Abstract
Background
In 2008, an outbreak of ovine footrot occurred in Norway. Dichelobacter nodosus isolates collected between 2008 and 2011 have been characterised. Isolates defined as virulent by the gelatin gel test (GG-test) were only found in sheep in Rogaland County, where the severe cases of footrot were registered. The majority (96%) of the virulent isolates belonged to serogroup A. It is suspected that they represent a newly introduced strain, and the aim of the present study was to investigate whether they are genetically similar. Sixty-one virulent isolates from sheep and 116 benign isolates from sheep, cattle and goats were included. Four GG-test virulent isolates from Danish sheep were also included. All isolates were genotyped by pulsed-field gel electrophoresis (PFGE) and by PCR for pgr variant determination.
Results
The Norwegian virulent isolates were assigned to 8 pulsotypes (PTs), while the benign isolates were assigned to 66 PTs. Thirty-seven (68.5%) of the 54, virulent, serogroup A isolates belonged to the same PT, and included isolates from 2008 through 2011. Isolates belonging to this PT were defined as the outbreak strain. The remaining virulent serogroup A isolates belonged to 4 PTs differing by ≤3 bands from the outbreak strain. Two virulent, Danish, serogroup A isolates differed by 2 bands from the Norwegian outbreak strain. All but 3 (95%) of the virulent isolates had the pgrA variant while 85% of the benign isolates had the pgrB variant.
Conclusion
This study provides evidence that the footrot outbreak in Norway in 2008 most likely was caused by new introduction and local spread of one virulent D. nodosus strain.
Keywords: Cow, Genotyping, Goat, Pulsed-field gel electrophoresis, PFGE, Serogroup converison, Sheep
Background
In 2008, footrot was diagnosed in Norwegian sheep for the first time in 60 years [1]. The infection is caused by Dichelobacter nodosus[2], and clinical signs range from interdigital dermatitis to complete loosening of the hoof horn. The severity of lesions depends on the virulence of the infecting bacterial strain, climatic conditions and the breed of sheep [3].
Following the outbreak, D. nodosus isolates from sheep, cattle and goats collected between 2008 and 2011 from different geographic locations in Norway were characterised with respect to virulence and serogroup [4]. Isolates that produced heat stable proteases, as shown by the gelatin gel test (GG-test) [5], were defined as virulent and were only found in sheep in the county of Rogaland where the most severe cases of footrot were registered. Interestingly, 96% of these isolates belonged to serogroup A [4]. Isolates defined as benign showed greater serogroup diversity and were more spread geographically [4]. There was also an association between laboratory-defined virulence and clinical manifestations in sheep [6,7]. An ongoing programme to control and eradicate ovine footrot, caused by virulent D. nodosus, was initiated in Norway in 2009 [8].
It is believed that the Norwegian footrot outbreak was caused by introduction and local spread of a virulent D. nodosus strain belonging to serogroup A. To confirm this it was necessary to genotype the isolates. It is possible that the infection was introduced through import of sheep. Import of live animals to Norway is restricted, but a few sheep imports were registered in the years prior to the outbreak in 2008, mainly from Denmark [9].
A few studies have used genotyping for molecular epidemiological investigations of D. nodosus[10-13]. Pulsed field gel electrophoresis (PFGE) is discriminatory and reproducible, and has been used for typing D. nodosus[10,13]. Typing based on the proline-glycine repeat (pgr) gene in D. nodosus was described by Calvo-Bado et al. [14], who developed a PCR assay to discriminate between the two pgr variants pgrA and pgrB.
In this study, a collection of D. nodosus isolates were genotyped by PFGE and pgr typing to find out whether the GG-test virulent, serogroup A isolates are genetically similar. The aim was to confirm whether the footrot outbreak in Norway was caused by local spread of one, single virulent D. nodosus strain.
Methods
Bacterial isolates
A total of 177 D. nodosus isolates from Norwegian sheep, cattle and goats were investigated (Table 1). The isolates were collected between 2008 and 2011, from 12 of the 19 Norwegian counties and 131 different farms. One hundred and thirty-one of the isolates were from Rogaland County, where the most severe cases of footrot had occurred.
Table 1.
