Abstract
Humans are exposed to low-dose ionizing radiation (LDIR) from a number of environmental and medical sources. In addition to inducing genetic mutations, there is concern that LDIR may also alter the epigenome. Such heritable effects early in life can either be positively adaptive or result in the enhanced formation of diseases, including cancer, diabetes, and obesity. Herein, we show that LDIR significantly increased DNA methylation at the viable yellow agouti (Avy) locus in a sex-specific manner (P=0.004). Average DNA methylation was significantly increased in male offspring exposed to doses between 0.7 and 7.6 cGy, with maximum effects at 1.4 and 3.0 cGy (P<0.01). Offspring coat color was concomitantly shifted toward pseudoagouti (P<0.01). Maternal dietary antioxidant supplementation mitigated both the DNA methylation changes and coat color shift in the irradiated offspring. Thus, LDIR exposure during gestation elicits epigenetic alterations that lead to positive adaptive phenotypic changes that are negated with antioxidants, indicating they are mediated in part by oxidative stress. These findings provide evidence that in the isogenic Avy mouse model, epigenetic alterations resulting from LDIR play a role in radiation hormesis, bringing into question the assumption that every dose of radiation is harmful.—Bernal, A. J., Dolinoy, D. C., Huang, D., Skaar, D. A., Weinhouse, C., Jirtle, R. J. Adaptive radiation-induced epigenetic alterations mitigated by antioxidants.
Keywords: agouti mice, DNA methylation, hormesis, reactive oxygen species
Since the discovery of ionizing radiation in 1895, the human health effects stemming from moderate to high-dose exposures have been well documented; however, the majority of human exposures occur in the low-dose range (<10 cGy). For example, low-dose ionizing radiation (LDIR) exposure from medical procedures has increased >7-fold since the early 1980s and now comprises nearly 50% of the average American's yearly radiation exposure (1). Along with medical radiation, nuclear energy production also raises the potential for large-scale human environmental exposures, such as occurred after the nuclear power plant accidents in Chernobyl in 1986 (2) and Fukushima Daiichi in 2011 (3). Nevertheless, the human health risks of LDIR are still estimated by extrapolation from the biological effects observed at high doses, according to the linear no threshold (LNT) risk assessment model (4).
High doses of ionizing radiation result in epigenetic modifications in adult mice (5). They also induce genomic instability and the bystander effect, a phenomenon in which nonirradiated cells exhibit radiation damage even though they are not directly exposed (6). Moreover, the bystander effect is dependent on epigenetic signaling, since targeted disruption of Dnmt1 and Dnmt3a in cultured cells eliminates the transmission of genomic instability (7). Growing evidence also supports a role for reactive oxygen species (ROS) in these epigenetic processes through the generation of oxidative stress (8). Whether epigenetic alterations also occur in vivo in response to LDIR has not been thoroughly investigated, especially during early development.
To determine whether dose-dependent epigenetic responses develop in animals in response to LDIR, we utilized the viable yellow agouti (Avy) mouse model (9). This unique mouse strain is an exquisitely sensitive biosensor for environmental agents that alter the fetal epigenome. Variable expression of the Avy metastable epiallele is controlled by epigenetic modifications, such as histone marks (10) and cytosine-phosphate-guanine (CpG) site methylation (11) that are established early in development in and around the cryptic promoter in a transgene upstream of the Agouti gene (Fig. 1A, C). Hypomethylation of this alternative promoter leads to inappropriate Agouti gene expression in all tissues in Avy mice. This not only leads to a yellow coat color, it also antagonizes the melanocortin 4 receptor (MC4R) in the hypothalamus, causing the animals to become obese and develop diabetes and cancer at a high frequency. In contrast, the incidence of these diseases is markedly reduced in pseudoagouti (brown) offspring that develop when this promoter is hypermethylated (11–13).
Figure 1.
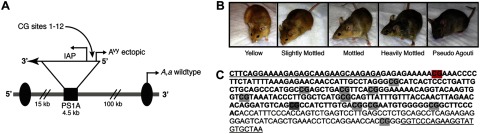
Avy locus and resulting mouse coat colors. A) Agouti gene encodes for a paracrine-signaling molecule that produces either black eumelanin (a) or yellow phaeomelanin (A) from the wild-type promoter (arrow labeled A,a wild type; refs. 9, 11). Agouti expression during follicle development results in brown (agouti) wild-type animals. Avy allele resulted from a spontaneous contraoriented insertion of an intracisternal A particle (IAP) into pseudoexon PS1A upstream of the wild-type promoter. This insertion carries a cryptic promoter (arrow labeled Avy ectopic) controlled by the methylation of upstream CpG sites. B) Level of CpG methylation at the Avy locus results in the formation of distinct coat color phenotypes. Hypermethylation of the cryptic IAP promoter results in brown, pseudoagouti offspring, hypomethylation results in yellow offspring, and mottled mice are epigenetic mosaic offspring. C) Amplified IAP sequence (bold font) contains 11 CpG sites in the ectopic promoter and 1 CpG site within the downstream 3′ genomic sequence (regular font). The first CpG site (highlighted in red) was not measured due to technical limitations associated with our sequencing approach. The eighth CpG site (highlighted in dark gray) was measured on the reverse strand.
