Abstract
Arp2/3 is a protein complex that nucleates actin filament assembly in the lamellipodium in adherent cells crawling on planar 2-dimensional (2D) substrates. However, in physiopathological situations, cell migration typically occurs within a 3-dimensional (3D) environment, and little is known about the role of Arp2/3 and associated proteins in 3D cell migration. Using time resolved live-cell imaging and HT1080, a fibrosarcoma cell line commonly used to study cell migration, we find that the Arp2/3 complex and associated proteins N-WASP, WAVE1, cortactin, and Cdc42 regulate 3D cell migration. We report that this regulation is caused by formation of multigeneration dendritic protrusions, which mediate traction forces on the surrounding matrix and effective cell migration. The primary protrusions emanating directly from the cell body and prolonging the nucleus forms independent of Arp2/3 and dependent on focal adhesion proteins FAK, talin, and p130Cas. The Arp2/3 complex, N-WASP, WAVE1, cortactin, and Cdc42 regulate the secondary protrusions branching off from the primary protrusions. In 3D matrices, fibrosarcoma cells as well as migrating breast, pancreatic, and prostate cancer cells do not display lamellipodial structures. This study characterizes the unique topology of protrusions made by cells in a 3D matrix and show that these dendritic protrusions play a critical role in 3D cell motility and matrix deformation. The relative contribution of these proteins to 3D migration is significantly different from their role in 2D migration.—Giri, A., Bajpai, S., Trenton, N., Jayatilaka, H., Longmore, G. D., Wirtz, D. The Arp2/3 complex mediates multigeneration dendritic protrusions for efficient 3-dimensional cancer cell migration.
Keywords: 3D environment, collagen I matrix, matrix deformation
The arp2/3 complex is the major actin-nucleating factor that induces the formation of the intracellular dendritic filament network that shapes the lamellipodial protrusive leading edge of motile cells on conventional 2-dimensional (2D) substrates (1). The Arp2/3 complex is known to have very little biochemical activity when present on its own, but its activity is greatly increased in presence of nucleation promoting factors like WASP, WAVE, and cortactin and Rho GTPases Cdc42 and Rac1 (2). Cdc42 binds to the GTPase binding domain of the WASP family protein N-WASP (3, 4). This relieves N-WASP from its autoinhibited confirmation and activates the Arp2/3 complex (5). Rac activates the Arp2/3 complex by signaling through another WASP-related protein WAVE (1).
Although the role of the Arp2/3 complex in chemotaxis is somewhat controversial, two recent reports have shown that the Arp2/3 complex mediates the formation of lamellipodium and random-walk cell motility on flat substrates (6, 7). However, during development and in the context of disease including cancer and inflammation, cells typically migrate in a 3-dimensional (3D) microenvironment (8, 9). Whether and how the Arp2/3 complex regulates protrusion activity and still plays a role in cell motility in the more physiological case of a 3D matrix has not been determined.
In 3D matrices, mesenchymal cells often display dendritic protrusions (10–13); however, detailed structural and functional characterization of these protrusions is still lacking. Moreover, to our knowledge, no known specific regulators of protrusions of cells in matrix have been identified. Here we classify cell protrusions based on their time-dependent spatial location in the cell. Our results show that while the protrusions emerging directly from the cell body and prolonging the nucleus (which we call mother protrusions) are specifically regulated by focal adhesion proteins FAK, talin, and p130Cas, the formation of dendritic protrusions (daughter protrusions) that stem from mother protrusions are regulated by the Arp2/3 complex and associated proteins N-WASP, WAVE1, cortactin, Cdc42, and VASP. The rate of generation (not the length) of daughter protrusions, and associated degree of branching from the mother protrusions, predicts cell speed in 3D matrices. These multigeneration dendritic protrusions are structurally and functionally distinct from well-characterized invadopodia that stem from the basal surface of cancer cells placed on the surface of soft gels and promote local invasion but do not seem to mediate cell migration.
MATERIALS AND METHODS
Cell culture
Human fibrosarcoma HT1080 cells [American Type Culture Collection (ATCC), Manasssas, VA, USA] were cultured in DMEM (Mediatech, Manasssas, VA, USA) supplemented with 10% (v/v) FBS (Hyclone Laboratories, South Logan, UT, USA), and 0.005% (w/v) gentamicin (Quality Biological, Gaithersburg, MD, USA). Human breast carcinoma MDA-MB-231 cells (ATCC) were cultured in DMEM supplemented with 10% FBS. Human prostate cancer E006AA cells (a generous gift from Prof. John Isaacs, Johns Hopkins University School of Medicine) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% FBS and 100 U penicillin/100 μg streptomycin per milliliter of medium. Human pancreatic adenocarcinoma SW1990 cells (ATCC) were cultured in RPMI 1640 medium supplemented with 10% FBS and 100 U penicillin/100 μg streptomycin per milliliter of media. HT1080 cells transfected with shRNAs (see below) were grown in medium containing 1 μg/ml puromycin. The cells were maintained at 37°C and 5% CO2 in a humidified incubator during cell culture and during live-cell microscopy.
Depletion of proteins with shRNAs
The lentivirus vector was generated by cotransfecting the shRNA construct with two other packaging plasmids, pMD.G VSV-G and pCMVΔR8.91 (encoding Gag, Pol, Tat, and Rev) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Briefly 293T cells at around 80% confluence were transfected with a mixture of 6 μg of lentiviral shRNA construct, 8 μg of pCMVΔR8.91, and 1 μg of pMD.G VSV-G. The conditioned medium containing the lentivirus was harvested 48 h post-transfection and filtered through a 0.4-μm filter (Millipore, Billerica, MA, USA) to remove cell debris.
For transduction, HT1080 cells were grown to 50–60% confluence in a 6-cm cell culture dish. Medium (2 ml) containing lentivirus was mixed with 1 ml of fresh medium containing protamine sulfate (final concentration 10 μg/ml) and added to HT1080 cells. After 8 h incubation, the medium containing the viruses was replaced with fresh medium containing 1 μg/ml puromycin for selection. shRNA constructs targeting various genes were purchased from Sigma. Five different shRNA sequences targeting different regions were chosen. After lentiviral-mediated transduction, Western blots were performed, and only shRNAs showing more than 85% knockdown were used for subsequent studies. They include the following: Arp2/3 sh36499 GCTGGCATGTTGAAGCGAAATC, Arp2/3 sh36501 CTACCACATCAAGTGCTCTAAC; N-WASP sh123061 GCACAACTTAAAGACAGAGAAC, N-WASP sh123062 CAGGAAACAAAGCAGCTCTTTC; cortactin sh40273 CGGCAAATACGGTATCGACAAC; Cdc42 sh299931 CCTGATATCCTACACAACAAAC; Cdc42 sh299932 CAGATGTATTTCTAGTCTGTTC; WAVE1 sh122995 CGCCGTATTGCTGTTGAATATC; WAVE1 sh122998 GCTAAGCATGAACGCATTGAAC.