Dichelobacter nodosus isolates from Norway included in the study, with respect to host species, virulence, number of farms and fim A variant (serogroup A-M)
| No. of isolates | No. of farms | A | B | C | D | E | F | G | H | I | M | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virulent isolates sheep |
61 |
52a |
54 |
1 |
1 |
0 |
2 |
0 |
0 |
2 |
1 |
0 |
| Benign isolates sheep |
79 |
68a |
26 |
13 |
21 |
0 |
2 |
0 |
5 |
8 |
4 |
0 |
| Benign isolates cattle |
34 |
16 |
11 |
8 |
3 |
1 |
4 |
0 |
2 |
2 |
1 |
2 |
| Benign isolates goats |
3 |
2 |
1 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| Total | 177 | 131a,b | 92 | 24 | 25 | 1 | 8 | 0 | 7 | 12 | 6 | 2 |
aFrom 4 sheep flocks both virulent and benign isolates were included.
bFrom 3 farms isolates from both sheep and cattle were included.
One hundred and fifty-five of the isolates have been described previously [4] and were selected to represent all Norwegian farms from which D. nodosus had been cultured before the end of 2011. Twenty-eight of them were from 10 farms that had been sampled 2–3 times with 3–15 month intervals and where isolates from all sampling occasions were included.
Twenty-two of the isolates were collected in a recent study of infectious foot diseases in Norwegian cattle [15,16].
All the isolates had been confirmed as D. nodosus and characterised with respect to virulence and serogroup as described previously [4] (Table 1). Isolates shown to produce heat stable or heat labile proteases by the GG-test were defined as virulent and benign, respectively, regardless of the clinical manifestations in the animals.
The 61 virulent Norwegian isolates included in the study were all from sheep in Rogaland County. Fifty-four of them belonged to serogroup A. The 116 benign isolates were from sheep, cattle and goats and 12 different counties.
Four isolates from 3 Danish sheep flocks were also included. These isolates were collected in 2009 [17] and had also been characterised as described previously [4]. All 4 isolates were virulent and belonged to serogroup A (n = 2) or E (n = 2).
Pulsed-field gel electrophoresis
All isolates were analysed by PFGE essentially as described by Buller et al. [10], but with the following modifications: Isolates were cultured on 2% TAS (trypticase-arginine-serine) agar for 2–3 days, and suspended in 300 μl wash buffer [10]. The agarose plugs were washed 5 times in Tris-EDTA (TE) buffer. Prior to restriction, plugs were washed twice with 0.6 ml TE at 25°C for 20 min. DNA was restricted with 50 U of ApaI (Sigma-Aldrich, St Louis, MO) in a 100-μl volume containing restriction buffer and 0.1 mg/ml Bovine serum albumin (BSA) (Sigma-Aldrich) for 3 h at 30°C. An additional 10 U ApaI was added halfway through the incubation period. The reference strain ATCC® 27521 was prepared as described above and included on each gel. Lambda ladder (New England Biolabs, Ipswich, MA) was used as size standard. Restriction fragments were separated in a CHEF-DR® III electrophoresis system (Bio-Rad, Hercules, CA) at 6 V/cm, with a pulse time of 0.2 to 25 s for 24 h.
Differentiation of banding patterns was performed using BioNumerics (version 6.1; Applied Maths, Kortrijk, Belgium) and visual inspection. PFGE bands larger than the smallest ladder fragment (48.5 kb) were included in the analysis. A unique banding pattern was defined as a pulsotype (PT). Pairwise similarity coefficients were calculated using the Dice formula, and the dendrogram was created using the unweighted pair group method with arithmetic averages. Optimisation and position tolerance settings were set at 1.5.
PCR for pgr
All isolates were tested for the presence of pgrA and pgrB by PCR as described by Calvo-Bado et al. [14]. DNA was extracted from 48 h HEPES-TAS broth cultures using a nucliSENS® easyMAG™ extractor (bioMèrieux, Boxtel, The Netherlands) following the manufacturers’ instructions. Extracted DNA was stored at −20°C.
