Abstract
Human T cell leukemia virus type 1 (HTLV-1) preferentially infects CD4+ T cells and these cells play a central role in HTLV-1 infection. In this study, we investigated the global gene expression profile of circulating CD4+ T cells from the distinct clinical status of HTLV-1-infected individuals in regard to TAX expression levels. CD4+ T cells were isolated from asymptomatic HTLV-1 carrier (HAC) and HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) patients in order to identify genes involved in HAM/TSP development using a microarray technique. Hierarchical clustering analysis showed that healthy control (CT) and HTLV-1-infected samples clustered separately. We also observed that the HAC and HAM/TSP groups clustered separately regardless of TAX expression. The gene expression profile of CD4+ T cells was compared among the CT, HAC, and HAM/TSP groups. The paxillin (Pxn), chemokine (C-X-C motif ) receptor 4 (Cxcr4), interleukin 27 (IL27), and granzyme A (Gzma) genes were differentially expressed between the HAC and HAM/TSP groups, regardless of TAX expression. The perforin 1 (Prf1) and forkhead box P3 (Foxp3) genes were increased in the HAM/TSP group and presented a positive correlation to the expression of TAX and the proviral load (PVL). The frequency of CD4+FOXP3+ regulatory T cells (Treg) was higher in HTLV-1-infected individuals. Foxp3 gene expression was positively correlated with cell lysis-related genes (Gzma, Gzmb, and Prf1). These findings suggest that CD4+ T cell activity is distinct between the HAC and HAM/TSP groups.
Introduction
Human T cell leukemia virus type 1 (HTLV-1) was the first human retrovirus isolated.1 HTLV-1 is the etiologic agent of HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP),2,3 adult T cell leukemia/lymphoma (ATLL),4,5 and several inflammatory diseases including polymyositis, arthropathy, infective dermatitis, uveitis, and Sjőgren's syndrome.6–10
The disease manifestations occur in 2–5% of infected individuals, and most individuals remain asymptomatic throughout life.11,12 The mechanisms that lead to disease in infected patients are not fully understood and a number of factors such as genetic, demographic, environmental, and others have been suggested to contribute to disease development.13–15
HAM/TSP is a chronic progressive inflammatory disorder of the central nervous system (CNS) characterized by slow progressive spastic paraparesis, bladder disorder, weakness of the lower limbs, and less conspicuous sensory signs.16 The clinical course of HAM/TSP varies among patients, but is more common in females.17–19 Among several HTLV-1 genes, a transcriptional activator Tax may play a major role in the development of HAM/TSP, regulating multiple cellular responses by protein–protein interactions with various host cell factors. Moreover, Tax has been shown to disrupt cell cycle and DNA repair checkpoint, inactivate several tumor suppressors, and stimulate cell growth, while protecting against apoptosis.20,21
Although HTLV-1 is known to infect a wide range of human and nonhuman cells in vitro, HTLV-1 preferentially infects CD4 T cells, which become the main reservoir of HTLV-1.22 It is also known that CD4 T cells predominate in the spinal cord mononuclear infiltrate of HAM/TSP patients.23 In this way, in the early stages of the inflammatory process, cytokines such as interleukin (IL)-1α, tumor necrosis factor (TNF)-α, and interferon (IFN)-γ are spontaneously secreted by these cells.24 Thus, the potential contribution of CD4 T cells to the pathogenesis of this inflammatory disease as well as their expansion in HAM/TSP is more efficient due to the chronic antigenic stimulation that occurs in these individuals.25,26
The aim of this work was to identify genes differentially expressed among healthy control (CT), asymptomatic HTLV-1 carrier (HAC), and HAM/TSP patients in CD4+ T cells isolated from these individuals. We showed that the global gene expression profile of circulating CD4+ T cells from HTLV-1 individuals harboring distinct clinical status was altered regardless of TAX expression levels. Considering that most HTLV-1-infected individuals remain asymptomatic throughout life we explored genes that could be involved with HAM/TSP development.
Materials and Methods
HTLV-1-infected individuals and healthy controls
A total of 47 peripheral whole blood samples from HTLV-1-infected individuals were obtained and divided into two groups: the asymptomatic HAC group comprised of 26 individuals and the HAM/TSP group comprised of 21 symptomatic patients. Healthy controls (CT; n=28) had no previous or current infectious diseases caused by blood-borne pathogens. Samples of the CT individuals were recruited from the Regional Blood Center of Ribeirão Preto, São Paulo, Brazil, and HTLV-1-infected individuals were recruited from the Neurology Department of the Clinical Hospital of the Medical School of the University of São Paulo, Ribeirão Preto, São Paulo, Brazil.
The study was approved by the Institutional Ethics Committee (process number 3083/2007) and all individuals signed an informed consent before enrollment. All HTLV-1-infected individuals were evaluated for clinical status according to the criteria previously described for ATLL and HAM/TSP.27 To be included in this study, the HTLV-1-infected individuals must have serological (rp21e-enhanced EIA; Cambridge Biotech) and molecular [conventional long-term repeat (LTR) and TAX polymerase chain reaction (PCR)] confirmation of the HTLV-1 infection.
All HTLV-1-infected and CT individuals included in this study presented negative serology for other relevant blood-borne pathogens including hepatitis B virus, hepatitis C virus, human immunodeficiency virus, Chagas disease, and syphilis. Hemogram and CD4 and CD8 T cell counts were performed for all individuals.
Molecular diagnosis and proviral load
Genomic DNA was extracted using the Super Quick Gene DNA isolation kit (Analytical Genetic Testing Center–AGTC, Denver, CO) following the manufacturer's instructions. The in house PCR (LTR and Tax regions) tests were as previously described.28 The proviral load was quantified using TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA) and specific primers of the HTLV-1 Tax region. The reaction was composed of 500 ng of genomic DNA, 6.25 μl TaqMan Universal PCR Master Mix, 5 μM of the forward primer (5′-CCC ATC GAT GGA CGC GT-3′), 5 μM of the reverse primer (5′-CTC CTT CCC CAC CCA GAG AA-3′), and 5 μM of the specific probe (5′-FAM-CGG CTC AGC TCT ACA G-3′-MGB). Human β-actin (Actb) was used as the endogenous control (TaqMan Gene Expression Assays–Hs03023880_g1) (Applied Biosystems, Foster City, CA). Serially diluted (from 105 to 101 copies) DNA from HTLV-1-infected MT-2 cells was used for generating standard curves for the HTLV-1 Tax gene. Real-time PCR was performed in duplicate for all DNA standards and samples using the ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA) with the following conditions: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s/60°C for 1 min. Proviral load was calculated using the following formula: (copy number of Tax)/(copy number of β-actin)×2×100,000.
CD4+ T cell isolation
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll Hypaque PLUS (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). CD4+ T cells were then isolated from PBMCs by positive selection using immunomagnetic beads (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) and the purity of the CD3+CD4+ T cells was confirmed by flow cytometry using the surface markers anti-CD4-FITC and anti-CD3-PE (FACSCalibur, Becton & Dickinson, San Jose, CA).
