Abstract
We studied the prevalence of the omp50 gene and the Omp50 protein in Campylobacter strains. Immunodetection assays and DNA-DNA hybridizations showed that most C. coli strains tested were negative and most C. jejuni and C. lari strains tested were positive. A PCR assay was developed, using the omp50 gene as a species-specific target. We propose a combination of a hippurate test and an omp50 assay to perform identification of Campylobacter species.
Among campylobacters, the C. jejuni and C. coli species are the most frequently associated with human enteritis, but C. lari is now recognized as a significant human pathogen (10). These three species, closely related by phylogenetic and genetic criteria (17), are difficult to differentiate (13). C. lari was originally differentiated from the other species by nalidixic acid susceptibility, but some nalidixic acid-sensitive C. lari strains have been described, and resistance to nalidixic acid and fluoroquinolones has emerged among other Campylobacter species. Although the hippurate hydrolysis test (11) discriminates C. jejuni from the other species, C. jejuni hippurate-negative strains and false-positive strains exist (6, 11, 13). Thus, there is a need for new molecular techniques for differentiating Campylobacter species in clinical samples. Multiplex PCRs have been developed to identify C. jejuni and C. coli (6), but a recent study showed that identification of C. jejuni remains unclear (13).
Porins are pore-forming proteins allowing exchanges of hydrophilic compounds across the outer membranes of gram-negative bacteria. They are involved in adaptation of many bacteria to their environment (5, 8, 14). Porin genes can be used to identify different species, and their expression can be associated with different ecological niches (4, 7). To date, two porins have been characterized for C. jejuni: the major outer membrane protein (2) and Omp50 (3). Previously, we described the purification and the characterization of Omp50 channel activity (3). We observed that Omp50 was not ubiquitous in Campylobacter strains, but a fragment was obtained by PCR using omp50 primers. Herein, the omp50 locus was characterized in more detail.
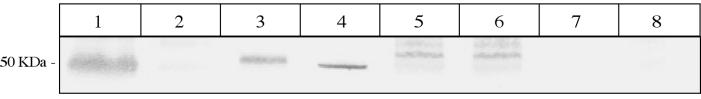
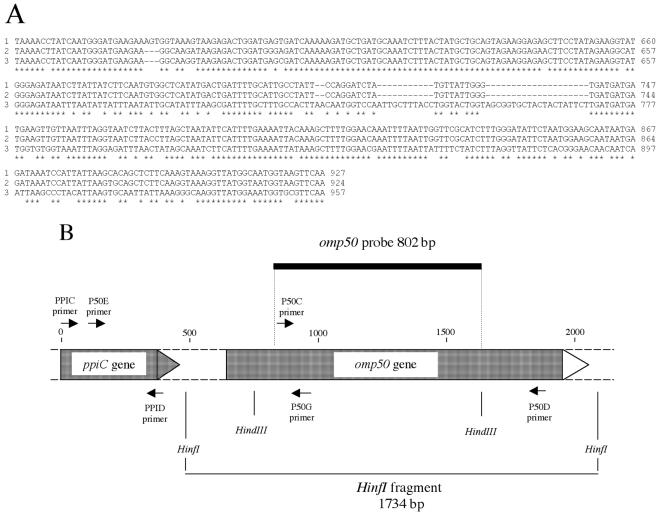
We analyzed a set of 104 strains, including 95 Campylobacter strains and 9 other strains (Table 1). Using the ApiCampy identification system (Biomérieux, Marcy L'Etoile, France), we obtained 39 C. jejuni strains, 51 C. coli strains, and 5 C. lari strains (Table 1). All strains were analyzed by using immunodetection assays using Omp50 polyclonal antibodies; a representative experiment is presented in Fig. 1. Results are detailed in Table 2. We used the C. jejuni NCTC 11168 strain as a positive standard and the C. coli 96C1 and 96C12 strains as negative standards because these strains have already been characterized (3). As expected, a 50-kDa signal