Abstract
Mitochondria have specialized ribosomes that have diverged from their bacterial and cytoplasmic counterparts. We have solved the structure of the yeast mitoribosomal large subunit using single-particle electron cryo-microscopy. The resolution of 3.2 Ångstroms enabled a nearly complete atomic model to be built de novo and refined, including 39 proteins, 13 of which are unique to mitochondria, as well as expansion segments of mitoribosomal RNA. The structure reveals a new exit tunnel path and architecture, unique elements of the E site and a putative membrane docking site.
Mitochondria are organelles in eukaryotic cells that play a major role in metabolism, especially the synthesis of ATP. During evolution, mitochondria have lost or transferred most of their genes to the nuclei, significantly reducing the size of their genome (1). In yeast, all but one of the few remaining protein genes encode subunits of respiratory chain complexes, whose synthesis involves insertion into the inner mitochondrial membrane along with incorporation of prosthetic groups (2). For the translation of these genes, mitochondria maintain their own ribosomes (mitoribosomes) and translation system. The mitochondrial ribosomal RNA and several tRNAs are encoded by the mitochondrial genome, whereas all but one of its ribosomal proteins are nuclear encoded and imported from the cytoplasm. Mitoribosomes have diverged greatly from their counterparts in the cytosol of bacterial and eukaryotic cells and also exhibit high variability depending on species (Table S1) (3). Several genetic diseases map to mitoribosomes (4). In addition, the toxicity of many ribosomal antibiotics, in particular aminoglycosides, is thought to be due to their interaction with the mitoribosome (5).
Mitochondrial translation in the yeast Saccharomyces cerevisiae (6) has been used as a model to study human mitochondrial diseases (7). The 74S yeast mitoribosome has an overall molecular weight of 3 MDa, or 30% higher than that of their bacterial counterpart. It consists of a 54S large subunit (1.9 MDa) and a 37S small subunit (1.1 MDa). High-resolution crystal structures have been solved for bacterial (8, 9) and eukaryotic ribosomes (10-12), as well as for an archaeal large subunit (13). In comparison, the only structures to date for mitoribosomes come from electron cryo-microscopy (cryo-EM) reconstructions at 12-14 Å (14-16).
The very low yields of mitoribosomes from most tissues have impeded their crystallization. However, the advent of high-speed direct electron detectors (that allow accounting for particle movement during imaging) (17, 18), and improved algorithms for the classification and alignment of particles (19), allowed us to determine the structure of the large subunit of the yeast mitoribosome (hereafter referred to as “54S”) to an overall resolution of 3.2 Å, resulting in a nearly complete stereochemically refined atomic model.
Structure determination
Ribosomes purified from yeast mitochondria were contaminated with cytoplasmic ribosomes and free 54S (Fig. S1A). By the use of maximum-likelihood image classification techniques (20) it was possible to sort these classes of particles into separate groups (Fig. S1B). A preliminary reconstruction of the 74S mitochondrial ribosome yielded a density map with overall resolution of 3.6 Å. However, the small subunit was found to adopt a wide range of orientations with respect to the large subunit, resulting in markedly worse resolution for this region (~5 Å). Attempts to further classify the mitoribosomes into separate groups failed to yield maps that were suitable for de novo model building of the small subunit. We focused our subsequent efforts on the large subunit only. Density for this region was improved further by reconstructions that combined the large subunit regions of particles consisting of the 74S mitoribosome with those of the free 54S subunit, in a refinement in which the small subunit was masked away. The resulting total of 47,1 24 particles yielded an overall resolution of 3.2 Å by the “gold standard” FSC=0.143 criterion (Fig. S2A) Scheres and Chen, 2012, Nat Methods, 9, 853-4}.