Microarray analysis
Twelve samples underwent microarray analysis, as follows: four CT, four HAC, and four HAM/TSP samples. The criterion used to select HTLV-1-infected individuals was TAX expression levels in PBMCs as assessed by flow cytometry. Two samples had high TAX expression and two samples had low TAX expression for both groups (Table 1). All samples had purity above 90%. We considered high TAX expression levels those with values higher than 1.39% (obtained by the median of all HTLV-1-infected individuals). In these samples, proviral load correlated with TAX expression (data not shown). Total RNA was isolated with TRIzol Reagent (Invitrogen, Carlsbad, CC) and purified with the RNeasy Mini Kit (Qiagen, Hilden, Germany).
Table 1.
Descriptive Characteristics of Human T Cell Leukemia Virus Type 1-Infected Individuals Included in the Microarray Analysis
| Clinical status | Gender | Age (years) | TAX expression in CD4+ (%) | Leukocytes global count (cells/mm3) | Absolute count of CD4/CD8 (reason) | Purity (%) CD4+ |
|---|---|---|---|---|---|---|
| HAC 08 | F | 26 | 8.87 | 5,600 | 1251/760 (1.65) | 96.87 |
| HAC 16 | F | 55 | 0.14 | 7,100 | 708/237 (2.98) | 95.22 |
| HAC 19 | F | 33 | 0.16 | 4,800 | 1166/274 (4.26) | 95.02 |
| HAC 21 | M | 46 | 3.32 | 6,700 | 1225/558 (2.20) | 95.31 |
| HAM 01 | F | 55 | 0.44 | 6,000 | 717/378 (1.90) | 94.94 |
| HAM 09 | M | 64 | 3.66 | 3,700 | 468/355 (1.32) | 96.31 |
| HAM 14 | F | 61 | 2.97 | 6,000 | 665/256 (2.60) | 93.82 |
| HAM 17 | F | 55 | 0.54 | 8,600 | 579/272 (2.13) | 93.62 |
Characteristics according to gender, age, TAX expression in CD4+ T cells, leukocyte global count, absolute count of CD4/CD8, and purity of CD4+ T cells.
HTLV-1, human T cell leukemia virus type 1; HAC, asymptomatic HTLV-1 carrier; HAM/TSP, HTLV-1-associated myelopathy/tropical spastic paraparesis; F, female; M, male.
One color microarray-based gene expression analysis (Quick Amp Labeling) (Agilent, Santa Clara, CA) was done according to the manufacturer's instructions. The microarray was scanned with an Axon GenePix 4000B Microarray Scanner (Molecular Devices, Sunnyvale, CA) and data were processed using Feature Extraction software version 9.5.1 (Agilent, Santa Clara, CA). Quality control and array normalization were done in R environment at (www.r-project.org) with the Agi4x44PreProcess bioconductor package (http://bioconductor.org/). Normalization and filtering were done following the Agi4x44Pre-Process instructions. Genes differentially expressed were identified based on a log2-fold change of 2-fold at least and a statistically significant level using the t-test (p value<0.005). The expression profiles of the differentially expressed genes were determined by cluster analysis based on the k-means method using Euclidean distance (Genesis 1.7.5). Ingenuity Pathway Analysis (IPA) was used to evaluate the microarray data for relevant biological themes within the differentially expressed genes. Microarray data have been deposited in the NCBI Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) (GEO ID: GSE38537).
Real-Time RT-PCR
Total RNA was reverse transcribed (RT) using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) following the manufacturer's instructions. Differentially expressed genes were validated by a real-time PCR technique using TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA). The PCR amplification and fluorescence data collection were performed with ABI 7500 Sequence Detection (Applied Biosystems, Foster City, CA).
The following genes were validated: granzyme A (Gzma) (Hs00196206_m1), granzyme B (Gzmb) (Hs00188051_m1), perforin 1 (Prf1) (Hs00169473_m1), chemokine (C-X-C motif ) receptor 4 (Cxcr4) (Hs00237052_m1), paxillin (Pxn) (Hs01104424_m1), forkhead box P3 (Foxp3) (Hs01085834_m1), and interleukin 27 (IL27) (Hs00377366_m1).
The housekeeping genes β-actin (Actb) (4326315E), glyceraldehyde-3-phosphate dehydrogenase (Gapdh) (4310884-E), β2-microglobulin (B2m) (4333766-0710013), and ribosomal protein L13a (Rpl13a) (185720330-7)29 were used to normalize sample loading (Applied Biosystems). The 2−ΔΔCt method was used to calculate the relative expression levels.30 All the reactions were duplicated.
Flow cytometry analysis
TAX expression was detected in PBMCs. Cells were cultured in RPMI 1640 (Sigma-Aldrich, Saint Louis, MO) containing 10% fetal calf serum (HyClone, Logan, UT) and 20 nM concanamycin A (Sigma-Aldrich, St. Louis, MO) for 12 h under 5% CO2 at 37°C.
A total of 2.5×105 cells were stained for surface markers anti-CD4-PE, anti-CD8-PerCP, and anti-CD3-APC (Becton & Dickinson, San Jose, CA). For intracellular analysis, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min.
Fixed cells were washed with PBS containing 4% normal goat serum (NGS) (Sigma) and then washed with PBS containing 0.1% Triton X-100 (Sigma) for 10 min at room temperature. Permeabilized cells were washed and resuspended in PBS/4% NGS containing an anti-HTLV-I TAX monoclonal antibody (mAb) (Lt-4; IgG3), antihuman-PRF1-clone δG9, antihuman-GZMB-clone GB11 (Becton & Dickinson, San Jose, CA), or an isotype control mAb (Southern Biotechnology Associates, Birmingham, AL) for 30 min at room temperature. For intracellular TAX detection, cells were stained with Alexa Fluor 488-labeled goat antimouse IgG3 (Invitrogen, Carlsbad, CA) for 30 min at room temperature.
For CXCR4 detection, 150 μl of whole blood and 5 μl of antibodies anti-CD4-FITC, anti-CD3-PerCP, and anti-human-CXCR4-PE-clone 12G5 (Becton & Dickinson, San Jose, CA) were used. The reaction was incubated for 15 min and then 1 ml of FACS Lysing Solution (1×) (Becton & Dickinson, San Jose, CA) was added. All samples were analyzed on a FACSCalibur flow cytometer (Becton & Dickinson, San Jose, CA).
For FOXP3 expression, isolated PBMCs were immunostained with a saturating concentration of anti-CD25-FITC, anti-CD4-PerCP, and anti-CD3-APC for 15 min at room temperature. Then 1 ml of FACS Lysing Solution (1×) (Becton & Dickinson, San Jose, CA) was added. Cells were permeabilized using FACS Permeabilizing Solution 2 (Becton & Dickinson, San Jose, CA) and stained intracellularly for 10 min with antihuman-FOXP3-PE-clone 259/C7. Finally, the cells were washed with PBS and analyzed on a FACSCalibur flow cytometer (Becton & Dickinson, San Jose, CA).