For the PCR, HotStarTaq Master Mix (QIAGEN GmbH, Hilden, Germany) was used with 5 μM of each primer. Amplification was performed on an MJ Research DNA Engine Dyad® (Bio-Rad) with an initial denaturation step of 95°C for 2 min followed by 31 cycles of 95°C for 1 min, 60°C (pgrA) or 55°C (pgrB) for 1 min, 72°C for 2 min, and a final extension time of 72°C for 7 min. PCR products were analysed by electrophoresis in a 1% agarose gel stained with GelRed™ (Biotium, Hayward, CA).
DNA from control strains of D. nodosus were kindly provided by Dr. L.A. Calvo-Bado, Department of Biological Sciences, University of Warwick, Coventry, UK: VCS1703A as positive control for pgrA, and C305 as positive control for pgrB.
Statistical analyses
Descriptive statistics were performed using Microsoft Excel 2010 (Microsoft Corporation, Redmond, WA). Further statistical analyses were performed using GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla, CA).
Fisher´s exact test [18] was used to investigate whether there was a difference between isolates defined as benign versus isolates defined as virulent with respect to the proportions of isolates having the pgrA or pgrB gene. Results were considered significant when p < 0.05.
Results
PFGE
The 177 Norwegian D. nodosus isolates were divided into 74 different pulsotypes (PTs), consisting of 5–11 bands. Between 1 and 40 isolates were assigned to each PT. The benign isolates were assigned to 66 PTs that clustered with >58% similarity and virulent isolates were assigned to 8 PTs that clustered with >72% similarity (Figure 1). Virulent and benign isolates belonged to different PTs.
Figure 1.
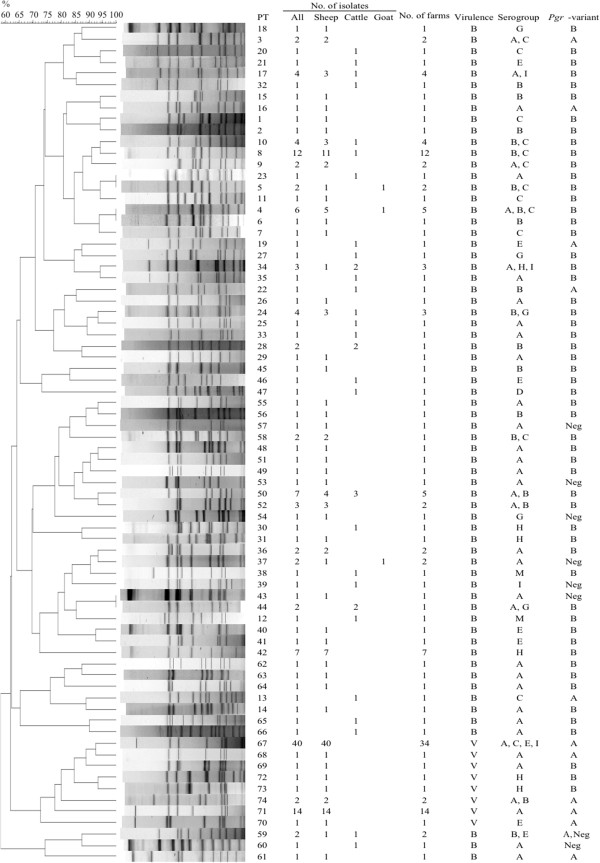
Dendrogram of Dichelobacter nodosus isolates from Norway analysed by Pulsed-field gel electrophoresis (PFGE), including one isolate from each pulsotype (PT). The assigned PT number and the total number of isolates assigned to each PT are indicated to the right, and for each PT the number of isolates from each species, number of farms from which they were collected, and the virulence, serogroup and proline-glycine repeat (pgr) variant of the isolates are listed.
The 54 virulent, serogroup A isolates were divided into 5 PTs (Table 2), and the majority (n = 37) belonged to PT 67. This PT included isolates collected from 2008 through to 2011. The other virulent, serogroup A isolates belonged to 4 different PTs that differed by ≤3 bands from PT 67 (Table 2). Three of the 7 virulent, non-serogroup A isolates also belonged to PT 67, while the remaining 4 isolates belonged to 4 PTs that differed from PT 67 by 2–4 bands.
Table 2.