A scrambled shRNA sequence was used as a control, CCTAAGGTTAAGTCGCCCTCGC (Addgene plasmid 1864; Addgene, Cambridge, MA, USA). Western blots were performed as described previously. The blots were incubated overnight at 4°C with the following antibodies: rabbit anti-human p34 (1:1000 in 5% milk; Millipore), rabbit anti-human N-WASP (1:1000 in 5% milk; Cell Signaling Technology, Danvers, MA, USA), rabbit anti-human cortactin (1:1000 in 5% milk; Cell Signaling Technology), rabbit anti-human Cdc42 (1:1000 in 5% milk; Cell Signaling Technology), and goat anti-β-actin (1:2500 in 5% milk; Santa Cruz, Santa Cruz, CA, USA). Depletion of talin, p130Cas, Vasp, and FAK was conducted as described previously (15).
Immunofluorescence microscopy
To visualize the subcellular localization of Arp2/3 and associated proteins, cells were plated on collagen I-coated 35-mm glass-bottom cell culture dishes. The next day, cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 10 min, blocked with 10% goat serum for 1 h at room temperature, and stained for nuclear DNA, Arp2/3 (p34, 1 μg/ml; Millipore), WAVE1 (1 μg/ml; Cell Signaling Technology), N-WASP (1 μg/ml; Cell Signaling Technology), cortactin (1 μg/ml; Cell Signaling Technology), and Cdc42 (1 μg/ml; Cell Signaling Technology).
Fluorescent micrographs of cells on 2D substrates were collected using a Cascade 1K CCD camera (Roper Scientific, Trenton, NJ, USA) mounted on a Nikon TE2000 microscope with a ×60 oil-immersion lens (Nikon, Tokyo, Japan). For immunofluorescence in 3D, cells were embedded in 3D collagen as mentioned below (3D collagen I matrix). After 24 h, cells were fixed with 4% formaldehyde for 30 min and permeabilized with extraction buffer consisting of 0.1% Triton-X 100 (v/v) for 30 min. Cells were then incubated with primary antibody [same antibodies as mentioned above, anti-phospho-myosin heavy chain 2A (Ser1943; Millipore), anti-α-tubulin (Abcam, Cambridge, MA, USA), 5 μg/ml final concentration for all antibodies] overnight at 4°C and washed 5 times with PBS for 30 min each. Next, the cells were incubated with appropriate secondary antibodies, phalloidin, and DAPI for 2 h at room temperature, after which they were washed extensively with PBS (5× for 30 min each). Cells completely embedded inside collagen gels were then imaged ≥150 μm away from the bottom on a Nikon A1 confocal microscope using a ×60 water-immersion lens.
Lamellipodium quantification
Lamellipodia of cells growing in 2D substrates were quantified as described previously (16, 17). Briefly, cells were stained for F-actin, and fluorescent and phase-contrast images were taken randomly for ≥100 cells/condition. Cell boundaries were traced using NIS-Elements image analysis software (Nikon). Lamellipodia were identified by dense networks of F-actin fluorescence on the front edge of the cell's perimeter. The ratio of lamellipodia was calculated by dividing the length of the lamellipodia by the total circumference of the cell.
3D collagen I matrix
HT1080 cells were embedded in 2 mg/ml collagen I gel as described previously (15). Briefly, 18,000 cells suspended in 1:1 (v/v) ratio of cell culture medium and reconstitution buffer [0.2 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and 0.26 M NaHCO3 in distilled water] were mixed with appropriate volume of soluble rat-tail collagen I (BD Biosciences, San Jose, CA, USA) to obtain a final collagen I concentration of 2 mg/ml. A calculated amount of 1 M NaOH was added quickly, and the final solution was mixed well to bring the pH to ∼7. The cells suspension was added to a 24-well coverslip-bottom cell culture dish and immediately transferred to an incubator maintained at 37°C to allow polymerization. This density was chosen so as to minimize cell collisions. Fresh medium was added 5 h before imaging.
Protrusion activity and topology of matrix-embedded cells
Phase-contrast images of matrix-embedded cells were recorded 2 min apart for 16.5 h using a Cascade 1K CCD camera (Roper Scientific) mounted on a Nikon TE2000 microscope with a ×10 objective lens. For the characterization of protrusion topology, the movies were used to count the total number of mother protrusions, and the number of first-, second-, and third-generation protrusions generated by the cell (e.g., see Fig. 2A–D). The protrusions emanating directly from the cell body, even when split, were termed mother protrusions; protrusions originating from the mother protrusions were termed first-generation, and so on. Because many mother protrusions showed multigeneration protrusions, they were termed dendritic. The degrees of branching are defined as the ratios of the number of first-generation to the number of mother protrusions (see Fig. 2L, inset), the number of second-generation protrusions to the number of first-generation protrusions (see Fig. 2M, inset), and so on. The rate of formation of a protrusion (of any generation) is the number of protrusions that were born, grew, and died for a duration of 1 h. Mitotic cells were not included in the measurements.
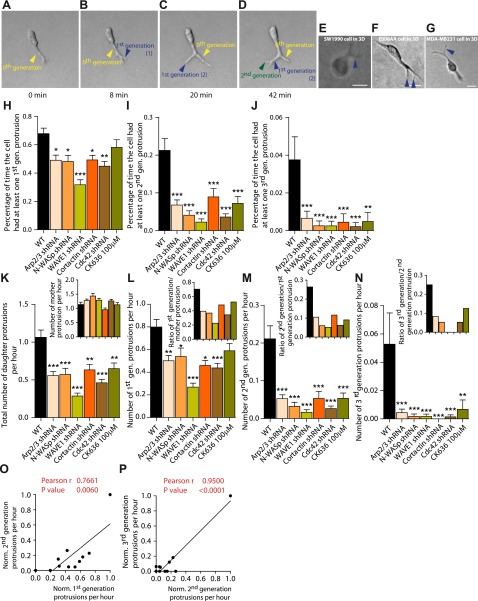
Figure 2.
Cells in matrix form multigeneration, dendritic pseudopodial protrusions. A–D) Dynamic formation of multigeneration, dendritic pseudopodial protrusions by a matrix-embedded cell. Arrows show the formation of a mother protrusion (yellow arrows; protrusions that stem directly from the cell body), as well as first- (blue arrows) and second-generation (green arrows) daughter protrusions, which stem from a mother protrusion. Times: 0 min (A), 8 min (B), 20 min (C), 42 min (D). E–G) Human pancreatic carcinomas (SW1910, E), human prostate carcinomas (E006AA, F), and human epithelial breast carcinomas (MDA-MB-231, G) also form daughter protrusions emanating from mother protrusions when embedded in a 3D collagen I matrix. Scale bars = 20 μm. H–J) Average fraction of time spent by matrix-embedded cells displaying ≥1 first-generation protrusion stemming from a mother protrusion (H), ≥1 second-generation protrusion (I), and ≥1 third-generation protrusion (J), and associated regulation by the Arp2/3 complex, N-WASP, WAVE1, cortactin, and Cdc42, as well as inhibition following cell treatment by 100 μM of the Arp2/3-complex-specific inhibitor CK636. K) Total number of daughter protrusions and mother protrusions (inset) generated per hour per cell (rates of formation). L–N) Number of first-generation protrusions (L), second-generation protrusions (M), and third-generation protrusions (N) generated per hour per cell. Insets: number of first-generation protrusions per mother protrusion (L), number of second-generation protrusions per first-generation protrusion (M), and number of third-generation protrusions per second-generation protrusion (N). O, P) Correlations between the rates of formation of first- and second-generation protrusions (O) and rates of formation of second- and third-generation protrusions (P). For all panels, cells were monitored for 16.5 h. For each condition, n = 3; ≥60 cells were probed. *P < 0.05, **P < 0.01, ***P < 0.001 vs. WT.