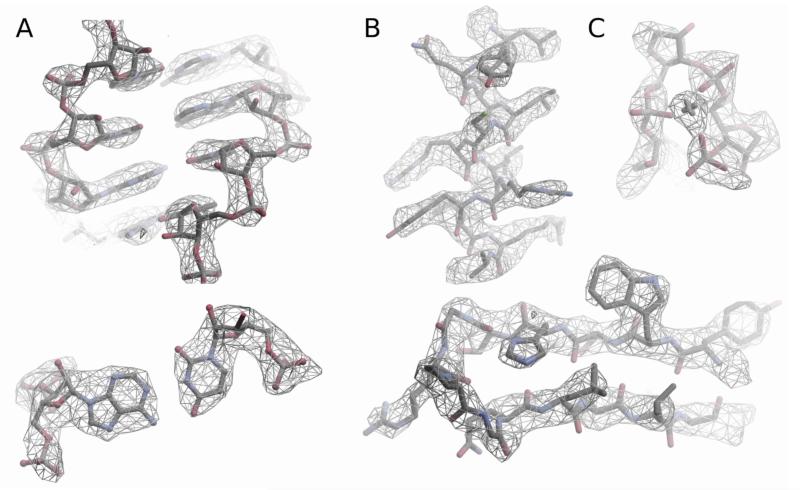
The local resolution (21) is better than 3 Å for most of the interior of the 54S subunit (Fig 1, S2B), but decreases towards the periphery, especially for elements such as the L1 and L7/L12 stalks which are often disordered even in crystal structures of the ribosome. Even at the surface, the resolution mostly appears to be better than 4 Å (Fig. S2C).
Figure 1.
Examples of the density in cryo-EM maps of the 54S subunit. A. RNA, showing a double helix (top) and a base pair (bottom) B. Alpha helix and a beta turn in proteins showing side chains C. Mg ion that coordinates a tight turn in RNA.
The high resolution for much of the molecule allowed us to construct a nearly complete model for the large subunit, including a third for which no previous structural homology was known. In some regions, the quality of the map allowed identification of amino acids directly from side chain densities without any additional information, leading to an unambiguous assignment of specific mitoribosomal proteins to their corresponding densities. For other regions, the identification of specific proteins required a combination of secondary structure prediction, homology modeling and molecular replacement (22). X-ray crystallographic tools such as Coot (23) and REFMAC (24) were modified for model building, refinement and cross validation (22). The final model (Fig. 2A) has good stereochemistry and agreement with the density (Table S2).
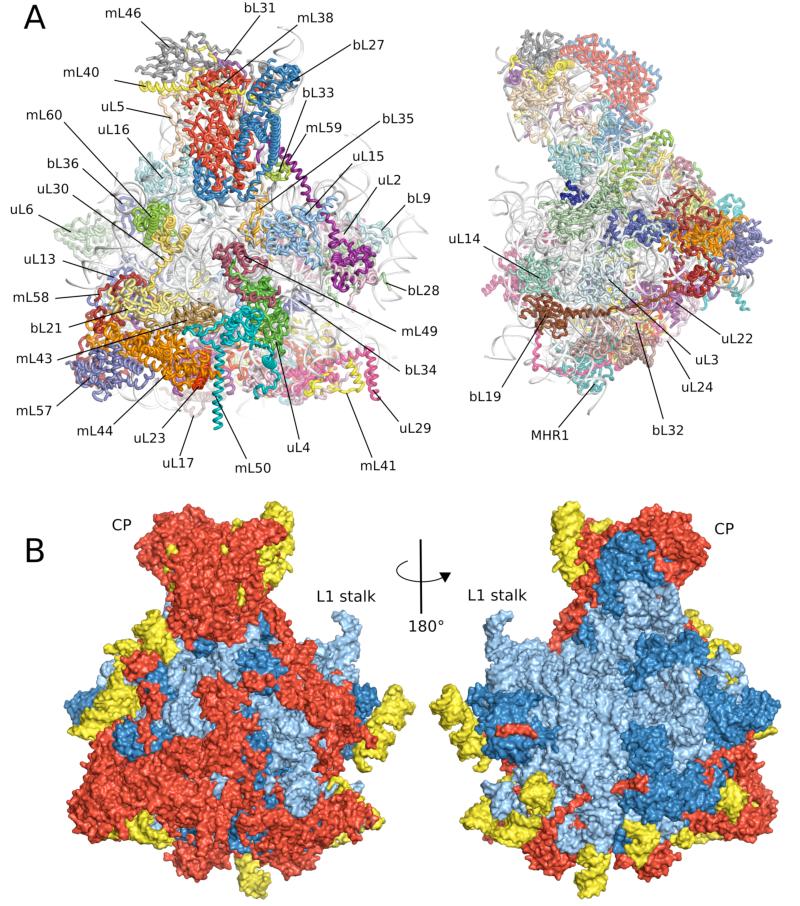
Figure 2.
