Abstract
Herpes B virus DNA was specifically amplified by PCR, targeting the regions that did not cross-react with herpes simplex virus (HSV). The amplified products, which were shown to be highly genetic polymorphisms among herpes B virus isolates, were identified by microplate hybridization with probes generated by PCR. The products immobilized in microplate wells were hybridized with the biotin-labeled probes derived from the SMHV strain of herpes B virus. The amplified products derived from the SMHV and E2490 strains of herpes B virus were identified by microplate hybridization. PCR products amplified from the trigeminal ganglia of seropositive cynomolgus macaques were identified as herpes B virus DNA. The utility of the PCR-microplate hybridization assay for genetic detection and identification of the polymorphic region of herpes B virus was determined.
Herpes B virus infection, caused by herpes B virus harbored in its natural host, Asian macaques, is a fatal encephalomyelitis in humans (6, 23, 26, 37). The primary means of transmission of herpes B virus to humans has been suggested to occur by direct and/or indirect contact with virus-contaminated secretion from the natural host (38). Forty-three cases of herpes B virus infection have been reported since the first reports by Sabin (7, 26). The infection has a high mortality if not treated by antiviral therapy in the early stages of infection. Thus, development of an assay is essential for prevention and early diagnosis of herpes B virus infection.
Herpes B virus is an alphaherpesvirus that shares some characteristics with the herpes simplex virus (HSV) (9). Both of the neurotropic viruses establish latency in the sensory nerve ganglia of their natural hosts (2, 34). Stress induces reactivation of the viruses from the latent state, resulting in shedding of infectious viruses from mucosal tissue (14, 35). In addition, herpes B virus shows strong serological cross-reactivity with HSV (5, 18). The genetic arrangement is almost identical between herpes B virus and HSV (8, 21, 22, 25), and the nucleotide sequences of the herpes B virus genes have been reported to show high homology with the corresponding HSV genes (1, 21, 22, 25). These similarities suggest that a sample from a patient with herpes B virus infection also contains HSV and that misidentification occurs in a diagnosis of the infection. In addition, HSV type 1 (HSV-1) was reported to be isolated and detected from a pet monkey and from white-faced monkeys with fatal infection, suggesting that a macaque, a natural host of herpes B virus, is infected with HSV (13, 30). Thus, accurate diagnosis of herpes B virus infection in both human and the natural herpes B virus host requires a specific assay to distinguish herpes B virus from the closely related HSV.
PCR was considered suitable for specific detection of herpes B virus. PCR was designed to amplify the herpes B virus target sequence, which is the most divergent in the genome sequence among HSV-1, HSV-2, and herpes B virus (4, 25). Furthermore, the amplified region was shown to have a highly genetic polymorphism among herpes B virus isolates derived from different natural host species (33). Thus, an applicable gene identification method is required to have the ability to identify the amplified products containing numerous point mutations. To overcome this difficulty, we developed a method of genetic identification by microplate hybridization with the probes generated by PCR.
MATERIALS AND METHODS
Virus.
The inactivated herpes B virus E2490 strain isolated from a rhesus macaque was provided by Ryozaburo Mukai (National Institute of Infectious Diseases of Japan). The SMHV strain of herpes B virus was isolated from a cynomolgus macaque (1). Three isolates of HSV-1 (strains K8, K200, and 198) and three isolates of HSV-2 (strains 79-29, 27, and 111) were obtained from the vesicular fluid of patients with HSV reactivation. Four human cytomegalovirus (HCMV) isolates (strains Towne, AD169, KH, and OK-1) and two simian CMV (SCMV) isolates (strains 68-1 and 1090K) were used in the present study (36). HSV-1 and HSV-2 isolates were propagated in Vero cells, and HCMV and SCMV isolates were propagated in human embryonic lung cells.
Preparation of templates.