Previous use of the Avy mouse model showed that early developmental exposures to methyl donors (14), genistein (15), bisphenol A (BPA; ref. 16), ethanol (17), and in vitro culturing (18) cause phenotypic changes in the offspring by altering the epigenome. Furthermore, these environmental exposures elicited epigenetic changes in other regions of the genome (16, 19). This study is the first to demonstrate that LDIR exposures during early development cause both dose- and sex-dependent epigenetically induced adaptive changes at the Avy locus that in part depend on a cellular oxidative stress response.
MATERIALS AND METHODS
Animal breeding and irradiation
Avy mice were obtained from an isogenic colony maintained by sibling mating and forced heterozygosity of the Avy allele for >200 generations, as described previously (14). At 7 wk of age, virgin black a/a females were placed on a phytoestrogen-free breeding diet, with 7% corn oil substituted for 7% soybean oil (TD·95092; Harlan Teklad, Madison, WI, USA). The dams were kept on the diet 1 wk before mating and throughout gestation and lactation. At 8 wk of age, female black a/a mice were bred to 8 wk-old Avy/a males of varying coat colors. Detection of a vaginal plug was considered gestational day 0.5. The intrauterine doses of 0.4 cGy (n=9 litters, 63 total offspring, 28 Avy/a offspring), 0.7 cGy (n=10 litters, 76 total offspring, 32 Avy/a offspring), 1.4 cGy (n=11 litters, 70 total offspring, 40 Avy/a offspring), 3.0 cGy (n=17 litters, 101 total offspring, 53 Avy/a offspring), and 7.6 cGy (n=12 litters, 79 total offspring, 41 Avy/a offspring) were delivered to pregnant dams at gestational day 4.5 with a Siemens MicroCT scanner (80 kVp, 500 μA; Siemens Medical Systems, Knoxville, TN, USA). The intrauterine doses were measured using a phantom mouse model and a Radcal ion dosimeter chamber (Radcal, Monrovia, CA, USA). To control for stress, control dams were sham-irradiated by being placed in the scanner for 1 min without being irradiated (n=14 litters, 90 total offspring, 47 Avy/a offspring).
Dams whose diet was supplemented with antioxidants (AOs) were fed diet TD.95092 containing 0.014 g/kg tert-butylhydroquinone (TBHQ), 0.006 g/kg seleno-l-methionine, 0.408 g/kg vitamin C, 0.0714 g/kg vitamin E, 0.0857 g/kg α-lipoic acid, and 0.1714 g/kg N-acetyl-l-cysteine (TD.10635; Harlan Teklad). The levels of AOs used in these studies were equivalent on a weight basis (g/kg) to the established maximum level of daily nutrient intake in humans that is likely to pose no adverse effects (20). During the AO study, 8 additional pregnant mice fed control diet were exposed to 3.0 cGy (n=8 litters, 51 total offspring, 27 Avy/a offspring) and 2 additional pregnant mice fed control diet were sham-irradiated (n=2 litters, 14 total offspring, 6 Avy/a offspring). These litters were added to the AO study, bringing the total number of litters exposed to 3.0 cGy with control diet to 25 (152 total offspring, 80 Avy/a offspring) and the total number of sham-irradiated litters to 16 (104 total offspring, 53 Avy/a offspring).
Animals used in this study were maintained in accordance with the Institute of Laboratory Animal Resources guidelines (21). They were treated humanely and with regard for alleviation of suffering. The study protocol was approved by the Duke University Institutional Animal Care and Use Committee.
Coat color analysis
Offspring were weaned on d 22 after birth and categorized into 1 of 5 coat color classes: yellow (<5% brown), slightly mottled (5-40% brown), mottled (∼50% brown), heavily mottled (60-95% brown), and pseudoagouti (>95% brown) by a single observer (A.J.B.; Fig. 1B).
DNA isolation and bisulfite treatment
Liver and tail tissue samples were collected from isogenic Avy/a offspring at 22 d after birth, and total genomic DNA was isolated using buffer ATL, proteinase K, and RNase A (Qiagen, Valencia, CA, USA), followed by phenol:chloroform extraction and ethanol precipitation. DNA (2.0 μg) was bisulfite treated using the EpiTect bisulfite kit (Qiagen) to allow for the conversion of unmethylated cytosines to uracil (read as thymine during PCR amplification), whereas the methylated cytosines remain unconverted (22).