Overview of the yeast 54S ribosomal subunit. A. Cytoplasmic and side views of final model with 21S rRNA colored in gray. B. Cytoplasmic and interface views colored showing regions conserved with bacteria (blue), found in both yeast and mammalian mitochondria (red) and present only in yeast mitochondria (yellow).
The model does not include surface exposed regions for which only partial density is visible, such as L1 and L7/L12 stalks. In addition, some of the mitochondria-specific rRNA expansions are highly mobile and thus could not be completely resolved. Therefore, we included only the parts for which clear density is apparent in appropriately sharpened maps. The relative orientation of the two subunits in the major class is shown in Fig. S3.
Overall Structure
Both protein and RNA moieties are significantly larger than their bacterial counterparts. From the total mass of 1.9 MDa, 0.4 MDa comes from mitochondria-specific proteins and extensions of known bacterial homologs, and 0.2 MDa from rRNA expansion segments (ES) with 1.3 MDa having structural equivalents in bacteria. Most of these additional elements occur on the surface of the conserved core (Fig. 2B), giving the 54S a markedly different overall shape from the large subunits of cytoplasmic ribosomes.
In particular, there is an expansion of the central protuberance (CP) and a new protuberance formed at the membrane-facing surface. In both cases the rRNA expansion serves as a scaffold for the addition of new proteins. Such progressive expansion shares principles with the development of modern ribosomes themselves from primordial ancestors (25). In addition to these changes at the surface, there is a substantial remodeling of the tunnel through which the nascent protein chain proceeds as it emerges from the ribosome.
rRNA Expansion Segments
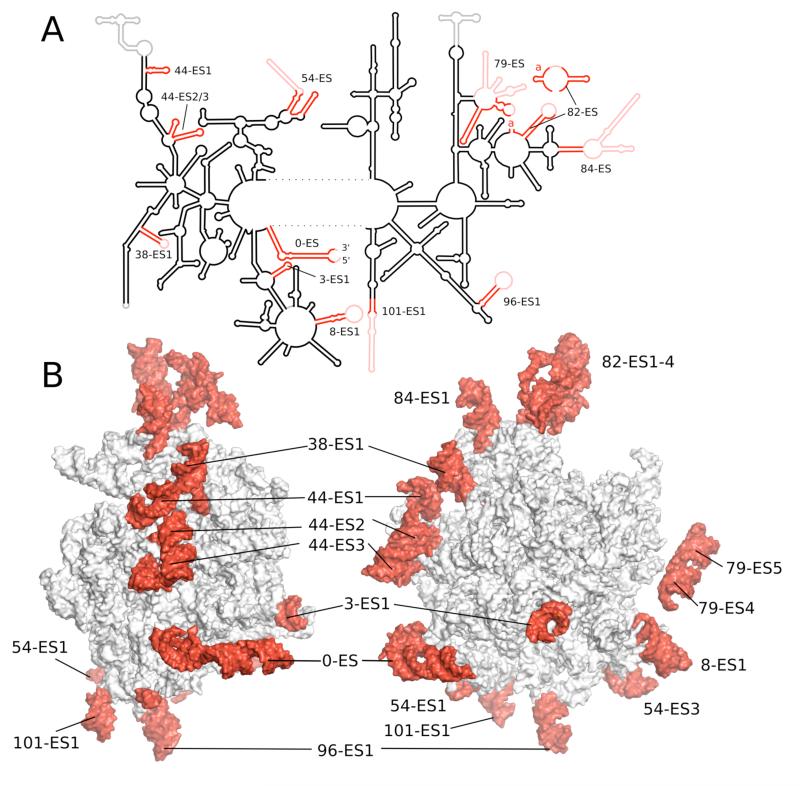
The 21S RNA of the 54S subunit contains 3,296 nucleotides, out of which 2,714 have been modeled in our structure. A secondary structure diagram was constructed using base-pairing information directly from the structure, supplemented by predictions for the unmodeled regions (Fig. 3 and S4-S5). Conserved rRNA helices are numbered corresponding to those in E.coli (26), with ESs denoted by the preceding conserved helix. Despite the lower sedimentation coefficient than E. coli 23S, there has been a considerable expansion of rRNA, resulting in 392 additional nucleotides spread over 11 ESs. Only a few insertions correlate with those of the eukaryotic ribosome. However, as in the eukaryotic case, many of the insertions are flexible expansion segments that project out from the core and are not well resolved in the map. It is not clear whether this similarity is a coincidental growth of complexity by neutral means (27) or whether both evolved independently under selective pressure for functional reasons.
