics. Current opinion in chemical biology. 2008;12:483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. Journal of proteome research. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 17.Chuang LS, Tan EH, Oh HK, Li BF. Selective depletion of human DNA-methyltransferase DNMT1 proteins by sulfonate-derived methylating agents. Cancer research. 2002;62:1592–1597. [PubMed] [Google Scholar]
- 18.Gospodinov A, Herceg Z. Shaping chromatin for repair. Mutation research. 2013;752:45–60. doi: 10.1016/j.mrrev.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Papamichos-Chronakis M, Peterson CL. Chromatin and the genome integrity network, Nature reviews. Genetics. 2013;14:62–75. doi: 10.1038/nrg3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boisvert FM, Lam YW, Lamont D, Lamond AI. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Molecular & cellular proteomics : MCP. 2010;9:457–470. doi: 10.1074/mcp.M900429-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, Lukas C, Bartek J, Andersen JS, Lukas J. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. The Journal of cell biology. 2010;190:731–740. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Molecular cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 25.Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. The Journal of biological chemistry. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- 26.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, Zhang H, Polakiewicz RD, Comb MJ. Profiling of UV-induced ATM/ATR signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bensimon A, Aebersold R, Shiloh Y. Beyond ATM: the protein kinase landscape of the DNA damage response. FEBS letters. 2011;585:1625–1639. doi: 10.1016/j.febslet.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wohlbold L, Fisher RP. Behind the wheel and under the hood: functions of cyclin-dependent kinases in response to DNA damage. DNA repair. 2009;8:1018–1024. doi: 10.1016/j.dnarep.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Molecular cell. 2012;46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Molecular & cellular proteomics : MCP. 2010;9:1314–1323. doi: 10.1074/mcp.M900616-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang SY, Chen DJ, Aebersold R, Shiloh Y. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Science signaling. 2010;3:rs3. doi: 10.1126/scisignal.2001034. [DOI] [PubMed] [Google Scholar]
- 33.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Molecular and cellular biology. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR., 3rd MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid communications in mass spectrometry : RCM. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 35.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. Journal of proteome research. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature biotechnology. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 37.Park SK, Yates JR., 3rd Census for proteome quantification. Current protocols in bioinformatics / editoral board, Andreas D Baxevanis … [et al] 2010;Chapter 13:Unit 13 12 11–11. doi: 10.1002/0471250953.bi1312s29. [DOI] [PubMed] [Google Scholar]
- 38.Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG, Wu C, Yates JR, 3rd, Su AI, Kelly JW, Wiseman RL. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell reports. 2013;3:1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 40.Mudunuri U, Che A, Yi M, Stephens RM. bioDBnet: the biological database network. Bioinformatics. 2009;25:555–556. doi: 10.1093/bioinformatics/btn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nature biotechnology. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 42.Moudry P, Lukas C, Macurek L, Neumann B, Heriche JK, Pepperkok R, Ellenberg J, Hodny Z, Lukas J, Bartek J. Nucleoporin NUP153 guards genome integrity by promoting nuclear import of 53BP1. Cell death and differentiation. 2012;19:798–807. doi: 10.1038/cdd.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, Meyer T, Cimprich KA. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Molecular cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Shaughnessy A, Hendrich B. CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochemical Society transactions. 2013;41:777–782. doi: 10.1042/BST20130027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nature structural & molecular biology. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 46.Kim JA, Haber JE. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1151–1156. doi: 10.1073/pnas.0812578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Molecular cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 48.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bukata L, Parker SL, D'Angelo MA. Nuclear pore complexes in the maintenance of genome integrity. Current opinion in cell biology. 2013 doi: 10.1016/j.ceb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Moudry P, Lukas C, Macurek L, Hanzlikova H, Hodny Z, Lukas J, Bartek J. Ubiquitin-activating enzyme UBA1 is required for cellular response to DNA damage. Cell cycle. 2012;11:1573–1582. doi: 10.4161/cc.19978. [DOI] [PubMed] [Google Scholar]
- 51.Kouranti I, Peyroche A. Protein degradation in DNA damage response. Seminars in cell & developmental biology. 2012;23:538–545. doi: 10.1016/j.semcdb.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 52.Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. The Journal of biological chemistry. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitcomb EA, Dudek EJ, Liu Q, Taylor A. Novel control of S phase of the cell cycle by ubiquitin-conjugating enzyme H7. Molecular biology of the cell. 2009;20:1–9. doi: 10.1091/mbc.E08-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Udeshi ND, Svinkina T, Mertins P, Kuhn E, Mani DR, Qiao JW, Carr SA. Refined preparation and use of anti-diglycine remnant (K-epsilon-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Molecular & cellular proteomics : MCP. 2013;12:825–831. doi: 10.1074/mcp.O112.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dantuma NP, Heinen C, Hoogstraten D. The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA repair. 2009;8:449–460. doi: 10.1016/j.dnarep.2009.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.