Statistical analysis
Data were analyzed in GraphPad PRISM, version 5.01 (GraphPad software California) using nonparametric statistical tests. The results from quantitative PCR and flow cytometry were analyzed using a one-tailed Mann–Whitney U-test. The correlations among different parameters were assessed with Spearman's rank correlation. In all these cases, statistically significant differences were considered when p values were ≤0.05.
Results
Clinical and demographic data
The clinical and demographic data of patients are shown in Table 2. A total of 75 individuals were included in our study of which 51 were female (68%). The mean onset age in the HAM/TSP group was 54.9 years (ranging from 37 to 74 years), which was higher than the CT (mean 46.5 years) and HAC groups (mean onset age 42.9 years). Proviral load was increased (1.8×) in the HAM/TSP group (p=0.0024) when compared to the HAC group (data not shown). TAX expression positively correlated with proviral load (r=0.7433; p<0.0001) and the percentage of CD4+ T cells expressing TAX was higher in the HAM/TSP group (data not shown). No differences were observed in leukocyte global counts and CD4/CD8 ratio among groups.
Table 2.
Clinical and Viral Characteristics of Human T Cell Leukemia Virus Type 1-Infected Individuals and Healthy Controls Enrolled in the Study
| CT (n=28) | HAC (n=26) | HAM/TSP (n=21) | |
|---|---|---|---|
| Age | 46.5 | 42.9 | 54.9 |
| Gender (F:M) | 2.5 | 1.4 | 3.2 |
| Proviral load mean (copy number/105 cells)a | — | 6,586 | 12,233b |
| TAX expression (%) | — | 1.1 | 2.5b |
| Leukocyte global count (cells/mm3) | 7,225 | 6,816 | 6,771 |
| CD4/CD8 ratio | 2.21 | 2.10 | 2.98 |
Proviral load copy number/105 cells.
p≤0.01 (unpaired t test).
Table shows average values. CT, healthy controls; HAC, asymptomatic HTLV-1 carrier; HAM/TSP, HTLV-1-associated myelopathy/tropical spastic paraparesis; F:M, female/male ratio.
Global gene expression in CD4+ T cells
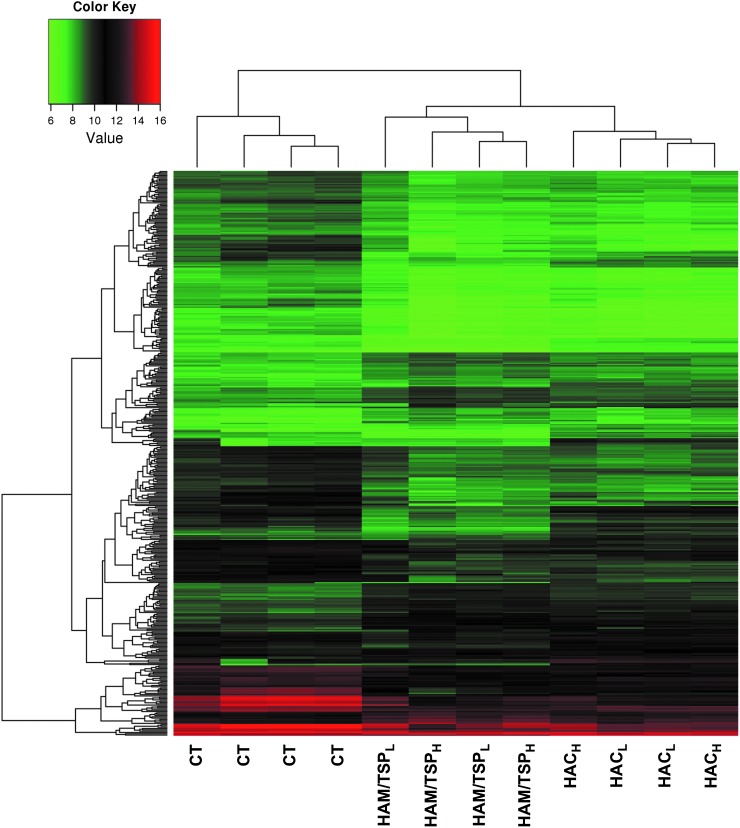
The microarray platform was tested with 12 individual samples divided according to patients' clinical status and TAX expression as follows: CT (n=4), HAC (n=4; composed of two samples with high TAX expression and two with low TAX expression), and HAM/TSP group (n=4; composed of two samples with high TAX expression and two with low TAX expression). We used a hierarchical clustering to group samples according to their gene expression levels. Dendrogram analysis showed that clustering of CD4+ T cells allowed a clear separation between CT and HTLV-1-infected individuals. We also observed that the HAC and HAM/TSP groups clustered apart (Fig. 1). However, HTLV-1 CD4+ T cell clustering did not correlate with TAX protein expression.
FIG. 1.
Gene expression heat map and dendrogram of CD4+ T cells. Hierarchical clustering of all detected genes showed a clear clustering of CT, HAC, and HAM/TSP samples using an average linkage and Euclidian distance metric. Expression values from all microarrays were used to group transcription profiles according to their similarities among groups. The heat map shows a distinct gene expression profile in the three groups analyzed. Rows indicate the relative levels of expression for a single gene, and columns show the expression level for a single sample. Green and red colors indicate those genes with lower and higher expression levels, respectively. CT, healthy control; HAC, asymptomatic human T cell leukemia virus type 1 (HTLV-1) carrier; HAM/TSP, HTLV-1-associated myelopathy/tropical spastic paraparesis; HACL, low TAX expression; HACH, high TAX expression; HAM/TSPL, low TAX expression; HAM/TSPH, high TAX expression.