Mean-square displacement (MSD) of cells in 2 and 3 dimensions
HT1080 cells were embedded in collagen I matrix, and low-magnification movies were collected, as described above. Single cells were tracked using Metamorph imaging software (Molecular Devices Corp., Sunnyvale, CA, USA). A custom MATLAB program (MathWorks, Natick, MA, USA) calculated the MSD for each cell using the x and y coordinates obtained from tracking data using the following equation: MSD = ⟨[x(t + Δt) − x(t)]2 + [y(t + Δt) − y(t)]2⟩. We note that this is the 2D projection of essentially 3D cellular movements in the matrix; i.e., we presumed that the movements of the cells were isotropic. To test this assumption, we verified that ⟨[x(t + Δt) − x(t)]2⟩ ≈ ⟨[y(t + Δt) − y(t)]2⟩; i.e., we assume that if the MSDs of the cell along arbitrarily chosen x and y axis are equal, then the cell's MSDs in the direction orthogonal z to the (x, y) plane is also equal to those in the x and y directions. Mitotic cells were not included in the measurements.
Imaging and analysis of 3D matrix traction
Cells were embedded in collagen matrix and plated in a 4-chambered dish (MatTek, Ashland, MA, USA). Cells were incubated for 24 h in a humidified incubator maintained at 37°C, and then 500 μl of fresh medium was added to each well 5 h before imaging. Using a 488-nm laser for illumination and choosing a set of filters designed to collect reflected light, a reflection-confocal image of the cell and its immediate surrounding gel was captured at an interval of 2 min for 2 h, in a CO2- and temperature-controlled environment using a Nikon A1 confocal microscope with a ×60 water-immersion lens. To ensure that the 3D behavior of pulling by the cell is incorporated in the analysis, up to 40 z planes were imaged at every time point. Moreover, in order to mitigate the possibility of the glass bottom affecting the local properties of the cell, it was ensured that the imaged cell was ≥150 μm above the glass bottom.
Images at each time point were projected along the z direction using the maximum-intensity module on NIS-Elements image analysis software (Nikon). Subsequently, each sequence of images was exported as tiff files and read into a MATPIV-based code to determine the instantaneous, whole-field deformation map of the imaged gel. A signal-to-noise ratio of 4 was used to eliminate false vectors, and interrogation areas were picked to be 16 × 16 pixels, with a targeted overlap of 75% between these areas. Having obtained the instantaneous deformation of the gel between every 2 successive frames, imaginary circular regions around the cell were picked to represent beads of diameter 10 pixels. Up to 14 such “computational” beads were placed and their trajectories tracked using the information about whole-field instantaneous deformation of the gel. The mean peak deformation and the mean net deformation of the imaginary beads were calculated for cells moving in one direction over the course of one 2-h movie.
Correlation analysis
To access correlation between various motility parameters (MSDs, protrusion topology in 2 and 3 dimensions), 11 data points were generated (1 data point from each shRNA treatment, 1 data point from control cells, and 1 data point from Arp2/3 inhibitor treatment). Each data point represents the average of the particular parameter considered.
Statistics
The mean ± se values were calculated and plotted using GraphPad Prism software (GraphPad, San Diego, CA, USE). One-way ANOVA test was performed to determine statistical significance, which is indicated in the graphs using a Michelin grade scale. Values of P < 0.05 were considered significant.
RESULTS
Transformed cells form multiple-generation dendritic protrusions in 3D matrix
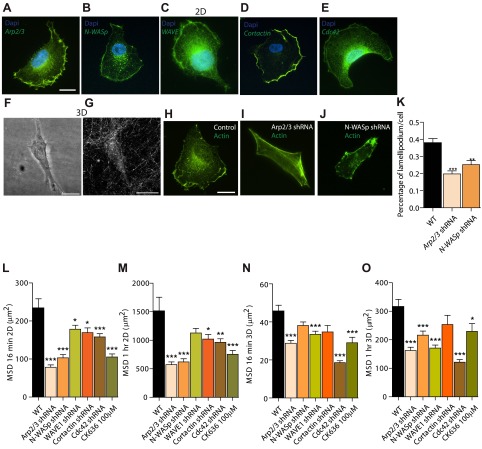
Wild-type HT0180 cells, a human fibrosarcoma cell line commonly used to study cell migration (17–20), were placed on flat collagen I-coated substrates. These cells formed a wide lamellipodium at their leading edge (Fig. 1A–E) and, as expected, the Arp2/3 complex and associated proteins, Arp2/3 activator N-WASP and WAVE1 (5, 21–24), N-WASP regulator cortactin (25–28), and N-WASP upstream effector Cdc42 (21, 22), localized in their lamellipodium (Fig. 1A–E). In contrast, and as observed previously by Yu and Machesky (29), immunofluorescence microscopy of these cells in 3D collagen I matrices showed Arp2/3, N-WASP, and cortactin localized to discrete puncta inside the protrusions and also in the cell body (Supplemental Fig. S2A–I, white arrowheads).
Figure 1.
Organization and role of Arp2/3 complex and associated molecules in cells on conventional 2D substrates. A–E) Arp2/3 complex (A), N-WASP (B), WAVE1 (C), cortactin (D), and Cdc42 (E) are localized primarily at the leading edge (lamellipodium) of motile cells placed on 2D collagen I-coated substrates. Human fibrosarcoma cells (HT1080) were stained with DAPI (nuclear DNA) and using antibodies against these proteins; images were obtained by immunofluorescence microscopy. F, G) Cells form no apparent wide lamellipodium when embedded in a 3D collagen I matrix; rather they form long pseudopodial protrusions that stem directly from the cell body and branch off into the matrix. Images of the HT1080 cell and its surrounding collagen I matrix were obtained by confocal phase contrast microscopy (F) and confocal reflection microscopy (G), respectively. H–K) Compared to control cells transfected with nontargeting shRNA (H), shRNA-induced depletion of the p34 subunit of the Arp2/3 complex (I), or N-WASP (J) induces the reduction of lamellipodium formation, as measured by the ratio of the length of lamellipodium marked by actin stain (phalloidin) and that of the cell periphery (method described in ref. 44); for each condition, n = 3 and a total of 100 cells were probed (K). **P < 0.01, ***P < 0.001 vs. wild type (WT). L–O) Regulation of 2D (L, M) and 3D (N, O) cell speed (measured as MSD at considered time lags; see Materials and Methods) by the Arp2/3 complex, N-WASP, WAVE1, cortactin, and Cdc42, as well as inhibition following cell treatment by 100 μM of the Arp2/3-complex-specific inhibitor CK636. MSDs were evaluated at time lags of 16 min (L, N), and 1 h (M, O).