A cloned 2.6-kb SalI-EcoRI fragment containing the US4, US5, and US6 genes from the SMHV strain (SMHV/pBV-DNA) was provided by Akio Yamada (National Institute of Infectious Diseases of Japan). Viral DNA from the E2490 strain was extracted and purified as previously described (11). Viral DNA from HSV-1, HSV-2, HCMV, and SCMV was extracted and purified from the infected cells according to a previously described procedure (11). Trigeminal ganglia from four healthy seropositive cynomolgus macaques were used as test samples. Both left and right ganglia were removed from the monkeys after rapid euthanasia following the intravenous injection of anesthesia. DNA was extracted and purified as described by Sambrook et al. (27). Purified DNA was stored at 4°C.
PCR.
Design of the primers for herpes B virus DNA amplification referred to as nucleotide sequences US4 through US6 as determined by Bennett et al. (1) and Smith et al. (33). The GenBank accession numbers for the sequences cited in the present study are AF082804 to AF082814, AF083210, and S48101. A partial 330-bp gene encoding HSV DNA polymerase was amplified with the primer pair KM-1 (5′-CAGTACGGCCCCGAGTTCGTGACCGGG-3′) and KM-2 (5′-GGCGTAGTAGGGCGGGGATGTCGCG-3′) as described by Kimura et al. (17). The 610-bp fragments encoding part of HCMV and SCMV VP25 were amplified with the sense primer CM-3 (5′-ACTCACAACATATTCGTTTGC-3′) and the antisense primer CM-2 (5′-TGTTCGGAAGTGATCGTGTTT-3′) as described by Meigata et al. (19). PCR was performed in 100 μl of 1× Ex Taq Buffer (Takara Shuzo, Shiga, Japan), 0.2 mM (each) deoxynucleoside triphosphates, 0.5 μM concentrations of each of the primers, 0.5 U of Ex Taq polymerase (Takara Shuzo), and the template. The reaction was carried out for 30 cycles of denaturation at 94°C for 2 min, annealing at 55°C for 3 min, and extension at 72°C for 4 min. The amplified products were separated by electrophoresis on 1.5% agarose gels, stained with ethidium bromide, and visualized under a UV transilluminator.
Microplate hybridization.
Microplate hybridization was performed as previously described (12, 15) with some modifications. Briefly, the heat-denatured PCR products were diluted by using 10-fold serial dilutions in coating buffer (1.5 M NaCl, 0.2 mM sodium phosphate [pH 7.4], 5 mM EDTA). The serially diluted products were allowed to adsorb to the microplate wells by incubation at 37°C for 3 h. Biotin-labeled probes were generated by PCR as described above, except for the use of Biotin 11-dUTP (Enzo Diagnostics, Inc.) in place of dTTP. Hybridization was performed at 42 and 56°C for 20 h. Hybridization signals were detected by binding streptavidin-conjugated β-galactosidase to biotinylated probes, followed by reaction with the 4-methylumbelliferyl-β-d-galactoside substrate. The amount of the fluorescent reactant was determined by measurement of the absorbance at 460 nm with a fluorometric microplate reader (Fluoroskan II; Labsystems, Tokyo, Japan) and calculated as fluorescence units (FU).
RESULTS
Design of primers of herpes B virus.
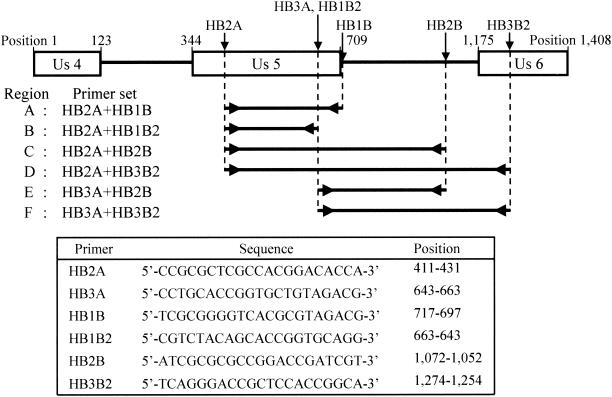
Two sense primers (HB2A and HB3A) and four antisense primers (HB1B, HB1B2, HB2B, and HB3B2) were designed in the present study (Fig. 1). HB2B is located on a noncoding intergenic region between the US5 and US6 genes, whereas HB3B2 is located on part of the open reading frame of the US6 gene, and the remaining primers are located on part of the open reading frame of the US5 gene. Six regions, A to F (Fig. 1), were generated by gene amplification combining a sense primer and an antisense primer.