Gene expression analysis
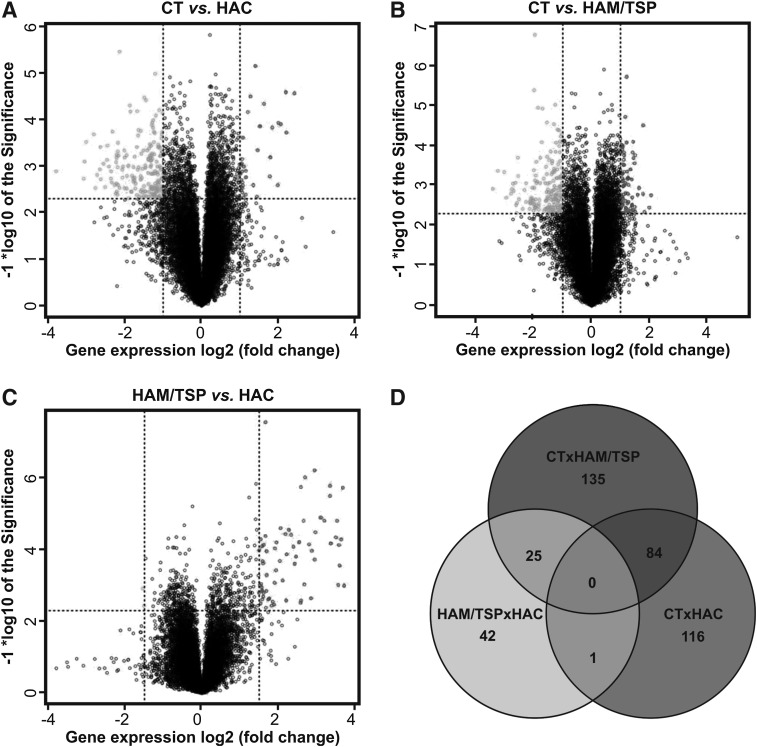
A total of 45,015 probes were screened by the Agilent microarray platform. The background correction, filtering, normalization, and summarization of probes were performed using the Agi4x44PreProcess package resulting in 27,061 distinct probes, from which only 19,668 remained in the subsequent analysis after the filtering of probes annotated as ribosomal proteins, or in sexual chromosomes or unannotated probes. In microarray analysis, genes were considered differentially expressed on the basis of a log2-fold change of 2-fold at least and a statistically significant level using a t-test (p value<0.005). We found 201 differently expressed genes between the CT and HAC groups (166 genes down-regulated and 35 genes up-regulated in the HAC group), 244 genes between the CT and HAM/TSP groups (165 genes down-regulated and 79 genes up-regulated in the HAM/TSP group), and 68 genes between the HAC and HAM/TSP groups (66 genes down-regulated and 2 genes up-regulated in the HAM/TSP group) (Fig. 2A–C). Additionally, we determined which genes were in common among these groups (Supplementary Tables S1–S6 (Supplementary Data are available online at www.liebertpub.com/aid) SUPPL TABLES S1–S6 list all genes shown in Venn diagrams. Only one differentially expressed gene (Tmeff2) was observed between HAM/TSP vs. HAC and CT vs. HAC analysis. We did not select this gene to study because it was not associated with inflammatory and infectious disease.
FIG. 2.
Differently expressed genes among the CT, HAC, and HAM/TSP groups. Differently expressed genes between the CT and HAC groups (A), CT and HAM/TSP groups (B), and HAC and HAM/TSP groups (C) represented by Volcano Plot. Overexpressed genes (right) are on the upper side and underexpressed genes are on the upper left side. Dots in the middle of the figure represent genes for which the expression showed no statistical difference (Euclidian distance – complete linkage). (D) Venn diagram representation of differentially expressed genes. Venn diagrams showed an overlap of genes that are commonly expressed in the three clinical states. CT, healthy control; HAC, asymptomatic HTLV-1 carrier; HAM/TSP, HTLV-1-associated myelopathy/tropical spastic paraparesis.
Genes related to cell migration were up-regulated in the HAM/TSP group
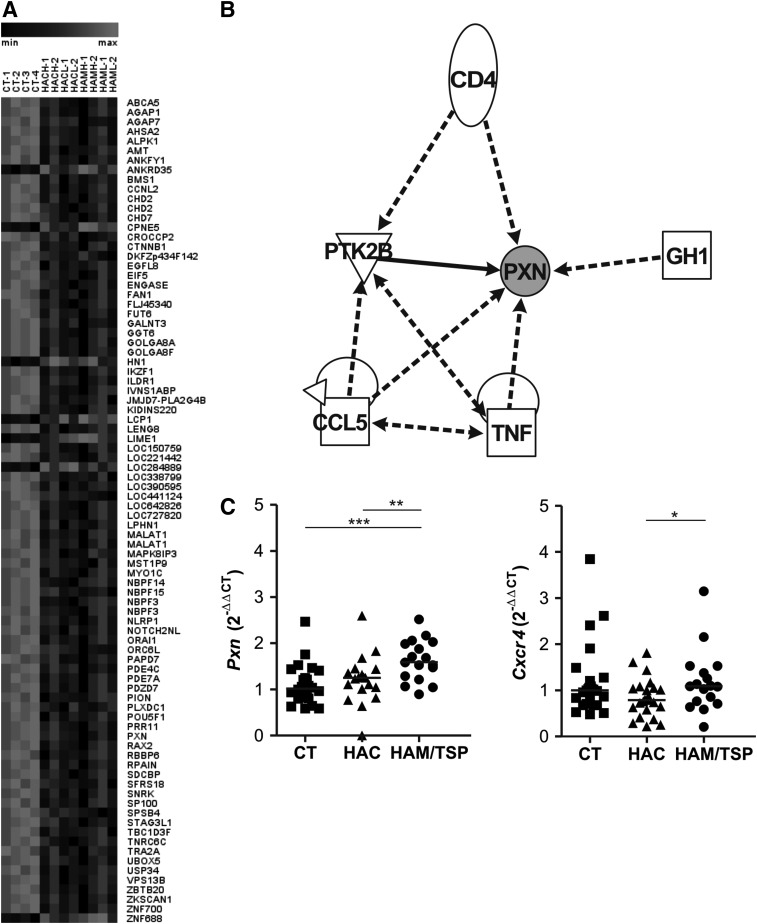
Eighty-four differentially expressed genes in common were observed between CT vs. HAC and CT vs. HAM/TSP analysis, with most of them overexpressed in the CT group (Fig. 3A). In silico analysis demonstrated that six genes, namely paxillin (Pxn), CD4, Ptk2b, Ccl5, Tnf, and Gh1, were represented in a network that is related to apoptosis, inflammatory and immunological diseases, cell–cell interaction, movement and repair of cell functions, and cell migration (Fig. 3B). The Pxn gene was also present in the Cxcr4 pathway that is responsible for cell migration. The global gene expression profile showed that Pxn was increased in the CT group compared to the HAC (fold change: 5.1, p=0.0913) and HAM/TSP groups (fold change: 8.2, p=0.0002). The Pxn and Cxcr4 genes were validated by quantitative real-time PCR (qPCR) and both of them showed an increased expression in HAM/TSP individuals (Fig. 3C). The level of Pxn gene expression was increased in HTLV-1-infected individuals (p=0.0022) (data not shown). Moreover, the CXCR4 protein was analyzed by flow cytometry among the groups, but no statistically significant difference was observed (data not shown).
FIG. 3.
Expression profiling of genes differentially expressed in HTLV-1. (A) Gene expression heat map of analyses of 84 differently expressed genes between the CT vs. HAC and the CT vs. HAM/TSP groups. (B) Schematic of molecular interactions. The network image was created using IPA software. The lines represent regulation of function between two genes. (C) Real-time PCR of Pxn and Cxcr4 mRNA in CD4+ T cells from CT, HAC, and HAM/TSP. The Mann–Whitney U-test was used to evaluate differences among groups (*p≤0.05; ** p≤0.01; ***p≤0.001). CT, healthy control; HAC, asymptomatic HTLV-1 carrier; HAM/TSP, HTLV-1-associated myelopathy/tropical spastic paraparesis.