Figure 3.
A. Schematic diagram of the secondary structure of 21S rRNA. B. Two views of tertiary structure showing mitochondria-specific ESs in red labeled after the conserved helix according to standard numbering (see text).
Mitochondria-specific protein elements
Of the 39 proteins built into the 54S, 26 share homology with bacterial ribosomes, 8 are present in both yeast and mammalian mitochondria and 5 are unique to yeast mitochondria (Table S3, Fig. S6). Following a recent proposal by the ribosome community, proteins universal to all domains of life have a “u” prefix and the bacterial numbering; those with only a bacterial homologue have a “b” prefix and bacterial numbering; and proteins unique to mitochondria have an “m” prefix. Bacterial ribosomal proteins L18, L20, and L25 are not present in the yeast mitoribosome. However a homolog of L35, which was previously thought not to be part o f the mitoribosome (28), was clearly identified in the maps. Mitochondrial homologous recombination protein 1 (MHR1) was also found to be a component of t he mitoribosome (28). Density is absent for mL53, mL54 and mL61 that were present in our sample as judged by mass spectrometry, indicating that they are disordered or dissociated during grid preparation.
Most of the homologous proteins are considerably larger, with an average mass of ~25 kDa compared to ~13.5 kDa for their bacterial counterparts (Table S3, Fig. S6). Similar to eukaryotic ribosomes, many of the extensions to bacterial proteins consist of long tails and loops extending from globular domains. For example, uL24 has approximately three times the number of residues and extends 115 Å across the surface of the mitoribosome (Fig. S7A). As in the eukaryotic ribosome (11, 12), many of these extensions contact other extensions and unique proteins and rRNA (Fig. 2) suggesting a role in the assembly of mitoribosome-specific components. Some of the flexible extensions are highly basic and probably assist rRNA folding by neutralizing negative charge, as suggested for bacterial and archaeal ribosomes (13, 29). Only the extensions to uL17 and bL27 form compact domains. uL17 is characterized by a four helix-bundle fold, whereas bL27 forms a domain with no homology to known structures (Fig. S7B); both domains interact with rRNA.
As a result of protein extensions, the number of protein-protein contacts in the 54S is not only larger than in bacteria, but also exceeds that observed in the eukaryotic large subunit (11). Each protein in the mitoribosome has on average 4.5 neighbors (Fig. S8), in comparison with only 1.5 in bacteria. Despite many more proteins overall, the number that contact only rRNA is halved compared to bacteria. Some of the new interactions include bridging different sites of the subunit (normally performed by 5S rRNA in cytoplasmic ribosomes), whereas others are involved in the architecture of the exit tunnel.
All of the 13 mitochondria-specific proteins interact with rRNA, including those that have domains not previously shown to have a role in binding nucleic acids. Of these, all except mL43 and mL49 are bound to rRNA ESs. Analysis of the unique proteins show that three of them have similarity with RNA-binding proteins (mL44, mL49 and mL57), two with DNA-binding proteins (mL58 and mL60), two with nucleotide-binding proteins (mL38 and mL46) and two with lipid-binding proteins (mL38 and mL50; Table S3). These proteins have lost their catalytic or ligand binding residues, and perform other roles in the mitoribosome.