Next, we systematically assessed whether these proteins regulated lamellipodium formation in cells on collagen-I-coated flat substrates. These experiments were conducted to determine whether cells that were shRNA depleted of these proteins displayed a motility phenotype on 2D substrates and, in turn, assess whether the role of Arp2/3 complex and binding partners in 3D motility could be simply extrapolated from the 2D case. All results presented in the study were verified using ≥2 different shRNA constructs (Supplemental Fig. S1A–E); results obtained with different shRNA constructs were pooled, as they were highly consistent. We found that shRNA-induced depletion of the major subunit p34 of the Arp2/3 complex (a positive control; refs. 6, 7), as well as N-WASP, significantly and consistently diminished the formation of the lamellipodium at the leading edge of cells moving on 2D substrates (Fig. 1H–K) and, as assessed by live-cell microscopy, reduced 2D cell speed (Fig. 1L, M). A simple assessment of cell speed consists of measuring MSDs of cells at different time scales, which here ranged between 2 min (the time between movie frames) and 16.5 h (the total duration of the movie). Short-time-scale cell speed (e.g., evaluated at 16 min) and long-time-scale cell speed (e.g., evaluated at 1 h) were both regulated by the Arp2/3 complex and associated proteins N-WASP, WAVE1, Cdc42, and cortactin (Fig. 1L, M). We note that the drastic reduction of lamellipodium formation by depletion of p34 and N-WASP (Fig. 1K), reduced, but did not completely prevent, cell migration on 2D substrates (Fig. 1L, M). For the cases examined here, correlation between 2D motility and lamellipodium formation was strong: cells showing little lamellipodium moved slowly, while cells showing extensive lamellipodium moved rapidly on 2D substrates (Fig. 1H–M).
Using the same cells as used in the above 2D studies, we determined whether the Arp2/3 complex and associated proteins still played a role in 3D cell migration for well-dispersed cells embedded in a matrix ≥400 μm away from the bottom substratum to avoid edge effects (30). The depletion of the Arp2/3 complex or associated proteins induced a robust phenotype of reduced 3D cell speed (Fig. 1N, O). Since shRNA-based depletion of p34 may not be complete, we examined the migration of matrix-embedded cells treated with specific Arp2/3 inhibitor CK636; these treated cells also showed greatly decreased cell speed, to an extent similar to that caused by shRNA depletion of p34, demonstrating a highly consistent phenotype of reduced 3D migration (Fig. 1N, O).
We asked whether regulation of cell speed by the Arp2/3 complex and associated proteins on 2D substrates was predictive of their regulatory role in 3D cell migration. On 2D collagen I-coated substrates, Arp2/3, N-WASP, WAVE1, cortactin, and Cdc42 regulated cell migration as in the 3D case; the presence of these proteins enhanced cell speed (Fig. 1L, M). However, close examination of the data showed that the extent of correlation between 2D and 3D motility parameters (MSDs) was limited (Pearson correlation coefficient ≪1; Supplemental Fig. S1F, G), suggesting a distinct mode of action for the Arp2/3 complex and associated proteins in 3D motility.
To determine the mechanism of Arp2/3-based regulation of 3D cell migration, protrusion morphology and dynamics of matrix-embedded cells were quantitatively assessed. However, unlike cells on collagen-coated surfaces, the same cells fully embedded inside a 3D collagen I matrix displayed no clear lamellipodia-like structure, as assessed by high-magnification microscopy (Fig. 1F and Supplemental Fig. S1J–L). Rather, cells showed a highly stretched body composed of an average of 1–2 major pseudopodial protrusions that stemmed from the cell body, prolonging the nucleus. The average thickness of these major protrusions was ∼5 μm, which is ∼10 times thicker than filopodial protrusions observed in cells on substrates (Fig. 1F, Supplemental Fig. S1J–L, and Table 1). Another important feature that sets apart pseudopodial protrusions from filopodia is the presence of microtubules inside these protrusions, whereas filopodia contain only filamentous actin (Supplemental Fig. S2M–O).
Table 1.
Morphological and functional differences between protrusions formed by cells in the 2D and 3D cases
| Type of protrusions | Width (μm) | Length (μm) | Number per cell at any time | Topology | F-actin organization | Arp2/3 | N-WASP | Regulatory role in cell motility |
|---|---|---|---|---|---|---|---|---|
| 2D case | ||||||||
| Lamellipodium | 20–50 | 10 | ∼1 | Thin and wide | Dendritic orthogonal meshwork + bundles | Yes | Yes | Yes |
| Filopodium | <1 | ∼1 | ∼10 | Cylindrical; no dendrites | Single core bundle | No | No | Yes |
| Invadopodia | 0.5–2 | >2 | 10–50 | Short and thin | Actin-associated proteins surround actin core | Yes | Yes | No |
| 3D case | ||||||||
| Mother protrusion | 2.5–10 | 10–60 | ∼1 | Cylindrical | Multiple cortical bundles | Yes | Yes | No |
| First-generation daughter protrusion | 2–5 | 10 | ∼0.5 | Cylindrical; dendrites | Multiple cortical bundles | Yes | Yes | Yes |
Differences in length and lateral dimensions, topology, F-actin organization, and regulatory roles played by lamellipodia-specific proteins Arp2/3 and N-WASP between pseudopodial protrusions formed by cells in 3D matrix and lamellipodial/filipodial/invapodial protrusions formed by the same cells on 2D substrates. Results compare WT HT1080 cells on collagen I-coated substrates and these cells inside a collagen I matrix.
Matrix-embedded cells developed 1 or 2 zeroth-generation mother protrusions, defined as the protrusions that extend directly from the nuclear area. Zeroth-generation protrusions branched into first-generation protrusions, which typically branched themselves further into second-generation protrusions (Fig. 2A–D). These second-generation protrusions rarely branched themselves further into third-generation daughter protrusions. Henceforth, zeroth-generation mother protrusions are referred to as mother protrusions, and all first-, second-, and third-generation protrusions are collectively referred to as daughter protrusions. We note that we made sure to use time-lapse movies as opposed to still pictures to identify and quantify protrusions. That way, we could follow the birth, growth, and retraction of all protrusions, helping to delineate mother from daughter protrusions. The fraction of time spent by wild-type (WT) cells in a matrix showing first- and second-generation protrusions was ∼70 and ∼22%, respectively (Fig. 2H, I); i.e., dendritic branching from mother protrusions was a common occurrence in HT1080 cells in 3D matrix.