IL27 gene expression is higher in HAC individuals
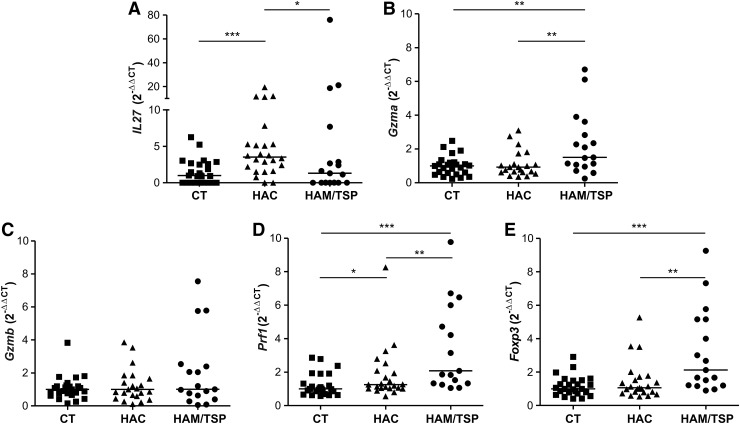
Forty-two differentially expressed genes exclusive between HAC and HAM/TSP analysis were found. These genes were related to cell–cell interaction, inflammatory response, and cellular immune response, which include interleukin 27 (IL27). A global gene expression profile showed that IL27 was increased in the HAC group compared to the HAM/TSP groups (fold change: 5.24). This result was validated by qPCR in which levels of IL27 gene expression were higher in the HAC group compared to the HAM/TSP group (p=0.0392) (Fig. 4A). Additionally, IL27 gene expression was higher in HTLV-1-infected individuals compared to the CT group (p=0.0019) (data not shown).
FIG. 4.
IL27, Gzma, Gzmb, Prf1, and Foxp3 gene expression by real-time PCR. Gene expression levels comparison of IL27 (A), Gzma (B), Gzmb (C), Prf1 (D), and Foxp3 (E) among the CT, HAC, and HAM/TSP groups. The Mann–Whitney U-test was used to evaluate differences among groups (*p≤0.05; **p≤0.01; ***p≤0.001). CT, healthy control; HAC, asymptomatic HTLV-1 carrier; HAM/TSP, HTLV-1-associated myelopathy/tropical spastic paraparesis.
HAM/TSP CD4+ T cells had higher expression of cell lysis-related genes
The 25 differentially expressed genes in common between CT vs. HAM/TSP and HAM/TSP vs. HAC analyses were evaluated for their participation in signaling pathways and only three pathways were observed, among them granzyme A (Gzma). Although the Gzma gene was not in accordance with the parameters of analyses (2-fold and p value 0.005), the global gene expression profile showed that Gzma was increased in the HAM/TSP group compared to the CT (fold change: 1.9, p=0.0038) and HAC groups (fold change: 1.9, p=0.0071). For that reason, this gene was selected for validation due to it being poorly studied on CD4+ T cells infected by HTLV-1 and due to its possible involvement in HAM/TSP development.
The levels of Gzma gene expression were evaluated (Fig. 4B) and we observed an increase in the HAM/TSP group compared to the CT (p=0.0038) and HAC (p=0.0071) groups. Therefore, we suggest that other genes related to cell lysis could also be increased in the HAM/TSP group, such as Gzmb and Prf1. Therefore, we analyzed Gzmb and Prf1 gene expression (Fig. 4C and D). The HAM/TSP group showed higher gene expression levels of Prf1 compared to the CT (p<0.0001) and HAC (p=0.0037) groups. Prf1 gene expression was increased (p=0.003) in HTLV-1-infected individuals when compared to the CT group (data not shown). No difference was observed in gene expression level of Gzmb among the three groups. GZMB and PRF1 proteins were analyzed by flow cytometry; however, no difference was observed among the CT, HAC, and HAM/TSP groups (data not shown).
It was reported that regulatory T cells (Treg) have cytolytic capacity and perforin/granzyme pathways are required for this activity.31 Foxp3 gene expression, one of the main Treg cell markers, was also evaluated (Fig. 4E). As expected, we notice an overexpression of Foxp3 in the HAM/TSP group compared to the CT and HAC groups (p=0.0003 and p=0.0016, respectively). Foxp3 gene expression was increased in HTLV-1-infected individuals (p=0.0128) compared to CT individuals (data not shown).
The lysis-related genes were correlated with Foxp3 expression and Prf1 and Foxp3 genes were correlated with TAX expression and proviral load (PVL)
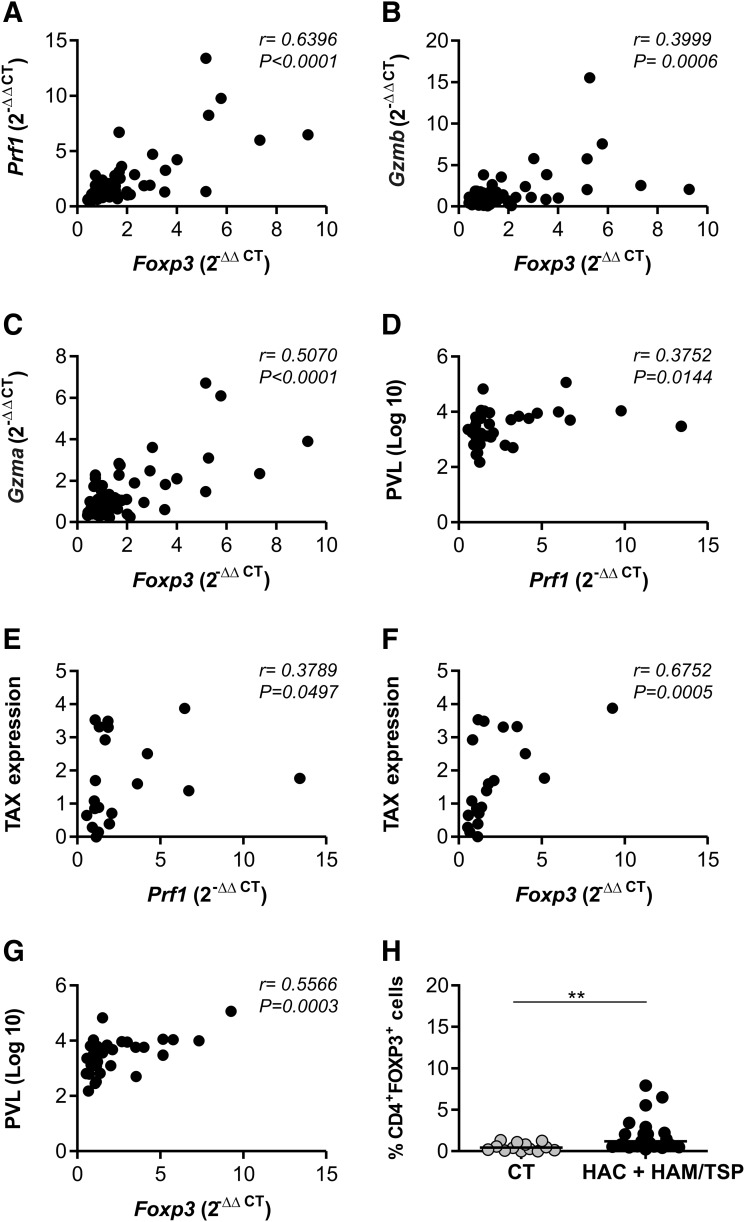
Since the Gzma, Prf1, and Foxp3 genes were shown to have an increased expression in HAM/TSP individuals, we thought these genes could be correlated. We performed Spearman's correlation test and verified that the Gzma, Gzmb, and Prf1 genes were positively correlated with Foxp3 gene expression (Fig. 5A–C). We evaluated whether all the genes analyzed in this study (Pxn, Cxcr4, IL27, Gzma, Gzmb, Prf1, and Foxp3 genes) were correlated with TAX viral protein expression and PVL values. However, only the Prf1 and Foxp3 genes presented a positive correlation with TAX expression and the PVL (Fig. 5D–G).