was observed for strain NCTC 11168, and no signal was observed for strains 96C1 and 96C12 (Fig. 1). These results showed that all C. lari strains had the Omp50 protein, all but two strains of C. jejuni had the Omp50 protein, and all but five C. coli strains had no Omp50 protein. The PCR fragments previously obtained by omp50 PCR by using primers P50C and P50D (3) were sequenced. The sequences of seven strains were analyzed. For the amplification fragments obtained from C. jejuni and C. lari, we found P50C and P50D primer sequences at each extremity and aligned the sequence with the omp50 gene of strain NCTC 11168 (15) (Sanger Center Web Site [www.sanger.ac.uk/Projects/C_jejuni]). We obtained 100% identities for C. jejuni NCTC 11168 strains, 96% identities for C. jejuni strain 79AH, and 93% identities for C. lari strain 96C15 (Fig. 2A). The omp50 genes of three other strains, including C. lari 96C22, were sequenced, and we observed that the two C. lari omp50 genes were closely related and that they were different from the C. jejuni omp50 genes because of two insertions (Fig. 2A). On the contrary, the PCR amplification fragment obtained from the C. coli strain 96C1 P50D primer sequence was not found, but the P50C primer sequence was found twice. Using BLAST (1), we found that the sequence was not homologous to the omp50 gene but showed 92% similarities with a nonrelevant open reading frame, the NCTC 11168 infB gene. We performed PCR amplification on 96C1 DNA with only the P50C primer and determined the sequence of the amplification fragment. The same sequence was obtained, confirming that it was a nonspecific amplification fragment.
TABLE 1.
Strains used in this study
| Genus and species | Strain name(s) | Origin (reference) |
|---|---|---|
| Campylobacter coli | 79BD, 79K, 85AN, 85AP, 85AN, 85N, 85W, 87AC, 87C, 87AGC | Humans |
| P26 | Human (12) | |
| 96C1, 96C12, 96C13, 96C25, 96C47, 96C151, 96C208, BKR20, BKR314, BKR431, BKR546, BKR548, BKR550, BKR571, BKR604, AA846, AA849, AA879, AA884, AA963 | Poultry | |
| AA1538, AA1541, AA1546, AA1549, AA1552, AA1575, AA1578, AA1635, AA1642, AA1649, VA1221, VA5511, VA5611, VA5812, VA6221, VA6321, VA6811, VA7541 | Swine | |
| CIP70.80T, CIP7054 | Pasteur Institute | |
| Campylobacter jejuni | 85H, 79AH, 85R, 79BM, 85AA, 85R, 85AF, 85AG, 85S, 85Z, 85X, 85D, 85AC, 85AJ, 87AF | Humans |
| 81176, NCTC11168 | Humans (9, 15) | |
| 96C6, 96C37, 96C101, 96C399, 96C545, 96C571, 96C579, 96C668, 96C672, BKR409, BKR540, BKR543, BKR589, AA728, AA800, AA922, AA940, AA943, AA1020, AA1621 | Poultry | |
| CIP103726, ATCC35560 | Pasteur Institute | |
| Campylobacter lari | 87AGL, 85AB, 86BJ | Humans |
| 96C15, 96C22 | Poultry | |
| Escherichia coli | C600, DH5α | Our laboratory collection |
| Salmonella enterica | CIP81.3, CIP105355 | |
| Enterobacter aerogenes | EA13048, EA15038 | |
| Pseudomonas aeruginosa | PA8839 | |
| Klebsiella pneumoniae | KP89, K80 |
FIG. 1.