FIG. 1.
Target regions of PCR and primers used in the present study. Arrows on the genetic map indicate the locations of each primer. Open boxes indicate open reading frames of the 3′ side of the US4 gene, the complete US5 gene, and the 5′ side of the US6 gene. Nucleotide positions in this figure are cited as the E2490 strain sequence numbers deposited in the GenBank database (accession no. AF083210).
Specificity of PCR.
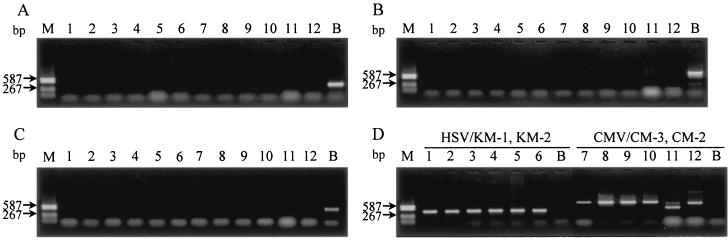
PCR was performed with five different virus templates (SMHV/pBV-DNA, HSV-1, HSV-2, HCMV, and SCMV). Amplification of the A, C, and E regions successfully generated products from SMHV/pBV-DNA but not from HSV-1, HSV-2, HCMV, and SCMV isolates (Fig. 2). Amplified products of the A, C, and E regions were also detected from the E2490 strain of herpes B virus (Fig. 3). In contrast, amplification of the B and D regions yielded products from SMHV/pBV-DNA and the HSV-1 and HSV-2 isolates, which showed almost the same size bands on agarose gels (data not shown). A gene amplification targeting the F region also resulted in a product from the HSV-2 27 strain, although the fragment showed a different size band than that from the herpes B virus strain.
FIG. 2.
Specific detection of herpes B virus by gene amplification of the A, C, and E regions (A, B, and C, respectively). (D) Amplification of HSV and CMV with each specific primer set (primer pairs KM1-KM2 and CM3-CM2, respectively). HSV-1 isolates are in lanes 1 to 3 (strains K8, K200, and 198, respectively); HSV-2 isolates are in lanes 4 to 6, (strains 79-29, 27, and 111, respectively); HCMV isolates are in lanes 7 to 10 (strains Towne, AD169, KH, and OK-1, respectively); and SCMV isolates are in lanes 11 and 12 (strains 68-1 and 1090K, respectively). Lanes B and M show SMHV/pBV-DNA and DNA standard size markers (DNA-Molecular Weight Marker V; Roche Diagnostics), respectively.
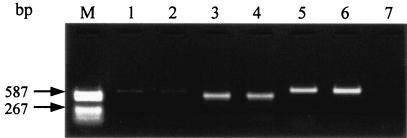
FIG. 3.
Agarose gel electrophoresis of the amplified products derived from the herpes B virus strains. The gene amplification was performed to yield the C-region products with the primer pair HB2A-HB2B. The amplified products derived from the seropositive monkey specimens are in lanes 1 to 4, the E2490 strain is in lane 5, and SMHV/pBV-DNA is in lane 6. The PCR reactant of the negative control is in lane 7. A difference in band size can be seen between the PCR products generated from the E2490 strain or SMHV/pBV-DNA and that from the seropositive monkeys (specimens 3 and 4 for lanes 3 and 4, respectively).
Sensitivity of PCR.
Amplification of the A, C, and E regions was performed with DNA templates of serial 10-fold dilutions of SMHV/pBV-DNA from 10 ng to 10 fg. The bands of amplified products on agarose gels were confirmed under a UV transilluminator. PCR of the C region could generate the products from 10 fg of the plasmid DNA, whereas the detectable limits of PCR of the A and E regions were 1 pg and 100 fg of the templates, respectively (data not shown).
Microplate hybridization identification of the amplified herpes B virus product.