Notably, dendritic protrusions were not unique to HT1080 cells and were also readily observed in human pancreatic carcinomas (SW1910; Fig. 2E), human prostate carcinomas (E006AA; Fig. 2F), and invasive human breast carcinomas (MDA-MB-231; Fig. 2G), when these cells were fully embedded inside a 3D matrix. These cells formed a wide lamellipodium when migrating on 2D substrates, but again showed no clear lamellipodial structures in 3D matrices (Fig. 1F and Supplemental Fig. S1J–L).
Arp2/3 complex and associated proteins mediate dendritic protrusion activity
Given the role of the Arp2/3 complex in nucleating dendritic actin assembly in vitro, we asked whether the Arp2/3 complex regulated the dendritic protrusive activity and topology of protrusions in cells in matrix. We found that the fraction of time during which p34-depleted cells displayed ≥1 first-generation, ≥1 second-generation, and ≥1 third-generation pseudopodial protrusion was reduced by 40, 75, and 87%, respectively, compared to control cells (Fig. 2H–J). The formation of daughter protrusions, but not mother protrusions, was drastically reduced (Fig. 2K, inset). The rate of formation of an nth-generation protrusion is defined here as the number of protrusions generated per hour. Moreover, the degrees of branching, i.e., the numbers of first-generation daughter protrusions per mother protrusion (Fig. 2L, inset), second-generation protrusions per first-generation protrusion (Fig. 2M, inset), and third-generation protrusions per second-generation protrusion (Fig. 2N, inset), were all significantly reduced.
This significant reduction of daughter branches and degree of branching from mother protrusions were confirmed by treating cells with the Arp2/3 complex inhibitor CK636. Cells treated with Arp2/3 inhibitor CK636 showed greatly decreased dendritic protrusive branching, to an extent similar to that caused by shRNA depletion of p34 (Fig. 2H–N). Notably, while the Arp2/3 complex mediated protrusive branching off from mother protrusions (Fig. 2K), it did not mediate the formation of mother protrusions (Fig. 2K, inset), which suggested distinct mechanisms for the formation of mother vs. daughter protrusions. These results show that a variety of human cancer cells in matrices, but not on flat substrates, feature highly dendritic protrusions with rates of formation of daughter protrusions and degree of branching tightly regulated by the Arp2/3 complex.
Biochemical data indicate that F-actin nucleating ability of the Arp2/3 complex is greatly enhanced by N-WASP, WAVE1, cortactin, and Cdc42 through direct or indirect binding interactions (24, 31–33). Here we found that the degree of protrusive branching was reduced in N-WASP-depleted cells to a similar extent as in p34-depleted cells (Fig. 2H–N). The fraction of time during which N-WASP-depleted cells displayed ≥1 first-generation, ≥1 second-generation, or ≥1 third-generation pseudopodial protrusion was reduced by 33, 85, and 98%, respectively, compared to control cells (Fig. 2H–J), similarly to the Arp2/3 complex. We observed a much larger effect in WAVE1-depleted cells, and the fraction of time during which the WAVE1-depleted cells displayed ≥1 first-generation, ≥1 second-generation, or ≥1 third-generation protrusion was reduced by 54, 89, and 94%, respectively, compared to control cells (Fig. 2H–J). The degree of protrusive branching from mother protrusions was decreased in Cdc42- and cortactin-depleted cells to a similar extent as in p34- and N-WASP-depleted cells (Fig. 2K–N). We also found that changes in the rates of formation of first-generation protrusions modulated by Arp2/3, N-WASP, WAVE1, cortactin, and Cdc42 correlated strongly with changes in the rates of formation of second-generation protrusions by the same molecules (Fig. 2O, P), which suggests that the same molecular mechanisms support the formation of first- and second-generation protrusions.
Finally, similarly to the Arp2/3 complex, N-WASP, WAVE1, cortactin, and Cdc42 did not play an important role in the formation of mother protrusions (Fig. 2K, inset), further suggesting distinct mechanisms for the formation of mother and daughter protrusions. These results suggest that perturbations of the Arp2/3 complex through its activators N-WASP and WAVE1, or through the N-WASP regulator cortactin and effector Cdc42, or through direct depletion, had the similar effect of specifically reducing protrusive dendritic branches.
Mother and daughter protrusions are differentially regulated
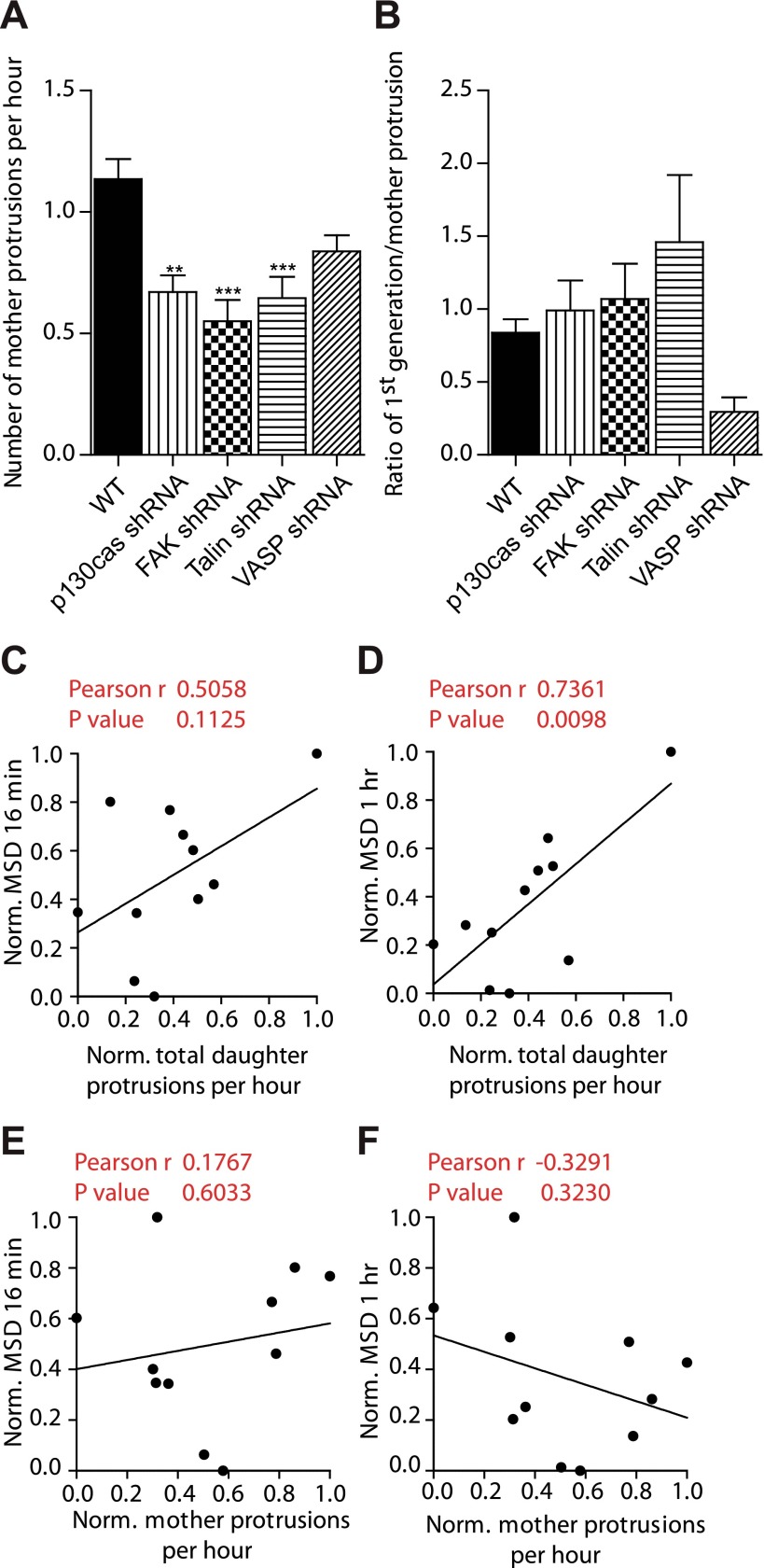
Since mother and daughter protrusions were differentially regulated, we asked whether one could identify molecules that specifically regulated mother protrusions, not daughter protrusions. A phenotypic screen revealed that scaffolding protein p130Cas and focal adhesion proteins talin and focal adhesion kinase regulated the formation of mother protrusions, but not the degree of branching from mother protrusions in matrix-embedded cells (Fig. 3A, B). Although not statistically significant, focal adhesion and lamellipodium protein VASP seemed to regulate both daughter and mother protrusions (Fig. 3A, B). Together, these results suggest that mother and daughter protrusions are molecularly distinct and differentially regulated. Moreover, through a highly consistent and robust phenotype, these results further demonstrate the specificity of the role of the Arp2/3 complex in mediating the formation of dendritic daughter protrusions in 3D matrix.