Species specificity of the Omp50 protein, determined by immunodetection assay using Omp50 polyclonal antibodies. Protein expression was analyzed on whole-cell lysates by gel electrophoresis, using a standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis protocol and a standard Western blotting protocol (5). For each gel, Coomassie blue staining was performed in order to assess the quality of samples and migration. For immunodetection assays, previously described Omp50 polyclonal rabbit antibodies were used (3). C. jejuni strain NCTC 11168 (lane 1) was used as a positive standard, and C. coli strains 96C1 (lane 2) and 96C12 (lane 7) were used as negative standards. Analyses of C. jejuni, C. coli, and C. lari strains are presented. The lanes represent the following strains: lane 1, C. jejuni NCTC 11168; lane 2, C. coli 96C1; lane 3, C. jejuni 85H; lane 4, C. jejuni 79AH; lane 5, C. lari 96C15; lane 6, C. lari 96C22; lane 7, C. coli 96C12; lane 8, C. coli CIP7054.
TABLE 2.
Prevalence of Omp50 protein studied by immunodection assays, PCR amplification, and Southern blot anlayses
| Strain group | No. of strains with indicated result
|
||||||
|---|---|---|---|---|---|---|---|
| Total | Immunodetection
|
PCR
|
Southern blotting
|
||||
| Positive | Negative | Positive | Negative | Positive | Negative | ||
| C. jejuni | 39 | 37 | 2 | 37 | 2 | 37 | 2 |
| C. coli | 51 | 5 | 46 | 5 | 46 | 5 | 46 |
| C. lari | 5 | 5 | 0 | 5 | 0 | 5 | 0 |
| Other strains | 9 | 0 | 9 | 0 | 9 | NDa | ND |
ND, not determined.
FIG. 2.
Genetic study of the omp50 locus. (A) Alignment of omp50 gene sequences of C. jejuni NCTC 11168 (lane 1), C. jejuni 79AH (lane 2), and C. lari 96C15 (lane 3). Only the more different parts, nucleotides 600 to 927 on the C. jejuni NCTC 11168 strains, are represented. (B) Primers and probe positions for PCR experiments and Southern blot analyses, respectively. Forward primer, →; reverse primer, ←; omp50 DNA probe,  .
.
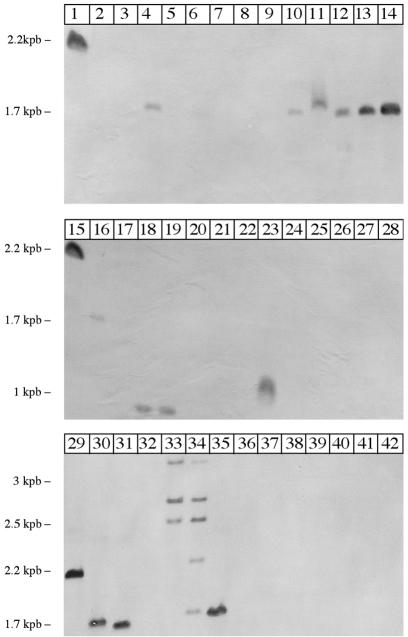
We hypothesized that the omp50 gene was absent from C. coli strains except the five which were positive in the immunodetection assay. We performed DNA-DNA hybridization analyses. P50C-P50D PCR amplification was performed on strain NCTC 11168, and a 1,083-bp fragment was obtained. The PCR product was restricted using HindIII, giving an 802-bp fragment, which was purified from agarose gel using the Wizard PCR Preps DNA purification system (Promega). The purification product was cloned using the pGEM-T vector sys-tem (Promega), and cloning was verified by sequencing. This plasmid was prepared using the Wizard Plus SV Miniprep system (Promega), restricted using BstZI to isolate the insert that was purified from agarose gel, and labeled by digoxigenin dUTP by using DIG High Prime DNA labeling (Roche Diagnostics, Indianapolis, Ind.). Southern blots were performed according to standard protocol, and hybridization, washes, and immunological detection were performed according to the manufacturer's instructions, with some modifications. Briefly, the genomic DNAs were restricted using HinfI, and the fragments were resolved on agarose gel and stained using ethidium bromide. The gel was washed three times in water, for 10 min in 0.25 N HCl, for 15 min in water, and for 20 min in 0.4 M NaOH. Blotting was performed overnight by capillary transfer. Hybridization was performed at 40°C for 30 min. Detection was performed with an enzyme immunoassay, using antidigoxigenin antibodies coupled with alkaline phosphatase, and an color detection system using Nitro Blue Tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (BCIP). As expected, C. jejuni strain NCTC 11168 showed a signal, while no signal was observed for C. coli strains 96C1 and 96C12 strains (Fig. 3). Results obtained for all strains are detailed in Table 2. We observed a signal on DNA-DNA hybridization for all the Campylobacter strains that were positive in the immunodetection assay. A great majority of the C. jejuni and C. lari strains showed only one signal, of 1.7 kbp, on Southern blots. On the contrary, all the five C. coli positive strains showed signals with different sizes or several fragments (Fig. 3), suggesting a polymorphism for Campylobacter strains at HinfI sites and omp50 homologous sequences in this species.