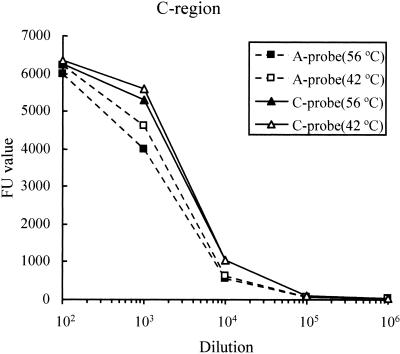
The C region of the PCR product derived from SMHV/pBV-DNA was used as both DNA samples and probes in this experiment. The products were diluted, and the resultant 10−2 to 10−6 dilutions were analyzed by microplate hybridization. Each diluted product was hybridized with the A- and C-region probes labeled with biotin and the resulting signal intensity was calculated as FU. The FU values were plotted against each dilution of the products. The resulting dilution curve was used to evaluate the identification of the amplified product. The hybridization curves, as shown in Fig. 4, were observed in conditions at both 42 and 56°C.
FIG. 4.
Dilution curves from serial dilutions of the amplified product of the C regions. FU values were obtained by hybridization with probes from the A (squares) and C (triangles) regions at 56°C (solid symbols) and 42°C (open symbols). Dilution curves obtained by using the probes from the A and C regions are shown by dotted and solid lines, respectively.
Genetic identification by microplate hybridization between heterogeneous amplified products.
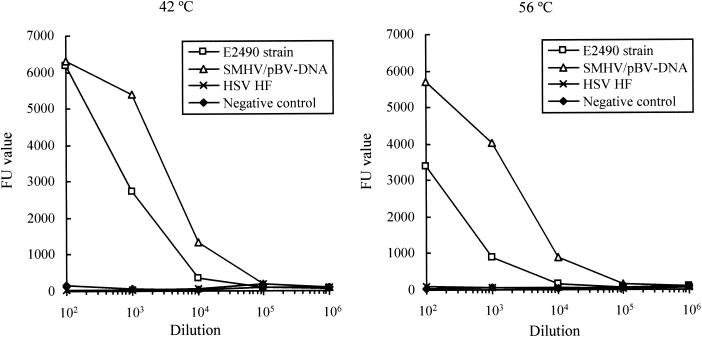
Heterogeneous hybridization was investigated by using the product amplified of the C region from the E2490 strain isolated from a rhesus macaque and the PCR product of the DNA polymerase gene derived from the HSV-1 HF strain. The products were hybridized with the C-region probe generated from SMHV/pBV-DNA. The nucleotide sequences of the C regions of the SMHV and the E2490 strains (GenBank accession no. S48101 and AF083210, respectively) share an 80.4% homology, while the product from HSV DNA polymerase shows no homology with the C region of the SMHV strain. Dilution curves obtained from hybridization of the E2490 and HSV HF strain are shown in Fig. 5. Specific hybridization signals were observed in dilution curves obtained from hybridization of the E2490 strain at both 42 and 56°C. In contrast, the FU values of the amplified products encoding the HSV HF strain DNA polymerase showed a curve almost identical to that of the negative control in hybridization at both 42 and 56°C.
FIG. 5.
Dilution curves of herpes B virus amplified products (E2490 strain and SMHV/pBV-DNA) and the HSV-1 HF strain obtained by PCR- microplate hybridization. Microplate hybridization with probes from the C regions was performed at 56°C (right) and 42°C (left). FU values are shown for E2490 (□), SMHV/pBV-DNA (▵), HSV HF (×), and negative control (⧫).
Application to seropositive monkey specimens.