The five pulsotypes (PT) to which the Norwegian, virulent, serogroup A Dichelobacter nodosus isolates belonged with respect to the band differences from PT 67, and the distribution of isolates with respect to the year of collection
|
PT |
Band diff. from PT67 |
No. of isolates |
||||
|---|---|---|---|---|---|---|
| Total (%) | 2008 | 2009 | 2010 | 2011 | ||
| 67 |
- |
37 (68.5) |
1 |
12 |
16 |
8 |
| 68 |
1 |
1 (1.9) |
0 |
0 |
1 |
0 |
| 69 |
2 |
1 (1.9) |
0 |
0 |
1 |
0 |
| 71 |
2 |
14 (25.9) |
0 |
0 |
10 |
4 |
| 74 | 3 | 1 (1.9) | 0 | 0 | 1 | 0 |
Twenty-one PTs included more than one isolate (Figure 1). Thirteen of these were only found in one county. Sixteen PTs included isolates belonging to different serogroups (Figure 1). Isolates from sheep (n = 140), cattle (n = 34) and goats (n = 3) were divided in to 52, 29 and 3 PTs, respectively. Twenty-six (18.6%) isolates from sheep and 10 (29.4%) isolates from cattle belonged to 7 PTs common to both species (Figure 1). The PTs of the 3 isolates from goats were all common to PTs from sheep.
The 4 Danish isolates were assigned to different PTs. The 2 serogroup A isolates belonged to PTs that differed from PT 67 by 2 bands (Figure 2). The 2 serogroup E isolates belonged to PTs that differed from PT 67 by 4 bands.
Figure 2.
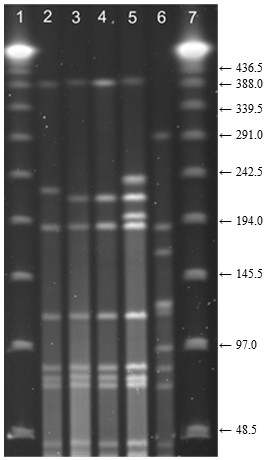
Pulsed-field gel electrophoresis (PFGE) of ApaI restricted, virulent Dichelobacter nodosus isolates belonging to serogroup A from Norway and Denmark. Lanes: 1 and 7, lambda ladder molecular size marker; 6, D. nodosus ATCC® 27521 reference strain; 2 and 5, isolates from Denmark; 3–4, isolates from Norway. Numbers to the right indicate molecular size (kilobases).
From 6 of the 10 farms from where isolates had been collected on 2–3 occasions, isolates from all samplings belonged to the same PT.
The 3 virulent, non-serogroup A isolates assigned to PT 67 were from 3 farms where a virulent, serogroup A isolate, belonging to PT 67 had also been cultured. It is possible that this represent cases of serogroup conversion. From one of the farms, a virulent, serogroup A isolate, a virulent, serogroup C isolate and a benign, serogroup C isolate were available for investigation. The 2 virulent isolates were identical (PT 67) while the benign isolate differed (Figure 3). No benign isolates were available from the 2 other farms.