FIG. 3.
Species specificity of the omp50 gene by Southern blot hybridization. C. jejuni strain NCTC 11168 (lanes 1, 15, and 29) was used as a positive standard, and C. coli strains 96C1 (lane 2) and 96C12 (lane 3) were used as negative standards. Analyses of C. jejuni, C. coli, and C. lari strains are presented. The lanes represent the following strains: lane 1, C. jejuni NCTC 11168; lane 2, C. coli 96C1; lane 3, C. coli 96C12; lane 4, C. jejuni 85H; lane 5, C. coli 96C13; lane 6, C. coli 96C25; lane 7, C. coli 96C47; lane 8, C. coli 96C151; lane 9, C. coli 96C208; lane 10, C. lari 96C15; lane 11, C. lari 96C22; lane 12, C. jejuni 96C6; lane 13, C. jejuni 96C37; lane 14, C. jejuni 96C101; lane 15, C. jejuni NCTC 11168; lane 16, C. coli AA800; lane 17, C. coli 96C12; lane 18, C. coli AA884; lane 19, C. coli AA884; lane 20, C. coliBKR20; lane 21, C. coli BKR431; 22, lane C. coli BKR314; lane 23, C. coli BKR550; lane 24, C. coli BKR548; lane 25, C. coli BKR571; lane 26, C. coli BKR604; lane 27, C. coli AA849; lane 28, C. coli AA963; lane 29, C. jejuni NCTC 11168; lane 30, C. jejuni 85H; lane 31, C. jejuni 79AH; lane 32, C. coli 87AGL; lane 33, C. coli 87C; lane 34, C. coli 87AC; lane 35, C. lari 86BJ; lane 36, C. coli 85W; lane 37, C. coli 85N; lane 38, C. coli 85AP; lane 39, C. coli 85AN; lane 40, C. coli 79K; lane 41, C. coli 79BD; lane 42, C. coli P26.
Southern blot analyses demonstrated the absence of the omp50 gene in C. coli strains, which may indicate a lack of the omp50 gene only or a larger genetic rearrangement. Using Artemis software (16), on the C. jejuni strain NCTC 11168 genome sequence, we found that the gene upstream of omp50 was the ppiC gene. PCR experiments were performed on the ppiC gene, using the specific primers PPIC and PPID (Fig. 2A), and gave positive results for all the Campylobacter strains (data not shown). To study the omp50 locus, PCR experiments using the forward primer P50E, located in the ppiC gene, and the reverse primer P50G, located in the omp50 gene (Fig. 2A), were performed on all omp50-positive strains, including the five C. coli strains, and gave amplification fragments for all of these strains (data not shown). This result suggested that if present, ppiC and omp50 genes are always contiguous.