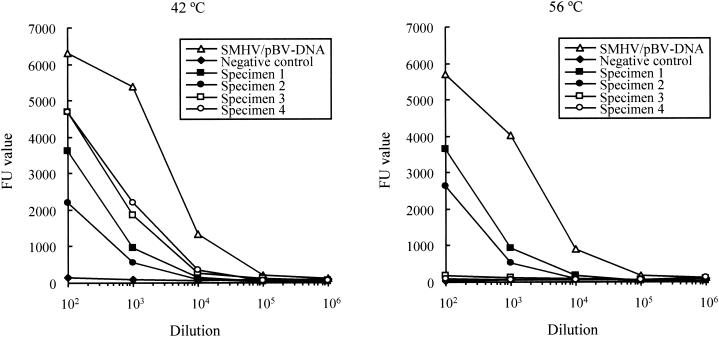
PCR-microplate hybridization was applied to the trigeminal ganglia of four seropositive cynomolgus monkeys. Amplified products of the C region were detected in all seropositive macaque specimens. The products generated from two seropositive specimens showed a smaller band size than the corresponding C regions of the SMHV and E2490 strains, whereas DNA bands from the other specimens showed the same size as those from the known herpes B virus strains (Fig. 3). Serial dilutions of the product were hybridized with the probe of the C region at 42 and 56°C. The specific hybridization signals were obtained from the serial dilutions of the PCR products from the two seropositive specimens, which were the same size as the SMHV and E2490 products, by hybridization at both 42 and 56°C (Fig. 6, specimens 1 and 2). In the experiment with products that had sizes different from those of the known herpes B virus strains, the FU values obtained from hybridization at 56°C were almost identical to those of the negative control over the entire range of dilutions, whereas the specific signals were obtained from hybridization at 42°C (specimens 3 and 4).
FIG. 6.
Dilution curves of herpes B virus amplified products of seropositive monkey specimens (specimens 1 to 4) and SMHV/pBV-DNA obtained by PCR-microplate hybridization. Microplate hybridization with probes from the C regions was performed at 56°C (right) and 42°C (left). FU values are shown for seropositive specimens (▪, •, □, and ○ for specimens 1 to 4, respectively), SMHV/pBV-DNA (▵), and a negative control (⧫).
DISCUSSION
Virus isolation in cell lines has been the standard method for diagnosis of a herpes B virus infection, even though contact with herpes B virus-contaminated specimens is extremely hazardous and requires a level 3 or higher biosafety containment facility (3). In order to avoid the danger of working with virus-contaminated specimens, sensitive and less-hazardous PCR methods of detection have been developed (31, 32). A specific herpes B virus detection method is also necessary in order to distinguish herpes B virus from the closely related HSV. In the present study, we designed PCR to amplify the regions of the US5 gene encoding glycoprotein J, which was shown to be the most divergent of the protein-coding genes in HSV-1, HSV-2, and herpes B viruses (4, 25) and the 3′-flanking noncoding intergenic region.
Herpesviruses have been reported to show intraspecies gene diversity among clinical isolates (20, 28, 29, 36). Thus, to investigate the specificity of the gene amplification, PCR was performed on templates from the HSV-1, HSV-2, HCMV, and SCMV isolates, which were confirmed to show a distinct genetic pattern in each herpesvirus. In gene amplification of the A, C, and E regions, no PCR products were detected from the isolates of the four species of herpesviruses. All amplified products, however, were detected from the herpes B virus E2490 and SMHV strains. PCR with the templates of serial 10-fold dilutions indicated that the gene amplification of the C region had the highest sensitivity in the products of the specific A, C, and E regions. The specificity and sensitivity of PCR suggest that amplification of the C region with the primer pair HB2A and HB2B is most suitable for the genetic detection of herpes B virus.
Microplate hybridization was performed with the A- or C-region probes generated by PCR, as reported previously on the development and application for diagnosis of varicella-zoster virus infection (10, 12, 15). Real-time PCR, however, a rapid genetic detection and identification method with oligonucleotide probes, has been developed for the diagnosis of herpes B virus infection (14, 24). The amplified region targeted in the present study showed a high degree of polymorphism among herpes B virus isolates (33), suggesting the presence of numerous point mutations. Thus, oligonucleotide probes were not likely to be available for the identification of variable amplified products because a point mutation has a great effect on the hybridization ability of an oligonucleotide probe, thus causing problems when we have limited gene information. In the present study, the full-length probe generated from the SMHV strain hybridized with PCR products derived not only from the SMHV strain but also from E2490 and seropositive specimen-derived strains, suggesting that the probe is useful for genetic identification of the herpes B virus.