Figure 3.
Daughter protrusions, not mother protrusions, regulate 3D cell speed through the Arp2/3, N-WASP, cortactin, and Cdc42 module. A, B) Total number of mother protrusions produced per hour (A) and degree of branching from mother protrusions (number of first-generation protrusions per mother protrusion; B) in FAK-, talin-, p130Cas-, and VASP-depleted cells. For all panels, cells were monitored for 16.5 h. For each condition, n = 3; ≥60 cells were probed for protrusion analysis. **P < 0.01, ***P < 0.001 vs. WT. C–F) Assessment of correlation between 3D cell speeds evaluated at time lags of 16 min (C, E) and 1 h (D, F) and the rates of formation of daughter protrusions (C, D) and mother protrusions (E, F).
What is the functional importance of dendritic daughter protrusions? Pearson assessment of the extent of correlation between 3D cell speed and the rate of generation of protrusions revealed that while the number of mother protrusions was not predictive of cell speed, 3D cell speed strongly correlated with the number of daughter protrusive branches. These results suggest that the rate of formation of daughter protrusions (Fig. 3C, D), not the rate of formation of mother protrusions (Fig. 3E, F), regulates cell migration in 3D matrix at both short and long time scales. Since the rates of formation of first- and second-generation protrusions correlate (Fig. 2O, P), this correlation with cell speed held for the rate of formation of first-generation protrusions and the rate of formation of second-generation protrusions.
Next, we asked whether the length of mother protrusions and/or daughter protrusions correlated with cell speed. We found that the length of daughter protrusions was not correlated with cell speed (Supplemental Fig. S1I). Similarly, the length of mother protrusions did not correlate with cell speed (Supplemental Fig. S1H). Together, these results suggest that the number of dendritic protrusions predicts cell speed in 3D matrices, which was not a result predictable from the conventional 2D case, because cellular protrusions have fundamentally different topology in 3D matrices (Figs. 1 and 2 and Table 1).
Arp2/3 complex and associated proteins regulate matrix traction
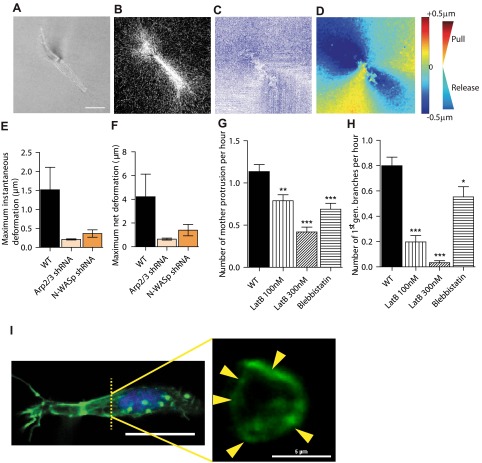
To further determine how the Arp2/3 complex modulated 3D cell migration, we asked whether changes in dendritic branching from mother protrusions by the Arp2/3 complex and associated proteins was accompanied by a differential ability of cells to apply traction forces on their surrounding matrix. Using time-resolved reflection confocal microscopy, the movements of collagen fibers in the vicinity of migrating cells were monitored (Fig. 4A–D). Customized PIV software allows us to track time-dependent local matrix deformation with high spatial resolution (Fig. 4C). Cells pulled on their surrounding matrix and then asymmetrically released the matrix for net movements (Fig. 4D). We found that the extent of deformation of the matrix was reduced on p34 depletion and N-WASP depletion (Fig. 4E, F).
Figure 4.
Arp2/3 and N-WASP mediate local matrix traction during 3D cell migration and actin architecture of cells. A–D) Particle (PIV) method used to map the time-dependent deformation field of the matrix generated by individual cells in a 3D matrix. Phase contrast (A) and reflection confocal micrograph (B) are simultaneously recorded every 2 min for 2 h to generate instantaneous strain fields of the matrix (C) and determine regions of matrix traction (red; matrix movement toward the cell) and matrix release from the cell (blue; movement away from the cell) (D). E, F) Average instantaneous deformation (E) and maximum deformation (F) of the matrix of fiduciary points located at ∼10 μm distance from the cell. G, H) Rate of formation of mother protrusions (G) and daughter protrusions (H) for control cells and cells treated with actin depolymerizing drug latrunculin B and myosin II inhibitor blebbistatin. *P < 0.05, **P < 0.01, ***P < 0.001 vs. WT. I) Architecture of the actin filament network in matrix-embedded cells, evaluated by confocal microscopy. Maximum confocal projection (left panel) shows the elongated morphology and side protrusions in cells in matrix. Cross sections of the same cell (right panel) reveal that the actin network in protrusions is constituted of cortical bundles. Arrows indicate distinct longitudinal actin filament bundles positioned at the periphery of cell protrusions.