FIG. 5.
Correlation between cell lysis-related genes and Foxp3 in CD4+ T cells, percentage of TAX expression, and proviral load in peripheral blood mononuclear cells (PBMCs). The Foxp3 expression in CD4+ T cells positively correlated with Prf1 (A), Gzmb (B), and Gzma (C) expression. A positive correlation between PVL and Prf1 (D), TAX expression and Prf1 (E), TAX expression and Foxp3 (F), and PVL and Foxp3 (G) was observed. The correlation was calculated by the Spearman method. (H) The percentage of FOXP3-expressing cells in the CD4+ population from PBMCs. The Mann–Whitney U-test was used to evaluate differences among groups (**p≤0.01). CT, healthy control; HAC, asymptomatic HTLV-1 carrier; HAM/TSP, HTLV-1-associated myelopathy/tropical spastic paraparesis; PVL, proviral load.
CD4+FOXP3+ cell percentage was increased in HTLV-1-infected individuals
The phenotype CD4+FOXP3+ is used to identify a major population of Tregs.32 Thus we analyzed CD4+FOXP3+ cell percentage in the CT, HAC, and HAM/TSP groups. The percentage of CD4+FOXP3+ cells was consistently higher (p=0.0017) in HTLV-1-infected individuals than in the CT group (Fig. 5H). The analyses have revealed an increase (8×) in the HAC group compared to the CT group (p=0.0016) and an increase (3×) in the HAM/TSP group compared to the CT group (p=0.0128) (data not shown).
Discussion
We demonstrated that the gene expression profile in CD4+ T cells differs between the HAC and HAM/TSP groups regardless of TAX expression. It has been well established that HAM/TSP individuals have an HTLV-1 PVL and TAX expression higher than asymptomatic individuals.33–36 These findings suggest that the risk of HTLV-1 inflammatory diseases strongly correlates with PVL.37 Here, we demonstrated that PVL and TAX expression of CD4+ T cells positively correlated in HTLV-1-infected groups, being higher in the HAM/TSP group. This observation reinforced the fact that these cells may contribute to the progression of HTLV-1-related diseases.25,33 Although several authors have published TAX expression and PVL quantification values in HTLV-1-infected individuals, the standard reference values for these parameters have not been well established yet. In our study, we measured the levels of TAX expression and by setting the median value we defined low and high TAX expression samples. Hierarchical clustering analysis showed that CT and HTLV-1-infected groups clustered separately. Here, we also show that CD4+ T cells from HAC and HAM/TSP samples clustered separately regardless of TAX expression. Thus, our results demonstrate that genes are differentially expressed according to individuals' HTLV clinical status in CD4+ T cells. However, it would be important to analyze gene expression between the HAC and HAM/TSP groups with similar levels of TAX expression and PVL.
It was reported that the global gene expression profile did not allow observation of two independent clusters for HAC and HAM/TSP; however, ATLL samples were suitably clustered independently.38 Another previous study did not find differences between the HAC and HAM/TSP groups when clustering CD4+ T cells.37 These findings could be explained by how the sampling was done. In our analysis, individual samples were submitted to microarray analysis.
We analyzed Pxn and Cxcr4 gene expression since they are involved in cell migration. In response to its binding ligand stromal cell-derived factor-1 (SDF-1), CXCR4 induces downstream signaling by several different pathways. SDF-1 binding to CXCR4 promotes actin polymerization to initiate cell motility. CXCR4 triggers the activation of the src family of protein tyrosine kinases and then the focal adhesion complexes such as RAFTK/Pyk2, focal adhesion kinase, Crk, and paxillin are phosphorylated and activated.39–41 The focal adhesion components paxillin and Crk play a critical role in the chemotactic signaling pathways.42
Although the Cxcr4 gene did not show deregulation in microarray analysis, the Pxn gene is a member of the Cxcr4 pathway. The Pxn gene was overexpressed in the CT group compared to the HTLV-1-infected group in microarray analysis. Intriguingly, after a validation process this gene was significantly up-regulated in the HTLV-1-infected group (HAC+HAM/TSP). The discordant results between microarray and qPCR techniques are explained by the inherent pitfalls of each technique.43–47 To confirm the gene expression results obtained from microarray analysis qPCR is frequently used as a validation tool. Furthermore, many different platforms exist for both microarray and qPCR analyses that have led to debates over which techniques produce the most exact measurements of gene expression.48–50 Additionally, we can also explain our discrepant results based on the fact that microarray and qPCR were not performed with the same number of samples.
CXCR4 is highly expressed by leukocytes in the immune and central nervous systems.51,52 In this study, Cxcr4 gene expression was up-regulated in the HAM/TSP group compared to the HAC group and no difference in CXCR4 protein levels by flow cytometry was observed among the studied groups. These different results between Cxcr4 mRNA and protein expression can be explained by the fact that there are posttranscriptional mechanisms that might control the translation of the protein. The presence of this posttranscriptional control is demonstrated in many studies, suggesting that there is a poor correlation between the mRNA and protein pools in eukaryotic cells.53–58
On the other hand, one previous study demonstrated that CXCR4 levels in HTLV-1-infected cell lines were lower than in noninfected cell lines.59 This could be explained by the type of cell samples used, since we are working with cells belonging to HTLV-1-infected individuals, in which genetic susceptibility should be taken into account. It was also demonstrated that the inhibition of the SDF-1α-CXCR4 axis by the CXCR4 antagonist AMD3100 suppresses the migration of cultured cells from ATL patients and murine lymphoblastoid cells from HTLV-1 TAX transgenic mice. Therefore, these results show the association of the SDF-1alpha/CXCR4 interaction as one mechanism of leukemic cell migration and this may offer a new target as part of combination therapy for ATLL.60
HTLV-1-infected lymphocytes may cross the blood–brain barrier and may generate an intensive immune response leading to the development of HAM/TSP. All the aforementioned findings lead us to believe that there is a dynamic regulation of Cxcr4 expression, since the Pxn and Cxcr4 genes are overexpressed in the HAM/TSP group. This result suggests that this pathway could be deregulated in these patients and could lead to CD4+ T cell migration to the CNS. This way, our data suggest that the Pxn and Cxcr4 genes may be acting together in CD4+ T cells.