The PCR product generated from the E2490 strain was observed to hybridize with the probe of the SMHV strain under high- and low-stringency conditions. On the other hand, the amplified products from two of four seropositive specimens were hybridized with the SMHV strain probe only under the low-stringency conditions, a finding consistent with the previous results of microplate hybridization between HSV-1 and HSV-2 (16). These results suggest that the genetic distance between the PCR products of the SMHV strain and seropositive specimen-derived strains is greater than the distance between those of the E2490 and SMHV strains. The results of gel electrophoresis, in which the DNA bands of the seropositive specimens showed a size distinct from those of the SMHV or E2490 strains, support this suggestion. Thus, the results indicate the possibility of failure to identify the herpes B virus product by hybridization under the high-stringency conditions if the nucleotide sequence identity between the PCR product and the probe is <80%, the identity level of the C region between the SMHV and E2490 strain. In conclusion, hybridization under low-stringency conditions may be indispensable for correct identification of the PCR product.
In summary, we developed a PCR-microplate hybridization assay for the detection and identification of PCR products derived from herpes B virus; this assay is able to detect herpes B virus but not HSV. The assay will be helpful in diagnosing humans suspected to have been exposed to herpes B virus, even if they are infected with HSV. In addition, the assay is a powerful tool for detecting and identifying unknown or new herpes B virus genotypes in both natural and human hosts. The relationship between the FU values and the serial dilutions of the PCR products suggests that the PCR-microplate hybridization assay technique may be useful in quantifying the herpes B virus genome.
Acknowledgments
We thank to Ryozaburo Mukai and Akio Yamada for providing viral strains and recombinant plasmids, respectively.
This study was supported by a grant-in-aid for the Emerging and Re-emerging Disease project from the Ministry of Health, Labor, and Welfare of Japan.
REFERENCES
- 1.Bennett, A. M., L. Harrington, and D. C. Kelly. 1992. Nucleotide sequence analysis of genes encoding glycoproteins D and J in simian herpes B virus. J. Gen. Virol. 73:2963-2967. [DOI] [PubMed] [Google Scholar]
- 2.Boulter, E. A. 1975. The isolation of monkey B virus (Herpesvirus simiae) from the trigeminal ganglia of a healthy seropositive rhesus monkey. J. Biol. Stand. 3:279-280. [DOI] [PubMed] [Google Scholar]
- 3.Cohen, J. I., D. S. Davenport, J. A. Stewart, S. Deitchman, J. K. Hilliard, L. E. Chapman, et al. 2002. Recommendations for prevention of and therapy for exposure to B virus (Cercopithecine Herpesvirus 1). Clin. Infect. Dis. 35:1191-1203. [DOI] [PubMed] [Google Scholar]
- 4.Doran, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eberle, R., D. Black, and J. K. Hilliard. 1989. Relatedness of glycoproteins expressed on the surface of simian herpes-virus virions and infected cells to specific HSV glycoproteins. Arch. Virol. 109:233-252. [DOI] [PubMed] [Google Scholar]
- 6.Eberle, R., and J. Hilliard. 1995. The simian herpesviruses. Infect. Agents Dis. 4:55-70. [PubMed]
- 7.Engel, G. A., L. Jones-Engel, M. A. Schillaci, K. G. Suaryana, A. Putra, A. Fuetes, and R. Henkel. 2002. Human exposure to herpesvirus B-seropositive macaques, Bali, Indonesia. Emerg. Infect. Dis. 8:789-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrington, L., Wall, L. V. M., and D. C. Kelly. 1992. Molecular cloning and physical mapping of the genome of simian herpes B virus and comparison of genome organization with that of herpes simplex virus type 1. J. Gen. Virol. 73:1217-1226. [DOI] [PubMed] [Google Scholar]