Treatment of matrix-embedded cells with the F-actin depolymerizing drug latrunculin B and, to a lesser extent, with myosin II inhibitor blebbistatin reduced the formation of both mother and daughter protrusions (Fig. 4G, H). This suggests that actin filament assembly and acto-myosin contractility are required for the generation of mother and daughter protrusions. Immunofluorescence microscopy showed phospho-myosin IIa colocalized with actin fibers: regions with higher actin content featured higher content in phosphorylated myosin (Supplemental Fig. S2J–L, white arrowheads). Due to inherent technical difficulties, the actin filament structure of matrix-embedded cells could not be assessed by electron microscopy; however, high-resolution confocal microscopy of actin organizations revealed that actin filaments in both mother and daughter protrusions were concentrated at the cortex of the protrusions (Fig. 4I). To avoid any edge effects, we imaged cells ≥150 μm from the bottom of the dish. Confocal cross sections revealed the presence of thick filament bundles that followed the length of mother and daughter protrusions and were confined to and spatially dispersed along their cortex (Fig. 4I). The average width of these cortical bundles of F-actin was ∼1 μm. Actin distribution in these protrusions is different from that of a conventional filopodia in the 2D case, which typically contain a single core bundle of actin filaments (34). In the 3D case, protrusions contain multiple bundles of actin filaments, and they are located at the cell cortex (35).
DISCUSSION
The Arp2/3 complex and its activators play important roles in cell migration on 2D substrates (6, 7), but their roles in 3D migration have not been elucidated. The major finding of this paper is that the branched protrusions, in particular terminal daughter protrusions, displayed by cells in 3D matrices are specifically regulated by the Arp2/3 complex and its activators and are determinants of cell migration and matrix traction. Of course, this result does not mean that mother protrusions are not required for 3D motility, since they are required for the formation of the daughter protrusions. Rather, our data suggest that as soon a minimum number of mother protrusions is generated (seemingly ∼1.2/h; Fig. 2K, inset), then cell speed is set by the Arp2/3-, N-WASP-, WAVE1-, cortactin-, andCdc42-regulated degree of branching from these mother protrusions. Hence, the Arp2/3 complex regulates 3D cell motility by modulating the ability of cells in matrix to form dendritic protrusions.
It is important to note that although the depletion of Arp2/3, N-WASP, WAVE1, cortactin, and Cdc42 reduced cell speed in both 2 and 3 dimensions, the levels of effect were significantly different. This does not mean that these proteins regulate cell migration the same way: this is why we conducted a rigorous comparison that showed a poor global correlation between 2D and 3D migration speed, i.e., the relative roles of these proteins in 2D and 3D migration are significantly different when all these proteins are assessed together, as opposed to one at a time.
Another important observation is that cells in 3 dimensions showed no clear lamellipodium (Supplemental Fig. S1J–L), a prominent process for cell on 2D surfaces. While we observed no wide lamellipodium, it is still possible that the tips of side branches show small lamellipodium-like protrusions. However, such a structure would have to be smaller than a couple of micrometers. Moreover, terminal dendritic protrusions move along collagen fibers, which in the present conditions are only <120 nm in diameter (36, 37), which would not be able to support the formation of flat lamellipodial structures, unlike cells on 2D substrates.
We note that the multigeneration dendritic protrusions described in this work are structurally and functionally distinct from well-characterized finger-like structures formed by adjoining endothelial cells in 3D matrix and invadopodia formed at the basal surface of cells placed on the surface of soft gels (Table 1). This phenomenon, also known as cell sprouting, is well described in endothelial cells (38). The protrusions we describe here are part of the same cell. These dendritic protrusions are also distinct from invadopodia, which are actin and actin-related protein-rich membrane extensions that some invasive tumor cells grow from the ventral surface in contact with the extracellular matrix (39–43). Invadopodia have an average width of 0.5–2 μm and a length of ∼2 μm and, most notably, are not branched (43). Although Arp2/3 and N-WASP are associated with invadopodia (42), invadopodial structures differ from 3D pseudopodial protrusions in length (2 vs. ∼10–60 μm), topology (linear vs. dendritic), function (invasion vs. migration), location in the cell (basal vs. all around the cell surface), and protein content (F-actin and associated proteins vs. F-actin, associated proteins and microtubule) (Table 1).
Recent work suggests that cells in 3D matrices undergo two modes of migration: amoeboid (integrin- and MMP-independent gliding migration) and mesenchymal (integrin- and MMP-dependent migration) (17, 20, 44). However, for cancer cell migration in gels made of acid-extracted collagen I, as used here, MT1-MMP inhibition or silencing blocks invasive activity of HT1080 cells in 3D collagen I matrix, and we do not observe amoeboid migration (20, 45).
The presence of cortical actin filament bundles in both mother and daughter protrusions suggests that these actin bundles in the mother protrusions branch off into actin bundles in daughter protrusions, unlike core bundles in filopodia for cells on 2D substrates (Table 1 and Supplemental Fig. S1M). Therefore, the actin-based mechanism of formation of daughter protrusions from mother protrusions in cells in 3D matrix is distinct from the actin-based mechanism of formation of filopodia from lamellipodium in cells on 2D substrates, which stem from the mixed orthogonal network/bundles of the lamellipodium (34).
In sum, this study reveals that highly branched protrusions in cells in matrix, which are specifically regulated by the Arp2/3 complex and associated proteins, play a critical role in 3D cell motility.
Supplementary Material
Acknowledgments
This work was supported by U.S. National Institutes of Health (NIH) grants GM084204, CA143868, and CA85839.