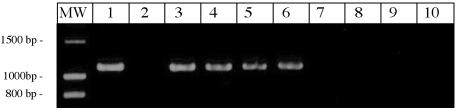
Based on these results, we tested different PCR conditions on three strains: the C. jejuni strain NCTC 11168 and the C. coli strains 96C1 and 96C12 as positive and negative standards, respectively. Primers P50C and P50D were used, and temperature and MgCl2 concentration gradients were tested (data not shown). Specific PCR conditions for omp50 amplification were obtained with a MgCl2 concentration of 1 mM, and hybridization steps were performed for 30 s at 58°C. An 1,100-bp amplification fragment was observed for strain NCTC 11168, and no signal was observed for strains 96C1 and 96C12 (Fig. 4). Using these conditions, we analyzed our set of Campylobacter strains. Results are detailed in Table 2, and a representative experiment is presented in Fig. 4. Amplification results were in agreement with Southern hybridization results, and all strains positive for hybridization showed a specific signal in PCR amplification. More than 90% of the C. coli strains were omp50 negative, 95% of the C. jejuni strains were omp50 positive, and 100% of the C. lari strains were omp50 positive.
FIG. 4.
Species specificity of the omp50 gene, determined by PCR amplification. C. jejuni strain NCTC 11168 (lane 1) was used as a positive standard, and C. coli strains 96C1 (lane 3) and 96C12 (lane 7) were used as negative standards. Analyses of C. jejuni, C. coli, and C. lari strains are presented. The lanes represent the following strains: lane 1, C. jejuni NCTC 11168; lane 2, C. coli 96C1; lane 3, C. jejuni 85H; lane 4, C. jejuni 96C37; lane 5, C. lari 96C15; lane 6, C. lari 96C22; lane 7, C. coli 96C12; lane 8, C. coli 96C13; lane 9, C. coli 96C25; lane 10, PCR negative standard, using H2O as a template.
The hippurate hydrolysis test discriminates C. jejuni from C. coli and C. lari (11), and we showed that the omp50 PCR assay differentiates C. coli from C. jejuni and C. lari. Thus, we combined results of the two tests (Table 3). We obtained 35 hippurate-positive and omp50-positive strains, all of which were C. jejuni strains. We obtained 46 hippurate-negative and omp50-negative strains, all of which were C. coli strains. We obtained two hippurate-positive and omp50-negative strains; these two strains were C. jejuni omp50 negative. We obtained 12 hippurate-negative and omp50-positive strains; five were C. coli omp50 positive, and two were C. jejuni hippurate negative. The five other hippurate-negative and omp50-positive strains were the five C. lari strains tested in this study.
TABLE 3.
Hippurate hydrolysis test and omp50 PCR assay
| Assay results | No. of strains with resulta
|
||
|---|---|---|---|
| C. jejuni | C. coli | C. lari | |
| Hippurate positive and omp50 positive | 35 | ||
| Hippurate positive and omp50 negative | 2 | ||
| Hippurate negative and omp50 positive | 2 | 5 | 5 |
| Hippurate negative and omp50 negative | 46 | ||
One hundred percent of hippurate positive, omp50 positive strains were C. jejuni; 36% of hippurate negative, omp50 positive strains were C. lari; and 100% of hippurate negative, omp50 negative strains were C. coli.
C. jejuni comprises two subspecies, C. jejuni subsp. jejuni and C. jejuni subsp. doylei. A recent study (13) with five C. jejuni subsp. doylei strains showed that identification of this taxon remains problematic. In the present study, the four C. jejuni subsp. doylei strains tested were correctly typed as C. jejuni strains. Hence, hippurate hydrolysis combined with the omp50 PCR assay may help in C. jejuni identification. All five C. lari strains tested in this study were hippurate negative and omp50 positive. This result suggest that hippurate hydrolysis combined with the omp50 PCR assay may also help in C. lari identification. Since all tested strains were positive for momp and ppiC (3; this study), a multiplex PCR based on these genes and on omp50 and hipO detection could be an efficient tool for rapid Campylobacter spp. identification.
Nucleotide sequence accession numbers.