We observed that the IL27 gene was up-regulated in the HAC group when compared to the CT and HAM/TSP groups. IL-27 is a member of the IL-12 cytokine family and has many functions in immune response such as induction of proliferation of naive specificity and also as an early product of activated antigen-presenting cells.61 Moreover, it has been shown that IL-27 is capable of regulating T helper subpopulations (Th1, Th2, and Th17) during acute and chronic infection and inflammatory processes.62–66 Previous studies have reported that IL-27 inhibits HIV-1 replication in T cells and macrophages and HCV replication.67–70 Furthermore, IL-27 may play an antiviral and antitumor role in ATLL and other lymphoid malignancies71 and it has gathered considerable attention in terms of its therapeutic application in immune disease. This way, IL-27 was shown to suppress experimental autoimmune encephalomyelitis during bone marrow stromal cell treatment.72 Other studies reported suppression of experimental autoimmune uveoretinitis (EAU),73 inflammation in the joint, and severe arthritis by administration of IL-27. In addition, IL-27 administered in vivo showed its antiinflammatory effects on a delayed type hypersensitivity model, demonstrating its therapeutic potential against some diseases of immune origin.74 However, IL-27 may be an effective therapeutic agent for HTLV-1-infected individuals in order to suppress the inflammation caused by retroviruses and consequently to decrease the risk of HAM/TSP development. Therefore, the specific role of this cytokine in HTLV-1 infection remains unclear.
The perforin/granzyme mechanism is mainly carried out by circulating white blood cells such as CTLs and natural killer (NK) cells. Several studies have shown that Treg cells may use the perforin/granzyme pathway as a system to suppress the function of immune cells by killing them.31,75,76 It was demonstrated that activated murine Treg cells suppressed B cell proliferation in a granzyme B- and perforin-dependent fashion.76 Likewise, the activated human Treg cells expressed granzyme A and/or B and could use the perforin pathway to cause autologous target cell death.75 The transcriptional factor FOXP3 is considered one of the main specific markers for Treg cells77 whose expression is a critical mediator for the development and the function of these cells.78 Treg cells are major effector cells during immune response by the perforin/granzyme pathway. Considering this, we selected Gzma, Gzmb, Prf1, and Foxp3 for gene expression analysis.
The level of Prf1 and Gzma gene expression was significantly higher in the HAM/TSP group compared to the CT and HAC groups. No differences were observed in intracellular PRF1 among the studied groups. Protein detection and quantification are potent tools since mRNA detection could not guarantee that protein will be translated or functional. Thus there could be posttranscriptional mechanisms that regulate the expression of these molecules. Moreover, measurement of granzyme and perforin mRNA is better than immunostaining of granzyme and perforin proteins as a correlate of cytotoxicity, because the granzyme and perforin proteins do not accumulate in the cell but are rapidly and continually discharged in the lytic granules.37 Therefore, all these considerations can explain our results.
Although it has been demonstrated that HTLV-1 has a tropism to Treg cells,79 reduced FOXP3 protein expression in HTLV-1-infected cells of HAM/TSP patients has been reported.80,81 However, in our analysis of Foxp3 gene expression, we also noticed a significant increase in the HAM/TSP group compared to the CT and HAC groups. We also observed a positive correlation among Gzma, Gzmb, and Prf1 to Foxp3. In addition to that, there was also a positive correlation between the Foxp3 gene and PVL and the Foxp3 gene and TAX expression. These data are consistent with the results from a previous report82 describing higher FOXP3+ expression in CD4+ T cells of individuals with HAM/TSP compared to healthy individuals. In addition, the researchers have also observed a positive correlation between FOXP3 and PVL and FOXP3 and TAX expression. We also found an increased frequency of CD4+FOXP3+ cells in HTLV-1-infected individuals, as reported previously.82,83 In our results, we showed that Foxp3 gene expression levels were increased in the HAM/TSP group compared to the HAC group, while the proportions of CD4+FOXP3+ cells were higher in the HAC group compared to the HAM/TSP group. This discordance may be due to differences between both applied techniques. In general, the sensitivity of real-time RT-PCR is higher than that of flow cytometry.84
Many studies on solid-organ transplant recipients (heart, kidney, and intestine) have demonstrated a direct correlation between Treg marker FOXP3 and the cytotoxic T cell effector molecules (granzyme B, perforin, and granulysin) in the process of acute rejection.85–89 Therefore, in our study we speculate that FOXP3 is correlated with the cytotoxic T cell effector molecules (Gzma, Gzmb, and Prf1) in HTLV-1 infection. One previous study suggested two possibilities for the role of Treg cells in HTLV-1 infection. First, FOXP3+ T cells have hyperproliferation ability90 and this could collaborate in the clonal expansion of HTLV-1-infected cells. Second, HTLV-1 can invade the immune system by direct infection of this immunosuppressive cell population. Consequently, HTLV-1 infection of FOXP3+ T cells can enable the virus to increase or maintain its proviral load and thus reach a state of persistent infection.91 With this in mind, we suggest another possibility for the role of Treg cells. Treg cells may use the perforin/granzyme pathway as a system to suppress the immune cells by killing them. This way, the increase of FOXP3+ T cells analyzed in HTLV-1 infection may contribute to immunodeficiency, which is observed in HTLV-1 infection.92
In a previous study, we reported differentially expressed genes in CD8+ T cells isolated from the same population as this study. Our results showed that patients with HAM/TSP have high expression levels of degranulation-related genes (Gzmh and Prf1) and of the cytoskeletal adaptor Pxn. We indicated that Gzmb and Zap70 genes were overexpressed in HTLV-infected individuals compared to the healthy control group. We also found that Ccl5 was higher in the HAM/TSP group compared to the HAC and CT groups. Therefore, our findings showed that CD8+ and CD4+ T cells from HAM/TSP patients have an inflammatory and active profile.93
In conclusion, we demonstrated that Pxn, Cxcr4, IL27, and Gzma gene expression in CD4+ T cells differs between the HAC and HAM/TSP groups regardless of TAX expression. Prf1 and Foxp3 genes are increased in the HAM/TSP group compared to the HAC group and present a positive correlation to the expression of TAX and PVL. We believe that the IL27, Pxn, Cxcr4, Gzma, Prf1, and Foxp3 genes are novel molecules that could play an important role in HTLV-1 infection. Moreover, Treg may be a future strategy for the treatment and prevention of HTLV-1-infected individuals. However, further studies are required to elucidate the molecular mechanisms of CD4+ T cell involved in HTLV-1 infection.