Avy locus amplification and methylation analysis
The cryptic promoter region upstream of the Agouti gene (Fig. 1A, C) was amplified from bisulfite-modified liver and tail tissue DNA in 25-μl PCR reaction volumes using 1.5 U of Platinum TaqDNA polymerase (Invitrogen, Carlsbad, CA, USA), 10 μmol of primers, 1.5 mM MgCl2, and 10 mM dNTPs (94°C×2 min; 94°C×30 s, 62°C×30 s, and 72°C×60 s for 40 cycles; 72°C×9 min). A T7 promoter tag was introduced to facilitate RNA transcription, which is necessary in the Sequenom EpiTYPER platform (Sequenom, San Diego, CA, USA) before base-specific cleavage occurs. We used forward primer Seq_Avy_FS_F1 (5′-AGGAAGAGAGTTTTAGGAAAAGAGAGTAAGAAGTAAGAGA-3′) and reverse primer Seq_Avy_FS_R1b (5′-CAGTAATACGACTCACTATAGGGAGAAGGCTTAACACATACCTTCTAAAACC-3′), creating a 336-bp product on the forward strand. Amplification of the reverse strand was also necessary to quantify the eighth CpG site (Fig. 1C). For the reverse strand, we used forward primer Seq_Avy_RS_F2 (5′-AGGAAGAGAGGAGGTTTAAGGATTTAGATTGGTGG-3′) and reverse primer Seq_Avy_RS_R2 (5′-CAGTAATACGACTCACTATAGGGAGAAGGCATCACTCCCTAATTACTACAACCCA-3′), creating a 199-bp product. Following amplification, in vitro RNA transcription was performed on the reverse strand of each amplicon, followed by base-specific cleavage and quantification with MALDI-TOF mass spectrometry, according to the Sequenom EpiTYPER platform. Each sample was run in triplicate, and individual CpG sites were averaged. Unmethylated, 50% methylated, and fully methylated DNA samples were also run to ensure assay quality. The 12 CpG sites studied on the Avy allele are located at nucleotide positions 132, 174, 206, 214, 220, 244, 265, 306, 319, 322, 334, and 425 of GenBank accession number AF540972.
Statistical analysis
The distribution of the 5 coat color phenotypes, a discrete variable categorized as yellow, slightly mottled, mottled, heavily mottled, or brown, between each exposure group and the control group was analyzed using a χ2 goodness-of-fit test with GraphPad Prism software (GraphPad Software, La Jolla, CA, USA). For analysis of DNA methylation, a continuous variable representing the percentage of cells methylated, triplicate runs were averaged, and a 3-factor repeated measures ANOVA was performed using StatView software (SAS, Cary, NC, USA). CpG site was designated as the repeated measure, and dose and sex the interacting factors. On determining significant interactions (ANOVA<0.05), post hoc analysis was performed using Fisher's protected least significant difference (PLSD) test.
RESULTS
Offspring statistics
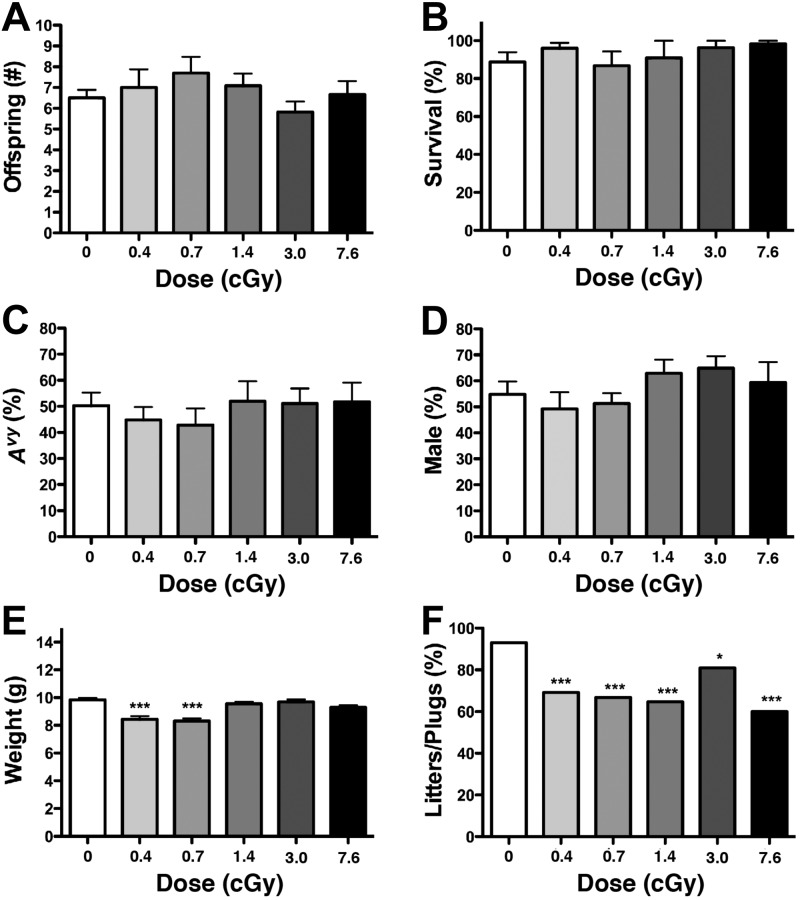
LDIR did not significantly influence litter size, offspring survival, genotypic ratio, or sex ratio (ANOVA, P>0.3; Fig. 2A–D). Average weanling weight was significantly decreased when compared with sham-irradiated controls (ANOVA, P<0.0001) but only in the 0.4- and 0.7-cGy exposure groups (Fig. 2E). LDIR also significantly reduced the percentage of plugged females with litters at all exposure groups (χ2, P<0.05; Fig. 2F).