The Central Protuberance (CP)
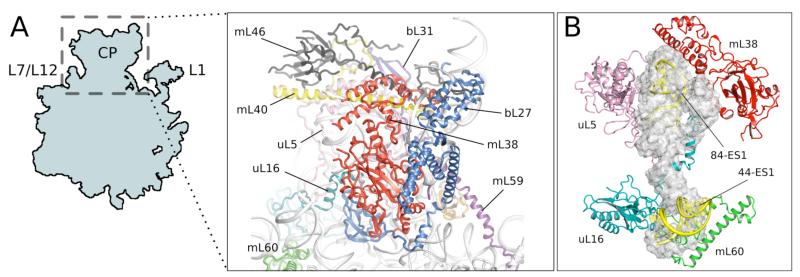
One site of major expansion is the CP, which because of its location is thought to facilitate communication between various functional sites of the ribosome in a manner that is not fully understood (30, 31). In bacterial, archaeal and eukaryotic ribosomes, the CP consists of a separate 5S rRNA core interacting with several ribosomal proteins and 23S rRNA.
Yeast 54S lacks 5S rRNA and the 5S RNA-binding proteins L18 and L25. It is extensively remodeled (Fig. 4A) and consists of mitochondria-specific proteins (mL38, mL40, mL46), RNA expansion clusters (82-ES, 84-ES1) and the extensions of the five homologous proteins (uL5, uL 16, bL27, bL31, bL33). These elements do not structurally mimic 5S rRNA, but occupy its location in bacterial ribosomes (Fig. 4B). The bottom “toe” of 5S rRNA is replaced by 44-ES1, which interacts with uL16 and mL60. The large extension of bL27 binds the 82-ES1-3 to form part of the CP, illustrating how expansions in both proteins and rRNA have been used to reform the CP (Fig. S7B). Despite the absence of 5S rRNA, the CP is triple the volume of the bacterial version. There is a larger network of interactions with the rest of the large subunit, and potential new contacts with the head of the small subunit (Fig. S3).
Figure 4.
Central protuberance (CP) of the 54S subunit. A. Proteins involved in the C P. B. Elements that replace 5S rRNA (gray) in the CP
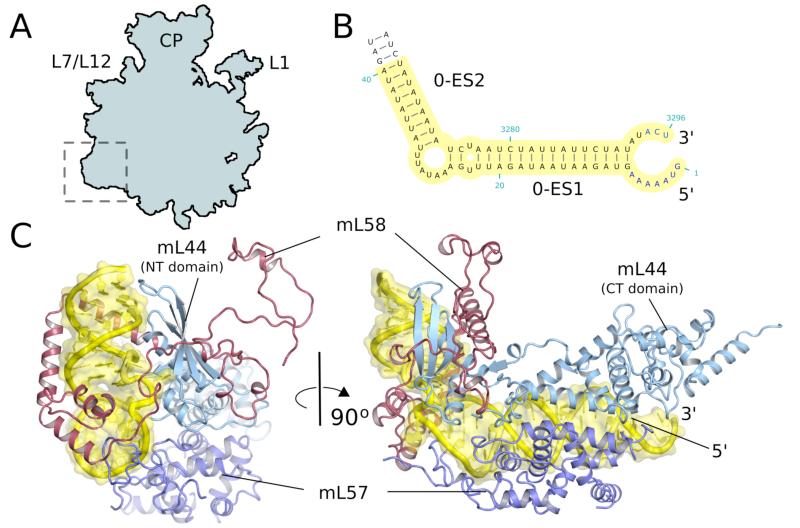
A membrane-facing protuberance
A second site enriched with mitochondria-specific proteins and rRNA is located adjacent to the mouth of the exit tunnel and thus must be close to the point of attachment of the ribosome to the translocon in the membrane (Fig. S9). This protuberance consists of mitochondria-specific mL43, 44, 50, 57, 58 (Fig. 2A), and rRNA expansion segments 0-ES1, 0-ES2, 44-ES3. mL44 and mL57, inactive members of the ribonuclease III family, form a heterodimer and bend the 0-ES by an angle of 90° forming an ‘L-shaped’ helix (Fig. 5B,C). The rest of the protuberance involves a second rRNA ES (44-ES3), the C-terminal part of mL50, N- and C-terminal extensions of uL22 and mL43, which binds predominantly to helix 46.