GenBank accession numbers for the complete omp50 gene sequences of C. jejuni 79AH and C. lari 96C15 strains are AJ582064 and AJ582067, respectively. (Data not shown: GenBank accession numbers for the complete omp50 gene sequences of C. jejuni strains 85H and F38011 and of C. lari strain 96C22 are AJ582066, AJ582065, and AJ582068 respectively.)
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bolla, J. M., E. Loret, M. Ladzunski, and J.-M. Pagès. 1995. Conformational analysis of the Campylobacter jejuni porin. J. Bacteriol. 177:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolla, J. M., E. Dé, A. Dorez, and J.-M. Pagès. 2000. Purification, characterization and sequence analysis of Omp50, a new porin isolated from Campylobacter jejuni. Biochem. J. 352:637-643. [PMC free article] [PubMed] [Google Scholar]
- 4.Cloeckaert, A., J. M. Verger, M. Grayon, J. Y. Paquet, B. Garin-Bastuji, G. Foster, and J. Godfroid. 2001. Classification of Brucella spp. isolated from marine mammals by DNA polymorphism at the omp2 locus. Microbes Infect. 3:729-738. [DOI] [PubMed] [Google Scholar]
- 5.Dedieu, L., J.-M. Pagès, and J. M. Bolla. 2002. Environmental regulation of Campylobacter jejuni MOMP-porin expression in E. coli monitored using green fluorescent protein. Appl. Environ. Microbiol. 68:4209-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denis, M., C. Soumet, K. Rivoal, G. Ermel, D. Blivet, G. Salvat, and P. Colin. 1999. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett. Appl. Microbiol. 29:406-410. [DOI] [PubMed] [Google Scholar]
- 7.Derrick, J. P., R. Urwin, J. Suker, I. M. Feavers, and M. C. Maiden. 1999. Structural and evolutionary inference from molecular variation in Neisseria porins. Infect. Immun. 67:2406-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferenci, T. 1999. Regulation by nutrient limitation. Curr. Opin. Microbiol. 2:208-213. [DOI] [PubMed] [Google Scholar]
- 9.Korlath, J. A., M. T. Osterholm, A. Judy, J. C. Forgang, and R. A. Robinson. 1985. A point-source outbreak of Campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 10.Lastovica, A. J., and M. B. Blaser. 2000. Clinical significance of Campylobacter and related species other than Campylobacter jejuni and C. coli, p. 89-120. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 11.Morris, G. K., M. R. Sherbeeny, C. M. Patton, H. Kodaka, G. L. Lombard, P. Edmonds, D. G. Hollis, and D. J. Brenner. 1985. Comparison of four hippurate hydrolysis methods for identification of thermophilic Campylobacter spp. J. Clin. Microbiol. 22:714-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moutinho-Fragoso, Guilhermina. 2001. Pesquisa e caracterizacao de toxinas em estirpes enteropatogenicas de campylobacter. Dissertation. Faculdade de farmacia da universidade de lisboa, Lisbon, Portugal.
- 13.On, S. L., and P. J. Jordan. 2003. Evaluation of 11 PCR assays for species-level identification of Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 41:330-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozawa, Y., and S. Mizushima. 1983. Regulation of outer membrane porin protein synthesis in Escherichia coli K-12: ompF regulates the expression of ompC. J. Bacteriol. 154:669-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkhill, J., B. Wren, K. Mungall, J. Ketley, C. Churcher, D. Basham, T. Chillinggworth, T. Feltwell, S. Holroyd, K. Jagels, A. Karlyshev, S. Moule, M. Pallen, C. Penn, M. Quail, M. Rajandream, K. Rutherford, A. van Vliet, S. Whitehead, and B. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 16.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualisation and annotation. Bioinformatics 10:944-945. [DOI] [PubMed] [Google Scholar]
- 17.Vandamme, P. 2000. Taxonomy of the family Campylobacteriaceae. p. 3-26. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.