Figure 2.
Effect of radiation on litter size at weaning (A), percentage of offspring survival to weaning (B), percentage of offspring with an Avy genotype (C), percentage of male offspring (D), offspring weight at weaning (E), and percentage of plugged females with litters (F). *P < 0.05, ***P < 0.0001; χ2, ANOVA.
Offspring coat color
At the lowest exposure dose of 0.4 cGy (n=28), offspring coat color distribution was not significantly altered from that of sham-irradiated control offspring (n=47; χ2, P=0.9; Fig. 3A). In contrast, exposure to 0.7 cGy (n=32), 1.4 cGy (n=40), 3.0 cGy (n=53), and 7.6 cGy (n=41) significantly shifted the coat color distribution of the Avy/a offspring toward heavily mottled and pseudoagouti (χ2; P=0.01, P=0.02, P=0.002, and P=0.04, respectively). Offspring irradiated with 0.7, 1.4, and 3.0 cGy had more than twice as many pseudoagouti animals as the sham-irradiated offspring (χ2, P<0.01 for each dose; Fig. 3A). At the highest dose of 7.6 cGy, there were significantly more heavily mottled animals than in the sham-irradiated offspring (χ2, P<0.05), whereas the incidence of pseudoagouti offspring was more comparable to that observed in the sham-irradiated control offspring (χ2, P>0.1). This indicated that at the highest radiation dose used in this study, the coat color distribution of the offspring was returning to that of the sham-irradiated controls. Offspring with yellow coat colors were concomitantly decreased with increasing radiation dose, becoming significant at 3.0 cGy (χ2, P<0.05) and 7.6 cGy (χ2, P<0.05; Fig. 3A). Interestingly, the coat color changes were more pronounced in the male than in the female offspring. This shift in the coat color distribution was significant at all radiation doses in males (χ2, P<0.05), whereas it was only significant at 0.7 cGy in females (χ2, P=0.01). Thus, the change in coat color distribution was significantly altered in response to LDIR not only in a dose-dependent but also in a sex-dependent manner.
Figure 3.
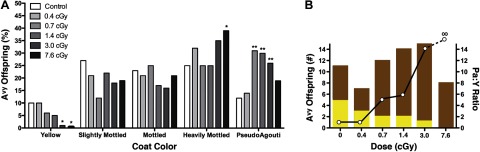
Effect of radiation on the coat color of Avy offspring. A) Percentage of yellow, slightly mottled, mottled, heavily mottled, and pseudoagouti Avy offspring at 0 cGy (n=47), 0.4 cGy (n=28), 0.7 cGy (n=32), 1.4 cGy (n=40), 3.0 cGy (n=53), and 7.6 cGy (n=41) was compared with that in the sham-irradiated offspring. *P < 0.05, **P < 0.01; χ2. B) Number of pseudoagouti (brown bars) and yellow (yellow bars) offspring at each radiation dose (left y axis) were used to calculate the pseudoagouti:yellow offspring ratio (open circles; right y axis) at each radiation dose.
The ratio of pseudoagouti to yellow offspring is ∼1 in the sham-irradiated offspring and those exposed to 0.4 cGy (Fig. 3B). This ratio increased to 14 for the offspring irradiated with 3.0 cGy, and it cannot be calculated at 7.6 cGy because there were no yellow offspring at this dose (Fig. 3B). Thus, the ratio of pseudoagouti to yellow offspring also increased markedly in a dose-dependent manner from that observed in the sham-irradiated animals.
DNA methylation at the Avy locus
LDIR had no significant effect on DNA methylation when averaged across the 11 measured CpG sites when the male and female offspring were grouped together (ANOVA, P=0.1), although exposure groups ≥0.7 cGy showed a trend toward increased methylation (Table 1).Further analysis of these data demonstrated that the radiation-induced CpG site-specific changes in DNA methylation were sex dependent (ANOVA, P=0.004). Across all exposure groups, female offspring displayed minimal CpG site-specific increases in DNA methylation compared with controls (data not shown). In contrast, among the male offspring DNA methylation in the liver increased significantly with dose at multiple CpG sites, maximizing at 1.4 and 3.0 cGy (ANOVA, P<0.05) with a 26 and 23% increase in methylation, respectively (Fig. 4A).
Table 1.