Figure 5.
A membrane facing protuberance. A. Site of expansion of the 54S relative to bacteria. B. ES0 of 21S rRNA serves as an anchor for the protuberance. C. Two views of the protuberance showing ho w mitochondria-specific proteins bind to the expansion segment.
Yeast mitoribosomes have been shown to be permanently associated with the inner mitochondrial membrane in both the translationally active and inactive state (32, 33). Moreover, mitoribosomes remain docked to the membrane even in the absence of the protein insertion machinery associated with the tunnel exit, indicating that additional and as yet uncharacterized anchors exist (34). The location of the membrane-facing protuberance makes it a candidate to be involved in the tethering of mitoribosomes to the membrane independent of nascent peptide insertion.
E-site tRNA
The yeast mitoribosomes co-purified with a tRNA bound in the exit (E) site. The density for the complete tRNA is relatively weak presumably due to a combination of partial occupancy and mixed species of tRNA (Fig. S10A). Nevertheless, we were able to model the acceptor stem including the conserved 3′ CCA. The terminal adenine intercalates between bases 2688 (E.coli : 2421) and 2689 (2422) in a manner identical to bacteria and eukaryotes (Fig. S10B). The tRNA appears to be stabilized by an additional contact not seen in bacteria. A C-terminal extension in bL33 creates a highly electropositive patch that stabilizes the phosphate backbone of the tRNA acceptor stem (Fig. S10C). The extension of bL33 is stabilized by a change in the rRNA path prior to helix 82-ES1, that is in itself partially stabilized by the unique protein mL59. Thus the E site of ribosomes from archaea (35), bacteria (9) and mitochondria (this work) all bind the terminal A76 in an essentially identical manner by intercalating between two purines of rRNA, while at the same time the rest of the E site has diverged among these classes of ribosomes.
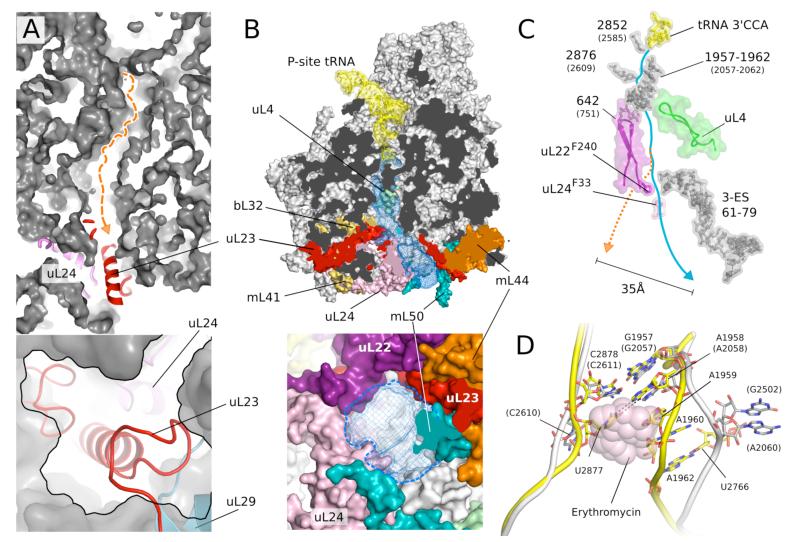
The exit tunnel
During translation, the growing nascent chain proceeds from the peptidyl transferase center (PTC) through a tunnel to the opposite end of the large subunit where it emerges ~100 Å away. Given its universality the tunnel is highly conserved among previously studied ribosomes. Despite this conservation, a superposition with bacterial ribosomes showed that the path of the bacterial tunnel is blocked in the 54S by a section of a long mitochondria-specific extension to uL23 that is well-anchored in the 54S on either side and thus unlikely to be displaced (Fig. 6A). Instead, a new tunnel path is seen that deviates from the bacterial one ~60 Å from the PTC (Fig. 6B). This is the only channel in the yeast mitoribosome that extends from the PTC to the exterior of the ribosome. The boundaries of the new tunnel are defined by extensions of uL22, uL23 and uL24, and rRNA 3-ES. The tunnel leads to a new exit site located roughly 35 Å away from where the bacterial tunnel would have emerged (Figs. 6B,C and S9), and is surrounded by two additional unique proteins, mL44 and mL50, which are also involved in forming the membrane-facing protuberance (Fig. 6B). This suggests that the attachment to a translocon-like entity for insertion of the nascent peptide into t he mitochondrial membrane must be very different in the 54S compared to bacterial ribosomes. In support of this idea, uL23, uL24, mL44 and mL50 were chemically found to cross-link with each other (36). The exit of the tunnel is also wider than in bacteria, possibly to allow co-translational assembly of oxidative phosphorylation complexes (34, 37).