Figure 3.
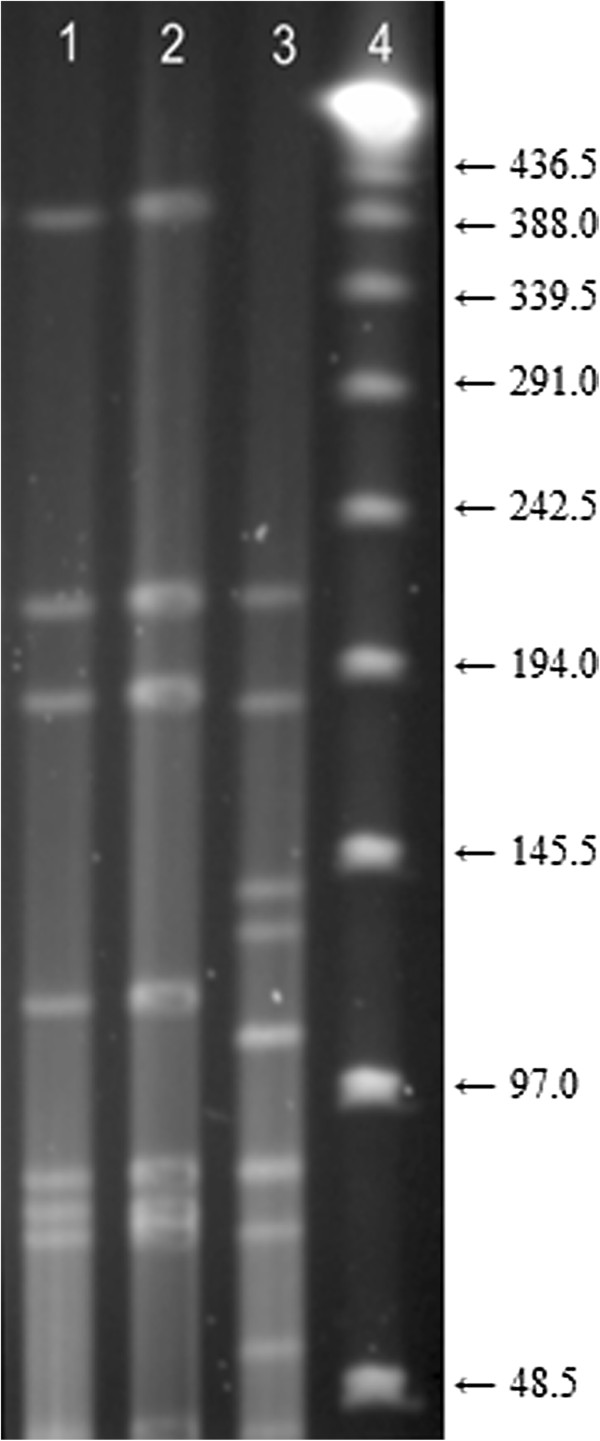
Pulsed-field gel electrophoresis (PFGE) of three ApaI restricted Dichelobacter nodosus isolates from one farm indicating a possible case of serogroup conversion. Lanes: 4, lambda ladder molecular size marker; 1, virulent, serogroup A isolate; 2, virulent, serogroup C isolates; 3, benign, serogroup C isolate. Numbers to the right indicate molecular size (kilobases).
PCR for pgr
Results from the pgr PCR are presented in Table 3. The pgrA variant was detected in 53 of the 54 virulent, serogroup A isolates and in 5 of the 7 virulent, non-serogroup A isolates. Five per cent of the isolates were negative for both pgrA and pgrB, with no other common features than being benign. The association between virulence, as defined by the GG-test, and pgr variant was statistically significant.
Table 3.
Distribution of pgr variant among virulent and benign isolates of Dichelobacter nodosus from Norway
| pgrA (%) | pgrB (%) | pgrA / pgrB negative (%) | |
|---|---|---|---|
| Virulent isolates |
58 (95.1) |
3 (4.9) |
0 (−) |
| Benign isolates |
8 (6.9) |
99 (85.3) |
9 (7.8) |
| Total | 66 (37.3) | 102 (57.6) | 9 (5.1) |
With only one exception, isolates within a PT had the same pgr variant (Figure 1). Three virulent isolates were pgrB-positive. They belonged to serogroup A (n = 1) or H (n = 2) and differed in pulsotype from PT 67 by 2 and 4 bands, respectively.
In the farm with a possible case of serogroup conversion, the 2 virulent isolates belonging to serogroup A and C both had the pgrA variant, while the benign serogroup C isolate had the pgrB variant.
The 4 virulent Danish isolates were all pgrA-positive.
Discussion
This study shows that the D. nodosus population in Norway, as a whole, is genetically diverse, but that isolates defined as virulent by the GG-test, belonging to serogroup A are genetically similar. It provides evidence that the outbreak of ovine footrot discovered in 2008, was caused by spread of one virulent strain of D. nodosus in the county of Rogaland.