Interactions of dose, CpG site, and sex on DNA methylation
| Parameter | n | P | Methylation (%) |
|---|---|---|---|
| Radiation dose to liver (cGy) | 234 | ||
| 0.0 | 47 | 33.1 ± 3.9 | |
| 0.4 | 26 | 30.6 ± 5.3 | |
| 0.7 | 32 | 41.3 ± 4.8 | |
| 1.4 | 40 | 42.7 ± 4.3 | |
| 3.0 | 51 | 43.8 ± 3.8 | |
| 7.6 | 38 | 42.6 ± 4.4 | |
| Dose | 0.1 | ||
| Dose × sex | 0.03* | ||
| Site × dose | 0.01* | ||
| Site × dose × sex | 0.004* |
Methylation values are means ± se.
P < 0.05, significant difference.
Figure 4.
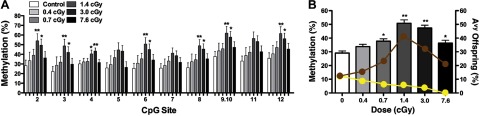
Effect of radiation on DNA methylation and the coat color of Avy male offspring. A) Percentage methylation ± se of 11 CpG sites at the Avy locus in liver tissue from male offspring exposed to 0.4 cGy (n=13), 0.7 cGy (n=22), 1.4 cGy (n=24), 3.0 cGy (n=29), and 7.6 cGy (n=19) compared with male sham-irradiated offspring (n=26). *P ≤ 0.05, **P < 0.01. B) Average percentage methylation ± se of the 11 CpG sites in male liver tissue at various radiation doses was compared with that in sham-irradiated offspring (ANOVA, P<0.0001). Yellow and brown circles depict the percentage of the male Avy offspring with yellow and brown coat colors, respectively. *P < 0.05, **P < 0.01; Fisher's PLSD.
These effects were also evident when DNA methylation was averaged across all CpG sites in male Avy offspring (ANOVA, P<0.0001; Fisher's PLSD, P≤0.05; Fig. 4B). Interestingly, DNA methylation in males returned close to the control level at 7.6 cGy, the highest radiation dose used in this study. This finding is consistent with the previously discussed observation that coat color distribution also appeared to be returning to that of the sham-irradiated controls at 7.6 cGy. Increased DNA methylation was similarly observed in the tail tissue of male offspring exposed to 1.4 and 3.0 cGy (ANOVA, P<0.05), but it returned to sham-irradiated levels at 7.6 cGy (data not shown). These results are consistent with previous findings that environmentally-induced alterations in methylation at the Avy metastable epiallele are tissue independent (14–16).
AO supplementation
Following AO supplementation, the coat color distribution of animals exposed to 3.0 cGy (n=51 Avy/a offspring) was not significantly different than sham-irradiated control offspring (n=53 Avy/a offspring; χ2, P=0.4; Fig. 5A). In contrast, non-AO-supplemented offspring exposed to 3.0 cGy (n=80 Avy/a offspring) had significantly lower percentages of yellow and higher percentages of pseudoagouti offspring than sham-irradiated control offspring. Thus, maternal AO supplementation reduced the pseudoagouti to yellow offspring ratio from 11:1 in offspring exposed to 3.0 cGy to 1:1 in offspring irradiated with 3.0 cGy and supplemented with AO; a ratio identical to that observed in sham-irradiated control offspring (Fig. 5B).
Figure 5.
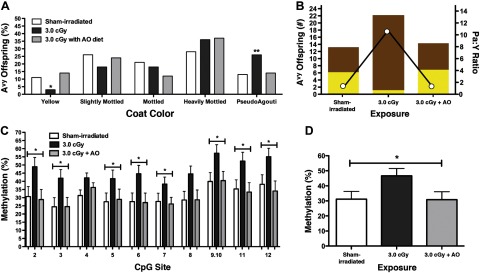
Effect of radiation and AO supplementation on DNA methylation and coat color of Avy offspring. A) Percentage of Avy offspring that were yellow, slightly mottled, mottled, heavily mottled, and pseudoagouti when the mothers were sham irradiated (n=53) and exposed to 3.0 cGy (n=80) or 3.0 cGy + AO (n=51). *P < 0.05, **P < 0.01; χ2. B) Number of pseudoagouti (brown bars) and yellow (yellow bars) offspring when the mothers were sham irradiated and exposed to 3·0 cGy or 3.0 cGy + AO (left y axis) were used to calculate the pseudoagouti:yellow offspring ratio (open circles; right y axis) for each exposure. C) Percentage methylation ± se of 11 CpG sites at the Avy locus in the liver tissue of male offspring sham-irradiated (n=27), and exposed to 3·0 cGy (n=38) or 3·0 cGy + AO (n=23). *P ≤ 0.05; ANOVA. D) Average percentage methylation ± se of 11 CpG sites at the Avy locus in the liver tissue of male offspring sham-irradiated (n=27), and exposed to 3.0 cGy (n=38) or 3.0 cGy + AO (n=23). *P ≤ 0.05; ANOVA.