D.W. acknowledges fruitful discussions with Dr. Sean X. Sun (Johns Hopkins Physical Sciences–Oncology Center, Johns Hopkins University). A.G. and D.W. designed the experiments; A.G., S.B.K., N.T., H.J., and D.W. conducted the experiments and analyzed the data; A.G. and D.W. wrote the article; A.G., D.W., and G.D.L. edited the article. The authors declare no conflicts of interest.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- 2D
- 2-dimensional
- 3D
- 3-dimensional
- MSD
- mean-square displacement
- WT
- wild type
REFERENCES
- 1. Pollard T. D., Borisy G. G. (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 2. Goley E. D., Welch M. D. (2006) The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726 [DOI] [PubMed] [Google Scholar]
- 3. Derry J. M., Ochs H. D., Francke U. (1994) Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell 78, 635–644 [DOI] [PubMed] [Google Scholar]
- 4. Miki H., Miura K., Takenawa T. (1996) N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 15, 5326–5335 [PMC free article] [PubMed] [Google Scholar]
- 5. Kim A. S., Kakalis L. T., Abdul-Manan N., Liu G. A., Rosen M. K. (2000) Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature 404, 151–158 [DOI] [PubMed] [Google Scholar]
- 6. Suraneni P., Rubinstein B., Unruh J. R., Durnin M., Hanein D., Li R. (2012) The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 197, 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu C., Asokan S. B., Berginski M. E., Haynes E. M., Sharpless N. E., Griffith J. D., Gomez S. M., Bear J. E. (2012) Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 148, 973–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wirtz D., Konstantopoulos K., Searson P. C. (2011) The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer 11, 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Konstantopoulos K., Wu P. H., Wirtz D. (2013) Dimensional control of cancer cell migration. Biophys. J. 104, 279–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grinnell F., Ho C. H., Tamariz E., Lee D. J., Skuta G. (2003) Dendritic fibroblasts in three-dimensional collagen matrices. Mol. Biol. Cell 14, 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rhee S., Jiang H., Ho C. H., Grinnell F. (2007) Microtubule function in fibroblast spreading is modulated according to the tension state of cell-matrix interactions. Proc. Natl. Acad. Sci. U. S. A. 104, 5425–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li A., Dawson J. C., Forero-Vargas M., Spence H. J., Yu X., Konig I., Anderson K., Machesky L. M. (2010) The actin-bundling protein fascin stabilizes actin in invadopodia and potentiates protrusive invasion. Curr. Biol. 20, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedl P., Wolf K. (2009) Proteolytic interstitial cell migration: a five-step process. Cancer Metast. Rev. 28, 129–135 [DOI] [PubMed] [Google Scholar]
- 14. Wolf K., Mazo I., Leung H., Engelke K., von Andrian U. H., Deryugina E. I., Strongin A. Y., Brocker E. B., Friedl P. (2003) Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 160, 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fraley S. I., Feng Y., Krishnamurthy R., Kim D. H., Celedon A., Longmore G. D., Wirtz D. (2010) A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 12, 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan A. Y., Coniglio S. J., Chuang Y. Y., Michaelson D., Knaus U. G., Philips M. R., Symons M. (2005) Roles of the Rac1 and Rac3 GTPases in human tumor cell invasion. Oncogene 24, 7821–7829 [DOI] [PubMed] [Google Scholar]
- 17. Yang C., Czech L., Gerboth S., Kojima S. I., Scita G., Svitkina T. (2007) Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PloS Biol. 5, 2624–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wolf K., Wu Y. I., Liu Y., Geiger J., Tam E., Overall C., Stack M. S., Friedl P. (2007) Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 9, 893–904 [DOI] [PubMed] [Google Scholar]
- 19. Zhou X., Rowe R. G., Hiraoka N., George J. P., Wirtz D., Mosher D. F., Virtanen I., Chernousov M. A., Weiss S. J. (2008) Fibronectin fibrillogenesis regulates three-dimensional neovessel formation. Genes Dev. 22, 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sabeh F., Shimizu-Hirota R., Weiss S. J. (2009) Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J. Cell Biol. 185, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rohatgi R., Ho H. Y., Kirschner M. W. (2000) Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J. Cell Biol. 150, 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M. W. (1999) The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97, 221–231 [DOI] [PubMed] [Google Scholar]
- 23. Machesky L. M., Insall R. H. (1998) Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8, 1347–1356 [DOI] [PubMed] [Google Scholar]
- 24. Shakir M. A., Jiang K., Struckhoff E. C., Demarco R. S., Patel F. B., Sotot M. C., Lundquist E. A. (2008) The Arp2/3 activators WAVE and WASP have distinct genetic interactions with Rac GTPases in Caenorhabditis elegans axon guidance. Genetics 179, 1957–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu H., Parsons J. T. (1993) Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J. Cell Biol. 120, 1417–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weaver A. M., Young M. E., Lee W. L., Cooper J. A. (2003) Integration of signals to the Arp2/3 complex. Curr. Opin. Cell Biol. 15, 23–30 [DOI] [PubMed] [Google Scholar]
- 27. Weaver A. M., Heuser J. E., Karginov A. V., Lee W. L., Parsons J. T., Cooper J. A. (2002) Interaction of cortactin and N-WASp with Arp2/3 complex. Curr. Biol. 12, 1270–1278 [DOI] [PubMed] [Google Scholar]
- 28. Weaver A. M., Karginov A. V., Kinley A. W., Weed S. A., Li Y., Parsons J. T., Cooper J. A. (2001) Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11, 370–374 [DOI] [PubMed] [Google Scholar]
- 29. Yu X., Machesky L. M. (2012) Cells assemble invadopodia-like structures and invade into matrigel in a matrix metalloprotease dependent manner in the circular invasion assay. PLoS ONE 7, e30605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fraley S. I., Feng Y., Wirtz D., Longmore G. D. (2011) Reply: reducing background fluorescence reveals adhesions in 3D matrices. Nat. Cell Biol. 13, 5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kowalski J. R., Egile C., Gil S., Snapper S. B., Li R., Thomas S. M. (2005) Cortactin regulates cell migration through activation of N-WASP. J. Cell Sci. 118, 79–87 [DOI] [PubMed] [Google Scholar]
- 32. Tapon N., Hall A. (1997) Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr. Opin. Cell Biol. 9, 86–92 [DOI] [PubMed] [Google Scholar]
- 33. Uruno T., Liu J., Zhang P., Fan Y., Egile C., Li R., Mueller S. C., Zhan X. (2001) Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat. Cell Biol. 3, 259–266 [DOI] [PubMed] [Google Scholar]
- 34. Vignjevic D., Yarar D., Welch M. D., Peloquin J., Svitkina T., Borisy G. G. (2003) Formation of filopodia-like bundles in vitro from a dendritic network. J. Cell Biol. 160, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khatau S. B., Bloom R. J., Bajpai S., Razafsky D., Zang S., Giri A., Wu P. H., Marchand J., Celedon A., Hale C. M., Sun S. X., Hodzic D., Wirtz D. (2012) The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci. Rep. 2, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Raub C. B., Suresh V., Krasieva T., Lyubovitsky J., Mih J. D., Putnam A. J., Tromberg B. J., George S. C. (2007) Noninvasive assessment of collagen gel microstructure and mechanics using multiphoton microscopy. Biophys. J. 92, 2212–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Christiansen D. L., Huang E. K., Silver F. H. (2000) Assembly of type I collagen: fusion of fibril subunits and the influence of fibril diameter on mechanical properties. Matrix Biol. 19, 409–420 [DOI] [PubMed] [Google Scholar]
- 38. Jakobsson L., Franco C. A., Bentley K., Collins R. T., Ponsioen B., Aspalter I. M., Rosewell I., Busse M., Thurston G., Medvinsky A., Schulte-Merker S., Gerhardt H. (2010) Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat. Cell Biol. 12, 943–953 [DOI] [PubMed] [Google Scholar]
- 39. Buccione R., Orth J. D., McNiven M. A. (2004) Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat. Rev. Mol. Cell Biol. 5, 647–657 [DOI] [PubMed] [Google Scholar]
- 40. Buccione R., Caldieri G., Ayala I. (2009) Invadopodia: specialized tumor cell structures for the focal degradation of the extracellular matrix. Cancer Metast. Rev. 28, 137–149 [DOI] [PubMed] [Google Scholar]
- 41. Clark E. S., Whigham A. S., Yarbrough W. G., Weaver A. M. (2007) Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67, 4227–4235 [DOI] [PubMed] [Google Scholar]
- 42. Weaver A. M. (2008) Invadopodia Curr. Biol. 18, R362–R364 [DOI] [PubMed] [Google Scholar]
- 43. Murphy D. A., Courtneidge S. A. (2011) The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huttenlocher A., Horwitz A. R. (2011) Integrins in cell migration. Cold Spring Harbor Perspect. Biol. 3, a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bloom R. J., George J. P., Celedon A., Sun S. X., Wirtz D. (2008) Mapping local matrix remodeling induced by a migrating tumor cell using three-dimensional multiple-particle tracking. Biophys. J. 95, 4077–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.