Figure 6.
The putative exit tunnel in the 54S subunit. A. The bacterial tunnel (top, path in orange) is blocked by a mitochondria-specific extension to uL23 (detail in bottom panel). B. The 54S tunnel (blue mesh) has been extensively remodeled by mitochondria-specific protein elements (colored) (top) resulting in a tunnel exit (bottom) that is substantially different from that in bacteria. In A and B the lower panels are 90 ° rotations of the upper panels. C. Conservation of elements that make up the entrance and top part of the tunnel. The constriction at the top is narrower, and two aromatic amino acids that line the tunnel make a close approach. The path of the bacterial tunnel (orange) would exit 35 Å away. D. Narrowing of the 54S tunnel entrance (yellow) due to an AU base pair not found in bacteria (white) that would cause a steric clash with macrolides such as erythromycin (pink).
The entrance to the tunnel just beyond the PTC is noticeably narrower than in all previously determined structures of the large subunit (Fig. 6D). In part this is due to a base pair between U2877 and A1958 that appears unique to yeast mitochondria and could only be identified on the basis of the structure. This pair brings opposing strands of 21S rRNA closer together by about 8 Å. This location is the binding site of macrolide antibiotics, which unlike many aminoglycosides do not bind to mitochondrial ribosomes (38) and thus do not have the same toxicity. A superposition of the macrolide erythromycin in its binding site shows that it would clash with the narrower tunnel entrance in mitochondria, so in yeast mitoribosomes there is an additional mechanism for resistance to these antibiotics beyond sequence variation of the binding pocket. Beyond the entrance, the tunnel is lined by conserved elements (Fig. 6C), including the constriction site formed by uL22 and uL4. In addition, near the point where the path diverges from the path of the bacterial tunnel, two aromatic residues of uL22 and uL24 narrow the tunnel to a diameter of ~9 Å (Fig. 6C).
This work shows that recent advances in cryo-EM can be used to determine structures of comparable quality to x-ray crystallography but from much smaller amounts of more heterogeneous material. It also reveals that this highly divergent ribosomal subunit differs substantially from those of bacteria, eukaryotes and archaea, with several unique features including an altered path of the exit tunnel.
Supplementary Material
Acknowledgements
We thank John Walker for co-initiating this project and his support in the early phase of the work, G. McMullan, X. Chen, C. Savva for help with data collection, I.S. Fernández and D. Tourigny for help with preliminary modeling, S. Peak-Chew and M. Skehel for mass spectrometry, J. Grimmett and T. Darling for help with computing, and R. Nicholls for help with ProSMART. This work was funded by grants from the UK Medical Research Council (MC_U105184332 to VR, MC_UP_A025_1013 to SHWS and MC_UP_A025_1012 to GM); a Wellcome Trust Senior Investigator award (WT096570), the Agouron Institute and the Jeantet Foundation (VR); fellowships from Human Frontiers Science Program (AA), EU FP7 Marie Curie (XB), FEBS (JLL), and EMBO (TH). Cryo-EM density maps have been deposited with the EMDB (accession number EM D-2566) and coordinates have been deposited with the PDB (entry code 3J6B ).