When compared with sham-irradiated controls (n=27), there were significant site-specific increases in liver DNA methylation of male offspring exposed to 3.0 cGy (n=38; P<0.05), but not in male offspring supplemented with AO and then irradiated (n=23; P>0.3; Fig. 5C). When DNA methylation was averaged across the 11 CpG sites, the increase in methylation observed in males exposed to 3.0 cGy was again shown to be reduced in the offspring of mothers supplemented with an AO diet and irradiated with 3.0 cGy (ANOVA, P<0.05; Fig. 5D). In contrast, there was no significant difference in the average DNA methylation in the liver tissue of sham-irradiated male offspring (n=27) and male offspring exposed to 3.0 cGy + AO (n=23; ANOVA, P>0.3). Tail tissue of offspring exposed to 3.0 cGy + AO was also examined and found to be similar to that of sham-irradiated male offspring (ANOVA, P>0.1; data not shown), but significantly decreased compared with male offspring exposed to 3.0 cGy (ANOVA, P<0.05; data not shown). These findings demonstrate that AO supplementation mitigates the DNA hypermethylation induced by LDIR, and support our postulate that oxidative stress is in part responsible for the epigenetic and phenotypic changes observed in response to LDIR.
DISCUSSION
The Avy agouti mouse model is a unique biosensor for detecting environmental exposures that alter the epigenome during gestation. Previously, this animal model has demonstrated that early nutritional (14, 15, 18) and chemical (16, 17) exposures induce persistent epigenetic changes at the Avy locus. Herein, we demonstrate that LDIR, a physical agent, also significantly alters DNA methylation and induces a positive adaptive phenotype in Avy offspring exposed during pregnancy to doses equivalent to those received for a chest or head CT scan.
High doses of radiation (i.e., 50–200 cGy) cause DNA hypomethylation in mouse liver (5) and in bystander spleen and skin tissues when only the brain is exposed (23). Hypomethylation at long interspersed element-1 (LINE-1) repetitive elements in rat spleen (24) and down-regulation of DNA methyl binding protein in mouse skin are also reported in response to high-dose radiation exposure (25). Moreover, studies that compared radiation sensitivities between males and females show sex-specific epigenetic effects in directly irradiated (26, 27) and bystander tissues (6), with males being affected more significantly than females. While previous investigations demonstrated that high doses of radiation reduce DNA methylation, often in a sex-dependent manner, we showed in this investigation that LDIR significantly increased DNA methylation at the Avy locus preferentially in male mice in a dose-dependent manner.
We also demonstrated that maternal AO supplementation mitigated the increase in DNA methylation and the concomitant shift in coat color observed in irradiated male Avy offspring. These findings support the postulate that LDIR increases DNA methylation at the Avy locus in part through the generation of ROS. This may help explain the unexpected increased rate of second primary tumors (SPTs) observed in head and neck cancer patients who received AO supplementation with radiation therapy (28). It also illustrates the potential difficulty in preventing SPTs during cancer treatment with the use of AOs in combination with radiation treatment. Thus, the efficacy of this combination therapy needs to be carefully evaluated in the future.
Hypomethylation of DNA at high doses of radiation and hypermethylation at low doses are indicative of a hormetic biphasic radiation dose response effect (29). Hypermethylation at the Avy locus increases the frequency of pseudoagouti offspring, which have lower risks of developing obesity, cancer, and insulin resistance in adulthood due to the loss of ectopic Agouti gene expression (11–13). Thus, LDIR during early gestation results in beneficial health effects in Avy mice. Hormesis in response to LDIR has been reported in the literature for decades (4, 30). The reported protective effects include increased apoptosis of damaged cells, radioadaptive protection, enhanced AO protection, removal of DNA lesions, immunological stimulation, and decreased disease rates (4, 30, 31). Our findings in the Avy mouse are likewise contrary to the assumption of the LNT risk assessment model that every dose of radiation is harmful. On the other hand, a recent epidemiological study of leukemia and brain cancer risk from childhood CT scans reports support of the LNT radiation risk model (32). It is important to note, however, that the observed cancer incidences in this study were not significantly above background at the doses we observed to be optimal for the induction of positive adaptive effects and hypermethylation at the Avy locus.
Although increased DNA methylation is advantageous at the Avy locus, hypermethylation is not always beneficial. The tumor suppressor genes, p16INK4a and Rassf1a, are hypermethylated and down-regulated in mouse skin exposed to UVB, leading to an increase in the incidence of skin cancer (33). Ethanol exposure likewise induces hypermethylation at the Avy locus, but it also leads to the enhanced development of craniofacial abnormalities (17). Thus, the response to LDIR may be loci-specific and have both beneficial and detrimental consequences. Future studies are needed to evaluate radiation-induced changes in other candidate genes, such as imprinted genes, as well as more broadly across the complete methylome. Furthermore, altered epigenetic programming can be inherited transgenerationally (19, 34), potentially further complicating human radiation risk assessment at low doses.