- 9.Hilliard, J. K., D. Black, and R. Eberle. 1987. Simian alphaherpesviruses and their relation to the human herpes simplex viruses. Arch. Virol. 109:83-102. [DOI] [PubMed] [Google Scholar]
- 10.Hiroshige, K., M. Ikeda, and R. Hondo. 2002. Detection of varicella-zoster virus DNA in tear fluid and saliva of patients with Ramsay Hunt syndrome. Otol. Neurotol. 23:602-607. [DOI] [PubMed]
- 11.Hondo, R., Y. Yogo, T. Kurata, and Y. Aoyama. 1987. Genome variation among varicella-zoster virus isolates derived from different individuals and from the same individuals. Arch. Virol. 93:1-12. [DOI] [PubMed] [Google Scholar]
- 12.Hondo, R., S. Ito, and S. Inoue. 1995. Titration of varicella-zoster virus DNA in throat swabs from varicella patients by combined use of PCR and microplate hybridization. Jpn. J. Med. Sci. Biol. 48:249-255. [DOI] [PubMed] [Google Scholar]
- 13.Huemer, H. P., C. Larcher, T. Czedik-Eysenberg, N. Nowotny, and M. Reifinger. 2002. Fatal infection of a pet monkey with human herpesvirus 1. Emerg. Infect. Dis. 8:639-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huff, J. L., R. Eberle, J. Capitanio, S. S. Zhou, and P. A. Barry. 2003. Differential detection of B virus and rhesus cytomegalovirus in rhesus macaques. J. Gen. Virol. 84:83-92. [DOI] [PubMed] [Google Scholar]
- 15.Inoue, S., and R. Hondo. 1990. Microplate hybridization of amplified viral DNA segment. J. Clin. Microbiol. 28:1469-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue, S., and R. Hondo. 1993. Type differentiation of herpes simplex virus by stringent hybridization of polymerase chain reaction products. Arch. Virol. 129:311-316. [DOI] [PubMed] [Google Scholar]
- 17.Kimura, H., M. Futamura, H. Kito, T. Ando, M. Goto, K. Kuzushima, M. Shibata, and T. Morishima. 1991. Detection of viral DNA in neonatal herpes simplex virus infections: frequent and prolonged presence in serum and cerebrospinal fluid. J. Infect. Dis. 164:289-293. [DOI] [PubMed] [Google Scholar]
- 18.Lees, D. N., A. Baskeville, L. M. Cropper, and D. W. Brown. 1991. Herpesvirus simiae (B virus) antibody response and virus shedding in experimental primary infection of cynomolgus monkeys. Lab. Anim. Sci. 41:360-364. [PubMed] [Google Scholar]
- 19.Meigata, K., R. Hondo, A. Fujiyama, M. Shinkai-Shibata, S. Itoh, Y. K. Kikuchi, Ando, N. Ichikawa, Y. Nomura, K. Watanabe, Y. H. Degawa, Beck, S. Tomikawa, T. Nagao, and H. Uchida. 1996. Titration of human cytomegalovirus (HCMV) DNA in urine by combined use of PCR and microplate hybridization in a renal transplant patient with HCMV pneumonitis. Jpn. J. Med. Sci. Biol. 49:121-127. [DOI] [PubMed] [Google Scholar]
- 20.Meyer-Konig, U., M. Haberland, von Laer, D., O. Haller, and F. T. Hufert. 1998. Intragenic variability of human cytomegalovirus glycoprotein B in clinical strains. J. Infect. Dis. 177:1162-1169. [DOI] [PubMed] [Google Scholar]
- 21.Ohsawa, K., D. H. Black, H. Sato, and R. Eberle. 2002. Sequence and genetic arrangement of the US region of the monkey B virus (Cercopithecine Herpesvirus 1) genome and primate herpesviruses. J. Virol. 76:1516-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohsawa, K., D. H. Black, H. Sato, K. Rogers, and R. Eberle. 2003. Sequence and genetic arrangement of the UL region of the monkey B virus (Cercopithecine Herpesvirus 1) genome and comparison with the UL region of other primate herpesviruses. Arch. Virol. 148:989-997. [DOI] [PubMed] [Google Scholar]