References
- 1.Kurland CG, Andersson SG. Origin and evolution of the mitochondrial proteome. Microbiol Mol Biol Rev. 2000;64:786. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ott M, Herrmann JM. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta. 2010;1803:767. doi: 10.1016/j.bbamcr.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal RK, Sharma MR. Structural aspects of mitochondrial translational apparatus. Curr Opin Struct Biol. 2012;22:797. doi: 10.1016/j.sbi.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smits P, Smeitink J, van den Heuvel L. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J Biomed Biotechnol. 2010;2010:737385. doi: 10.1155/2010/737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones CN, Miller C, Tenenbaum A, Spremulli LL, Saada A. Antibiotic effects on mitochondrial translation and in patients with mitochondrial translational defects. Mitochondrion. 2009;9:429. doi: 10.1016/j.mito.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Herrmann JM, Woellhaf MW, Bonnefoy N. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim Biophys Acta. 2013;1833:286. doi: 10.1016/j.bbamcr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Barrientos A. Yeast models of human mitochondrial diseases. IUBMB Life. 2003;55:83. doi: 10.1002/tbmb.718540876. [DOI] [PubMed] [Google Scholar]
- 8.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 9.Selmer M. Structure of the 70S Ribosome Complexed with mRNA and tRNA. Science. 2006;313:1935. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 10.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 11.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal Structure of the Eukaryotic 60S Ribosomal Subunit in Complex with Initiation Factor 6. Science. 2011;334:941. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 13.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 14.Sharma MR, et al. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 15.Mears JA, et al. A structural model for the large subunit of the mammalian mitochondrial ribosome. Journal of Molecular Biology. 2006;358:193. doi: 10.1016/j.jmb.2006.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK. Structure of a mitochondrial ribosome with minimal RNA. Proceedings of the National Academy of Sciences. 2009;106:9637. doi: 10.1073/pnas.0901631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai X-C, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheres SH. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012;415:406. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2013 doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.See supporting material in Science online,
- 23.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography. 2011;67:355. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox GE. Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol. 2010;2:1. doi: 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maly P, Brimacombe R. Refined secondary structure models for the 16S and 23S ribosomal RNA of Escherichia coli. Nucleic Acids Res. 1983;11:7263. doi: 10.1093/nar/11.21.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray MW, Lukes J, Archibald JM, Keeling PJ, Doolittle WF. Cell biology. Irremediable complexity? Science. 2010;330:920. doi: 10.1126/science.1198594. [DOI] [PubMed] [Google Scholar]
- 28.Smits P, Smeitink JAM, van den Heuvel LP, Huynen MA, Ettema TJG. Reconstructing the evolution of the mitochondrial ribosomal proteome. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 30.Dontsova OA, Dinman JD. 5S rRNA: Structure and Function from Head to Toe. Int J Biomed Sci. 2005;1:1. [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 32.Prestele M, Vogel F, Reichert AS, Herrmann JM, Ott M. Mrpl36 is important for generation of assembly competent protein s during mitochondrial translation. Mol Biol Cell. 2009;20:2615. doi: 10.1091/mbc.E08-12-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott M, et al. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 2006;25:1603. doi: 10.1038/sj.emboj.7601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keil M, et al. Oxa1-ribosome complexes coordinate the assembly of cytochrome C oxidase in mitochondria. J Biol Chem. 2012;287:34484. doi: 10.1074/jbc.M112.382630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruschke S, et al. Proteins at the polypeptide tunnel exit of the yeast mitochondrial ribosome. The Journal of biological chemistry. 2010;285:19022. doi: 10.1074/jbc.M110.113837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gruschke S, et al. Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J Cell Biol. 2011;193:1101. doi: 10.1083/jcb.201103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denslow ND, O’Brien TW. Antibiotic susceptibility of the peptidyl transferase locus of bovine mitochondrial ribosomes. Eur J Biochem. 1978;91:441. doi: 10.1111/j.1432-1033.1978.tb12696.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.