The mechanisms by which LDIR induces adaptive biological responses have remained enigmatic (35). We now provide evidence that in the Avy mouse, epigenetic alterations may be the memory system that results in hormesis at low doses of ionizing radiation. Moreover, the results of our AO study indicate that the cellular redox state early in development plays an important role in determining the methylation status at the Avy locus. Our findings not only have significant implications concerning the mechanism of hormesis, but they also emphasize the potential importance of this phenomenon in determining human risk at low radiation doses. Since the epigenome varies markedly between species, the effect of LDIR on the epigenome in multiple generations needs to be defined in humans. Epidemiological data alone will no longer suffice to assess our risk to clinically relevant doses of X-rays.
Acknowledgments
The authors thank Susan K. Murphy for critically reading this manuscript, Theodore Slotkin for assisting with statistical analysis, and Christopher Faulk for agouti mouse photography. The authors also thank Greta Toncheva and the Duke Radiation Dosimetry Laboratory for assistance in determining the intrauterine radiation doses.
Financial support from the U.S. Department of Energy (grant DE-FG02-10ER64931) and the Ester B. O'Keeffe Foundation is gratefully acknowledged.
The authors declare no conflicts of interest.
Footnotes
- AO
- antioxidant
- Avy
- viable yellow agouti
- BPA
- bisphenol A
- CpG
- cytosine-phosphate-guanine
- LDIR
- low-dose ionizing radiation
- LNT
- linear no threshold
- PLSD
- protected least significant difference
- ROS
- reactive oxygen species
- SPT
- second primary tumor
- TBHQ
- tert-butylhydroquinone
REFERENCES
- 1. Schauer D. A., Linton O. W. (2009) NCRP report no. 160, ionizing radiation exposure of the population of the United States, medical exposure–are we doing less with more, and is there a role for health physicists? Health Phys. 97, 1–5 [DOI] [PubMed] [Google Scholar]
- 2. Saenko V., Ivanov V., Tsyb A., Bogdanova T., Tronko M., Demidchik Y., Yamashita S. (2011) The Chernobyl accident and its consequences. Clin. Oncol. (R. Coll. Radiol.) 23, 234–243 [DOI] [PubMed] [Google Scholar]
- 3. Akiba S. (2012) Epidemiological studies of Fukushima residents exposed to ionising radiation from the Fukushima Daiichi nuclear power plant prefecture–a preliminary review of current plans. J. Radiol. Prot. 32, 1–10 [DOI] [PubMed] [Google Scholar]
- 4. Sanders C. (2009) Radiation Hormesis and the Linear-No-Threshold Assumption, Springer, New York [Google Scholar]
- 5. Tawa R., Kimura Y., Komura J., Miyamura Y., Kurishita A., Sasaki M. S., Sakurai H., Ono T. (1998) Effects of X-ray irradiation on genomic DNA methylation levels in mouse tissues. J. Radiat. Res. 39, 271–278 [DOI] [PubMed] [Google Scholar]
- 6. Koturbash I., Kutanzi K., Hendrickson K., Rodriguez-Juarez R., Kogosov D., Kovalchuk O. (2008) Radiation-induced bystander effects in vivo are sex specific. Mutat. Res. 642, 28–36 [DOI] [PubMed] [Google Scholar]
- 7. Rugo R. E., Mutamba J. T., Mohan K. N., Yee T., Chaillet J. R., Greenberger J. S., Engelward B. P. (2011) Methyltransferases mediate cell memory of a genotoxic insult. Oncogene 30, 751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Franco R., Schoneveld O., Georgakilas A. G., Panayiotidis M. I. (2008) Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 266, 6–11 [DOI] [PubMed] [Google Scholar]
- 9. Duhl D. M., Vrieling H., Miller K. A., Wolff G. L., Barsh G. S. (1994) Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 8, 59–65 [DOI] [PubMed] [Google Scholar]
- 10. Dolinoy D. C., Weinhouse C., Jones T. R., Rozek L. S., Jirtle R. L. (2010) Variable histone modifications at the A(vy) metastable epiallele. Epigenetics 5, 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jirtle R. L., Skinner M. K. (2007) Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 8, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yen T. T., Gill A. M., Frigeri L. G., Barsh G. S., Wolff G. L. (1994) Obesity, diabetes, and neoplasia in yellow A(vy)/- mice: ectopic expression of the agouti gene. FASEB J. 8, 479–488 [DOI] [PubMed] [Google Scholar]
- 13. Williams G., Harrold J. A., Cutler D. J. (2000) The hypothalamus and the regulation of energy homeostasis: lifting the lid on a black box. Proc. Nutr. Soc. 59, 385–396 [DOI] [PubMed] [Google Scholar]