All the Norwegian virulent isolates included in this study clustered together in a clonal manner in the dendrogram. Fifty-three of the 54 virulent, serogroup A isolates are likely to represent the outbreak strain or subtypes of this. Thirty-seven of these 53 isolates belonged to one pulsotype (PT 67), while the remaining 16 isolates belonged to 3 PTs that differed from PT 67 by 3 bands or less. The degree of similarity between these 4 PTs indicates that the isolates are closely related, genetically [19]. Although there is no exact documentation of an epidemiological link between all the isolates, they were all from one county with a high density of sheep farms. The combination of the epidemiological data and the phenotypic and genotypic similarites between these isolates in terms of virulence, serogroup, PT and pgr variant, strengthens the assumption that they are, indeed, genetically related. Isolates belonging to PT 67 were collected over all 4 years. The remaining 3 PTs were observed in isolates collected in 2010 or 2011. They may represent variants of PT 67 that have undergone a genetic event, which can occur over time. One of the virulent, serogroup A isolates (PT 69) differed only by 2 bands from PT 67, but had the pgrB variant. This could be a case of a genetic event involving the pgr region, or it may represent a strain not related to the outbreak strain.
Four of the virulent, non-serogroup A isolates differed from PT 67 by 3–4 bands and 2 isolates contained a different pgr variant. We believe that these isolates are not genetically related to the outbreak strain. Rather, they may represent virulent strains that were also imported, but did not spread to the same degree. This may be the result of epidemiological circumstances or strain differences in their ability to survive, spread and cause disease under Norwegian conditions.
Three possible cases of serogroup conversion were identified. Virulent, non-serogroup A isolates that were indistinguishable from PT 67 were found in 3 farms where virulent, serogroup A isolates were also found. From one of the farms a benign, serogroup C isolate, a virulent, serogroup C isolate and a virulent, serogroup A isolate were analysed. The 2 virulent isolates were indistinguishable by PFGE. We hypothesise that the benign, serogroup C isolate was the source of a new fimA gene to a virulent isolate, but this requires further investigations. Kennan et al. [20] demonstrated in vitro that serogroup conversion can occur in D. nodosus by natural transformation. It is likely that this can also happen under natural conditions. Serogroup conversion may also have occurred on the farms with benign isolates of different serogroups belonging to indistinguishable PTs.
The limited geographical spread of the virulent D. nodosus isolates is probably due to the national elimination programme and control measures initiated immediately after the outbreak in 2008 [8]. A general ban on movement of small ruminants between counties, which has been enforced since the 1970s to control maedi-visna and scrapie infection in Norway, is also likely to have contributed.
Results indicate that benign D. nodosus are endemic in Norway, and are likely to have been so for a long period of time. These isolates showed considerable genetic diversity and were from sheep, cattle and goats with generally mild disease [6,7]. Although the benign isolates were from 12 of the 19 Norwegian counties, most of them were from Rogaland County. This reflects the relatively few samples from outside Rogaland submitted to our laboratory for culturing of D. nodosus. The limited number of isolates from some counties may also be the reason why certain PTs were only found in one county.
Before the outbreak in 2008, the last registered case of ovine footrot in Norway was in 1948 [21]. The restricted movement of animals across counties for the last 40 years means that it is unlikely that the benign strains have spread through trade of live sheep during this period. They must have spread within the sheep population a long time ago, or other routes of transmission exist. Cattle are possible carriers as they are not subject to movement restrictions similar to sheep. In Norway co-grazing of cattle and sheep is practiced, and studies have shown that cross-infection between sheep and cattle may occur [16,22,23].
The correlation between pgr variant and the virulence of the D. nodosus isolates, as defined by the GG-test, is in agreement with previous findings [12,14]. Our results also strengthen previous suggestions that the pgrA gene may be associated with virulence in D. nodosus by coding for a protein possibly involved in adhesion to extracelluar matrix [14,24]. Nine isolates were negative for both pgrA and pgrB, which could indicate that other variants of the pgr gene exist.
Two of the Danish D. nodosus isolates differed by only 2 bands from the main Norwegian outbreak strain (PT 67). A few sheep imports from Denmark were registered in the years prior to 2008, and included imports to Rogaland County (not published). Sheep imported from Denmark could be the source of the new virulent D. nodosus strain, and hence the Norwegian outbreak. However, none of the isolates included in the study were from farms with known export to Norway, and without an epidemiological link it cannot be concluded.