- 23.Palmer, A. E. 1987. B virus, Herpesvirus simiae: historical perspective. J. Med. Primatol. 16:99-130. [PubMed] [Google Scholar]
- 24.Perelygina, L., I. Patrusheva, N. Manes, M. J. Wildes, P. Krug, and J. K. Hilliard. 2003. Quantitative real-time PCR for detection of monkey B virus (Cercopithecine Herpesvirus 1) in clinical samples. J. Virol. Methods 109:245-251. [DOI] [PubMed] [Google Scholar]
- 25.Perelygina, L., L. Zhu, H. Zurkuhlen, R. Mills, M. Borodovsky, and J. K. Hilliard. 2003. Complete sequence and comparative analysis of the genome of herpes B virus (Cercopithecine Herpesvirus 1) from a rhesus monkey. J. Virol. 77:6167-6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabin, A. B., and W. M. Wright. 1934. Acute ascending myelitis following a monkey bite, with isolation of a virus capable of reproducing the disease. J. Exp. Med. 59:115-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Sakaoka, H., T. Aomori, O. Honda, Y. Saheki, S. Ishida, S. Yamanishi, and K. Fujinaga. 1985. Subtypes of herpes simplex virus type1 in Japan: classification by restriction endonucleases and analysis of distribution. J. Infect. Dis. 152:190-197. [DOI] [PubMed] [Google Scholar]
- 29.Sakaoka, H., T. Kawana, L. Grillner, T. Aomori, T. Yamaguchi, H. Saito, and K. Fuginaga. 1987. Genome variations in herpes simplex virus type2 strains isolated in Japan and Sweden. J. Gen. Virol. 68:2105-2116. [DOI] [PubMed] [Google Scholar]
- 30.Schrenzel, M. D., G. K. Osborn, A. Shima, R. B. Klieforth, and G. A. Maallouf. 2003. Naturally occurring fatal herpes simplex virus 1 infection in a family of white-faced saki monkeys (Pithecia pithecia pithecia). J. Med. Primatol. 32:7-14. [DOI] [PubMed] [Google Scholar]
- 31.Scinicariello, F., R. Eberle, and J. K. Hilliard. 1993. Rapid detection of B virus (Herpesvirus simiae) DNA by polymerase chain reaction. J. Infect. Dis. 168:747-750. [DOI] [PubMed] [Google Scholar]
- 32.Slomka, M. J., D. W. Brown, J. P. Clewley, A. M. Bennett, L. Harrington, and D. C. Kelley. 1993. Polymerase chain reaction for detection of Herpesvirus simiae (B virus) in clinical specimens. Arch. Virol. 131:89-99. [DOI] [PubMed] [Google Scholar]
- 33.Smith, A. L., D. H. Black, and R. Eberle. 1998. Molecular evidence for distinct genotypes of monkey B virus (Herpesvirus simiae) which are related to the macaque host species. J. Virol. 72:9224-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vizoso, A. D. 1975. Recovery of Herpes simiae (B virus) from both primary and latent infections in rhesus monkeys. Br. J. Exp. Pathol. 56:485-488. [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner, E. K., and D. C. Bloom. 1997. Experimental investigation of herpes simplex virus latency. Clin. Microbiol. Rev. 10:419-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe, S., M. Shinkai, S. Hitomi, H. Kozuka, S. Kimura, K. Shimada, R. Hondo, and N. Yamaguchi. 1994. A polymorphic region of the human cytomegalovirus genome encoding putative glycoproteins. Arch. Virol. 137:11-121. [DOI] [PubMed] [Google Scholar]
- 37.Weigler, B. J. 1992. Biology of B virus in macaque and human hosts: a review. Clin. Infect. Dis. 14:555-567. [DOI] [PubMed] [Google Scholar]
- 38.Weigler, B. J., D. W. Hird, J. K. Hilliard, N. W. Lerche, J. A. Robert, and L. M. Scott. 1993. Epidemiology of Cercopithecine Herpesvirus 1 (B virus) infection and shedding in a large breeding cohort of rhesus macaques. J. Infect. Dis. 167:257-263. [DOI] [PubMed] [Google Scholar]