- 14. Waterland R. A., Jirtle R. L. (2003) Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23, 5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dolinoy D. C., Weidman J. R., Waterland R. A., Jirtle R. L. (2006) Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect. 114, 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dolinoy D. C., Huang D., Jirtle R. L. (2007) Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. U. S. A. 104, 13056–13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaminen-Ahola N., Ahola A., Maga M., Mallitt K. A., Fahey P., Cox T. C., Whitelaw E., Chong S. (2010) Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 6, e1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morgan H. D., Jin X. L., Li A., Whitelaw E., O'Neill C. (2008) The culture of zygotes to the blastocyst stage changes the postnatal expression of an epigentically labile allele, agouti viable yellow, in mice. Biol. Reprod. 79, 618–623 [DOI] [PubMed] [Google Scholar]
- 19. Feil R., Fraga M. F. (2011) Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 13, 97–109 [DOI] [PubMed] [Google Scholar]
- 20. Wambi C. O., Sanzari J. K., Sayers C. M., Nuth M., Zhou Z., Davis J., Finnberg N., Lewis-Wambi J. S., Ware J. H., El-Deiry W. S., Kennedy A. R. (2009) Protective effects of dietary antioxidants on proton total-body irradiation-mediated hematopoietic cell and animal survival. Radiat. Res. 172, 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Institute of Laboratory Animal Resources (1996) Guidelines for the Care and Use of Laboratory Animals, National Academy Press, Washington, DC [Google Scholar]
- 22. Grunau C., Clark S. J., Rosenthal A. (2001) Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 29, E65–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ilnytskyy Y., Koturbash I., Kovalchuk O. (2009) Radiation-induced bystander effects in vivo are epigenetically regulated in a tissue-specific manner. Environ. Mol. Mutagen. 50, 105–113 [DOI] [PubMed] [Google Scholar]
- 24. Koturbash I., Boyko A., Rodriguez-Juarez R., McDonald R. J., Tryndyak V. P., Kovalchuk I., Pogribny I. P., Kovalchuk O. (2007) Role of epigenetic effectors in maintenance of the long-term persistent bystander effect in spleen in vivo. Carcinogenesis 28, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 25. Koturbash I., Rugo R. E., Hendricks C. A., Loree J., Thibault B., Kutanzi K., Pogribny I., Yanch J. C., Engelward B. P., Kovalchuk O. (2006) Irradiation induces DNA damage and modulates epigenetic effectors in distant bystander tissue in vivo. Oncogene 25, 4267–4275 [DOI] [PubMed] [Google Scholar]
- 26. Kovalchuk O., Burke P., Besplug J., Slovack M., Filkowski J., Pogribny I. (2004) Methylation changes in muscle and liver tissues of male and female mice exposed to acute and chronic low-dose X-ray-irradiation. Mutat. Res. 548, 75–84 [DOI] [PubMed] [Google Scholar]
- 27. Koturbash I., Zemp F., Kolb B., Kovalchuk O. (2011) Sex-specific radiation-induced microRNAome responses in the hippocampus, cerebellum and frontal cortex in a mouse model. Mutat. Res. 722, 114–118 [DOI] [PubMed] [Google Scholar]
- 28. Bairati I., Meyer F., Gelinas M., Fortin A., Nabid A., Brochet F., Mercier J. P., Tetu B., Harel F., Masse B., Vigneault E., Vass S., del Vecchio P., Roy J. (2005) A randomized trial of antioxidant vitamins to prevent second primary cancers in head and neck cancer patients. J. Natl. Cancer Inst. 97, 481–488 [DOI] [PubMed] [Google Scholar]
- 29. Calabrese E. J., Baldwin L. A. (2002) Defining hormesis. Hum. Exp. Toxicol. 21, 91–97 [DOI] [PubMed] [Google Scholar]
- 30. Calabrese E. J. (2009) The road to linearity: why linearity at low doses became the basis for carcinogen risk assessment. Arch. Toxicol. 83, 203–225 [DOI] [PubMed] [Google Scholar]
- 31. Calabrese E. (2011) Improving the scientific foundations for estimating health risks from the Fukushima incident. Proc. Natl. Acad. Sci. U. S. A. 108, 19447–19448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pearce M. S., Salotti J. A., Little M. P., McHugh K., Lee C., Kim K. P., Howe N. L., Ronckers C. M., Rajaraman P., Sir Craft A. W., Parker L., de Gonzalez A. B. (2012) Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: a retrospective cohort study. Lancet 380, 499–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nandakumar V., Vaid M., Tollefsbol T. O., Katiyar S. K. (2011) Aberrant DNA hypermethylation patterns lead to transcriptional silencing of tumor suppressor genes in UVB-exposed skin and UVB-induced skin tumors of mice. Carcinogenesis 32, 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34. Anway M. D., Cupp A. S., Uzumcu M., Skinner M. K. (2005) Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308, 1466–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vaiserman A. M. (2011) Hormesis and epigenetics: is there a link? Ageing Res. Rev. 10, 413–421 [DOI] [PubMed] [Google Scholar]