Conclusions
Genotyping by PFGE shows that the virulent, serogroup A isolates from Rogaland County are genetically similar. This confirms that introduction and local spread of one D. nodosus strain caused the outbreak of ovine footrot in Norway in 2008. The fact that this virulent strain has had limited geographical spread increases the likelihood that the outbreak can be controlled and that this strain can be eradicated from Norway.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MG contributed to the design of the study, laboratory work (PFGE and pgr PCR), analysed the data, and drafted the manuscript. BK performed laboratory work (PFGE) and contributed to the writing of the manuscript. TLL contributed to the design of the study, data interpretation and to writing the manuscript. SV provided epidemiological information and contributed to the writing of the manuscript. HJJ contributed to the design of the study, analysing data and to writing of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Marianne Gilhuus, Email: marianne.gilhuus@vetinst.no.
Bjørg Kvitle, Email: bjorg.kvitle@vetinst.no.
Trine M L’Abée-Lund, Email: trine.labee-lund@nmbu.no.
Synnøve Vatn, Email: synnove.vatn@animalia.no.
Hannah J Jørgensen, Email: hannah.jorgensen@vetinst.no.
Acknowledgements
The Danish isolates were kindly provided by Øystein Angen, National Veterinary Institute, Technical University of Denmark. This work was financially supported by the Norwegian Research Council (project number 199422).
References
- Meling S, Ulvund MJ. In: Proceedings of the 7th International Sheep Veterinary Congress: 2009; Stavanger, Norway. Stuen S, Ulvund MJ, editor. Flock health visits in 17 sheep flocks in Rogaland; pp. 148–149. [Google Scholar]
- Beveridge WIB. Foot-rot in sheep: A transmissible disease due to infection with Fusiformis nodosus (n.sp.). Studies on its cause, epidemiology, and control. Melbourne: Council for Scientific and Industrial Research; 1941. [Bulletin No. 140] [Google Scholar]
- Stewart DJ. In: Footrot and foot abscess of ruminants. Egerton JR, Yong WK, Riffkin GG, editor. Boca Raton, FL: CRC Press; 1989. Footrot in sheep; pp. 5–45. [Google Scholar]
- Gilhuus M, Vatn S, Dhungyel OP, Tesfamichael B, L'Abée-Lund TM, Jørgensen HJ. Characterisation of Dichelobacter nodosus isolates from Norway. Vet Microbiol. 2013;56:142–148. doi: 10.1016/j.vetmic.2012.12.020. [DOI] [PubMed] [Google Scholar]
- Palmer MA. A gelatin test to detect activity and stability of proteases produced by Dichelobacter (Bacteroides) nodosus. Vet Microbiol. 1993;56:113–122. doi: 10.1016/0378-1135(93)90133-R. [DOI] [PubMed] [Google Scholar]
- Vatn S, Hektoen L, Høyland B, Reiersen A, Kampen AH, Jørgensen HJ. Elimination of severe footrot from the Norwegian sheep population - a progress report. Small Ruminant Res. 2012;56:11–13. doi: 10.1016/j.smallrumres.2012.04.012. [DOI] [Google Scholar]
- Vatn S, Hektoen L, Fredriksen B, Høyland B, Reiersen A, Jørgensen HJ. The 8th International Sheep Veterinary Congress: 2013; Rotorua, NZ. : New Zealand Veterinary Association; Elimination of footrot in Norway: observed clinical signs in sheep flocks infected with Dichelobacter nodosus; p. 149. [Google Scholar]
- Vatn S, Hektoen L, Høyland B, Kampen AH, Skarra TK. In: Proceedings of the 7th International Sheep Veterinary Congress: 2009; Stavanger, Norway. Stuen S, Ulvund MJ, editor. Surveillance, control and eradication of footrot in Norway; pp. 120–121. [Google Scholar]
- Statistics Norway. [ https://www.ssb.no/statistikkbanken/SelectVarVal/Define.asp?MainTable=UhArVareLand&KortNavnWeb=muh&PLanguage=0&checked=true]
- Buller NB, Ashley P, Palmer M, Pitman D, Richards RB, Hampson DJ. Understanding the molecular epidemiology of the footrot pathogen, Dichelobacter nodosus, to support control and eradication programs. J Clin Microbiol. 2010;56:877–882. doi: 10.1128/JCM.01355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghimire SC, Egerton JR. PCR-RFLP of outer membrane proteins gene of Dichelobacter nodosus: a new tool in the epidemiology of footrot. Epidemiol Infect. 1999;56:521–528. doi: 10.1017/S0950268899002290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell CL, Smith EM, Calvo-Bado LA, Green LE, Wellington EMH, Medley GF, Moore LJ, Grogono-Thomas R. Multiple locus VNTR analysis highlights that geographical clustering and distribution of Dichelobacter nodosus, the causal agent of footrot in sheep, correlates with inter-country movements. Infect Genet Evol. 2014;56:273–279. doi: 10.1016/j.meegid.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaria Z, Radu S, Sheikh-Omar AR, Mutalib AR, Joseph PG, Rusul G. Molecular analysis of Dichelobacter nodosus isolated from footrot in sheep in Malaysia. Vet Microbiol. 1998;56:243–250. doi: 10.1016/S0378-1135(98)00219-3. [DOI] [PubMed] [Google Scholar]
- Calvo-Bado LA, Green LE, Medley GF, Ul-Hassan A, Grogono-Thomas R, Buller N, Kaler J, Russell CL, Kennan RM, Rood JI, Wellington EMH. Detection and diversity of a putative novel heterogeneous polymorphic proline-glycine repeat (Pgr) protein in the footrot pathogen Dichelobacter nodosus. Vet Microbiol. 2010;56:358–366. doi: 10.1016/j.vetmic.2010.06.024. [DOI] [PubMed] [Google Scholar]
- Knappe-Poindecker M, Gilhuus M, Jensen TK, Klitgaard K, Larssen RB, Fjeldaas T. Interdigital dermatitis, heel horn erosion, and digital dermatitis in 14 Norwegian dairy herds. J Dairy Sci. 2013;56:7617–7629. doi: 10.3168/jds.2013-6717. [DOI] [PubMed] [Google Scholar]
- Knappe-Poindecker M, Gilhuus M, Jensen TK, Vatn S, Jørgensen HJ, Fjeldaas T. Cross-infection of virulent Dichelobacter nodosus between sheep and co-grazing cattle. Vet Microbiol. 2014;56:375–382. doi: 10.1016/j.vetmic.2014.02.044. [DOI] [PubMed] [Google Scholar]
- Angen Ø, Akkad NB, Worm R, Nymand A, Frosth S, Aspán A, Slettemeås JS, Jørgensen H, Klaas IC. Detektion af Dichelobacter nodosus og sanering for ondartet klovsyge i tre danske fårebesætninger. Dansk Vet Tidsskr. 2011;56:28–34. [Google Scholar]
- Altman DG. Practical statistics for medical research. London: Chapman and Hall; 1991. [Google Scholar]
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;56:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennan RM, Dhungyel OP, Whittington RJ, Egerton JR, Rood JI. Transformation-mediated serogroup conversion of Dichelobacter nodosus. Vet Microbiol. 2003;56:169–178. doi: 10.1016/S0378-1135(02)00359-0. [DOI] [PubMed] [Google Scholar]
- Øverås J. Smittsom klauvsjuke hos sau. Et tilbakeblikk på import og utbrudd i Norge. Småfenytt. 1994;56:29–33. [Google Scholar]
- Laing EA, Egerton JR. The occurrence, prevalence and transmission of Bacteroides nodosus infection in cattle. Res Vet Sci. 1978;56:300–304. [PubMed] [Google Scholar]
- Wilkinson FC, Egerton JR, Dickson J. Transmission of Fusiformis nodosus infection from cattle to sheep. Aust Vet J. 1970;56:382–384. doi: 10.1111/j.1751-0813.1970.tb15578.x. [DOI] [PubMed] [Google Scholar]
- Myers GS, Parker D, Al-Hasani K, Kennan RM, Seemann T, Ren Q, Badger JH, Selengut JD, Deboy RT, Tettelin H, Boyce JD, McCarl VP, Han X, Nelson WC, Madupu R, Mohamoud Y, Holley T, Fedorova N, Khouri H, Bottomley SP, Whittington RJ, Adler B, Songer JG, Rood JI, Paulsen IT. Genome sequence and identification of candidate vaccine antigens from the animal pathogen Dichelobacter nodosus. Nat Biotechnol. 2007;56:569–575. doi: 10.1038/nbt1302. [DOI] [PubMed] [Google Scholar]