Supplementary Material
Acknowledgments
The authors thank Rochele Azevedo for quantification of the proviral load, Patrícia Vianna Bonini Palma for flow cytometry analysis, Amelia Goes de Araujo for her assistance with the microarray technique, Maurício Cristiano Rocha Junior for help in writing the paper, and Prof. Charles Bangham for TAX expression analysis. They are also grateful to the patients. This work was supported by Fundação Hemocentro de Ribeirão Preto (FUNDHERP) and São Paulo Research Foundation (FAPESP-Process number: 2011/21740-7)
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Poiesz BJ, Ruscetti FW, Gazdar AF, et al. : Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA 1980;77(12):7415–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gessain A, Barin F, Vernant JC, et al. : Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet 1985;2:407–410 [DOI] [PubMed] [Google Scholar]
- 3.Osame M, Usuku K, Izumo S, et al. : HTLV-I associated myelopathy, a new clinical entity. Lancet 1986;1(8488):1031–1032 [DOI] [PubMed] [Google Scholar]
- 4.Uchiyama T, Yodoi J, Sagawa K, et al. : Adult T-cell leukemia: Clinical and hematologic features of 16 cases. Blood 1977;50(3):481–492 [PubMed] [Google Scholar]
- 5.Yoshida M, Seiki M, Yamaguchi K, et al. : Monoclonal integration of human T-cell leukemia provirus in all primary tumors of adult T-cell leukemia suggests causative role of human T-cell leukemia virus in the disease. Proc Natl Acad Sci USA 1984;81(8):2534–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaGrenade L, Hanchard B, Fletcher V, et al. : Infective dermatitis of Jamaican children: A marker for HTLV-I infection. Lancet 1990;336(8727):1345–1347 [DOI] [PubMed] [Google Scholar]
- 7.Mariette X, Agbalika F, Zucker-Franklin D, et al. : Detection of the tax gene of HTLV-I in labial salivary glands from patients with Sjogren's syndrome and other diseases of the oral cavity. Clin Exp Rheumatol 2000;18(3):341–347 [PubMed] [Google Scholar]
- 8.Mochizuki M, Ono A, Ikeda E, et al. : HTLV-I uveitis. J Acquir Immune Defic Syndr Hum Retrovirol 1996;13(Suppl 1):S50–56 [DOI] [PubMed] [Google Scholar]
- 9.Morgan OS, Rodgers-Johnson P, Mora C, et al. : HTLV-1 and polymyositis in Jamaica. Lancet 1989;2(8673):1184–1187 [DOI] [PubMed] [Google Scholar]
- 10.Nishioka K, Maruyama I, Sato K, et al. : Chronic inflammatory arthropathy associated with HTLV-I. Lancet 1989;1(8635):441. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan TA: Low-lying disordered states when ground-state long-range order exists; large-amplitude spin waves. Phys Rev B Condens Matter 1990;41(10):6882–6888 [DOI] [PubMed] [Google Scholar]
- 12.Murphy EL, Figueroa JP, Gibbs WN, et al. : Sexual transmission of human T-lymphotropic virus type I (HTLV-I). Ann Intern Med 1989;111(7):555–560 [DOI] [PubMed] [Google Scholar]
- 13.Catalan-Soares B, Carneiro-Proietti AB, and Proietti FA: Heterogeneous geographic distribution of human T-cell lymphotropic viruses I and II (HTLV-I/II): Serological screening prevalence rates in blood donors from large urban areas in Brazil. Cad Saude Publica 2005;21(3):926–931 [DOI] [PubMed] [Google Scholar]
- 14.Dourado I, Alcantara LC, Barreto ML, et al. : HTLV-I in the general population of Salvador, Brazil: A city with African ethnic and sociodemographic characteristics. J Acquir Immune Defic Syndr 2003;34(5):527–531 [DOI] [PubMed] [Google Scholar]
- 15.Matsuura E, Yamano Y, and Jacobson S: Neuroimmunity of HTLV-I infection. J Neuroimmune Pharmacol 2010;5(3):310–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Araujo AQ. and Silva MT: The HTLV-1 neurological complex. Lancet Neurol 2006;5(12):1068–1076 [DOI] [PubMed] [Google Scholar]
- 17.Vernant JC, Maurs L, Gessain A, et al. : Endemic tropical spastic paraparesis associated with human T-lymphotropic virus type I: A clinical and seroepidemiological study of 25 cases. Ann Neurol 1987;21(2):123–130 [DOI] [PubMed] [Google Scholar]
- 18.Lima MA, Bica RB, and Araujo AQ: Gender influence on the progression of HTLV-I associated myelopathy/tropical spastic paraparesis. J Neurol Neurosurg Psychiatry 2005;76(2):294–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagawa M, Izumo S, Ijichi S, et al. : HTLV-I-associated myelopathy: Analysis of 213 patients based on clinical features and laboratory findings. J Neurovirol 1995;1(1):50–61 [DOI] [PubMed] [Google Scholar]
- 20.Chang SC, Cheng JC, Kou YH, et al. : Roles of the AX(4)GKS and arginine-rich motifs of hepatitis C virus RNA helicase in ATP- and viral RNA-binding activity. J Virol 2000;74(20):9732–9737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor JM. and Nicot C: HTLV-1 and apoptosis: Role in cellular transformation and recent advances in therapeutic approaches. Apoptosis 2008;13(6):733–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richardson JH, Edwards AJ, Cruickshank JK, et al. : In vivo cellular tropism of human T-cell leukemia virus type 1. J Virol 1990;64(11):5682–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umehara F, Izumo S, Nakagawa M, et al. : Immunocytochemical analysis of the cellular infiltrate in the spinal cord lesions in HTLV-I-associated myelopathy. J Neuropathol Exp Neurol 1993;52(4):424–430 [DOI] [PubMed] [Google Scholar]
- 24.Umehara F, Izumo S, Ronquillo AT, et al. : Cytokine expression in the spinal cord lesions in HTLV-I-associated myelopathy. J Neuropathol Exp Neurol 1994;53(1):72–77 [DOI] [PubMed] [Google Scholar]
- 25.Goon PK, Igakura T, Hanon E, et al. : Human T cell lymphotropic virus type I (HTLV-I)-specific CD4+ T cells: Immunodominance hierarchy and preferential infection with HTLV-I. J Immunol 2004;172(3):1735–1743 [DOI] [PubMed] [Google Scholar]
- 26.Nose H, Kubota R, Seth NP, et al. : Ex vivo analysis of human T lymphotropic virus type 1-specific CD4+ cells by use of a major histocompatibility complex class II tetramer composed of a neurological disease-susceptibility allele and its immunodominant peptide. J Infect Dis 2007;196(12):1761–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Castro-Costa CM, Araujo AQ, Barreto MM, et al. : Proposal for diagnostic criteria of tropical spastic paraparesis/HTLV-I-associated myelopathy (TSP/HAM). AIDS Res Hum Retroviruses 2006;22(10):931–935 [DOI] [PubMed] [Google Scholar]
- 28.Pinto MT, Rodrigues ES, Malta TM, et al. : HTlV-1/2 Seroprevalence and coinfection rate in Brazilian first-time blood donors: An 11-year follow-up. Rev Inst Med Trop Sao Paulo 2012;54(3):123–129 [DOI] [PubMed] [Google Scholar]
- 29.Vandesompele J, De Preter K, Pattyn F, et al. : Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3(7):RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaffl MW: A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29(9):e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao X, Cai SF, Fehniger TA, et al. : Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity 2007;27(4):635–646 [DOI] [PubMed] [Google Scholar]
- 32.Ziegler SF: FOXP3: Of mice and men. Annu Rev Immunol 2006;24:209–226 [DOI] [PubMed] [Google Scholar]
- 33.Goon PK, Hanon E, Igakura T, et al. : High frequencies of Th1-type CD4(+) T cells specific to HTLV-1 Env and Tax proteins in patients with HTLV-1-associated myelopathy/tropical spastic paraparesis. Blood 2002;99(9):3335–3341 [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto K, Higuchi I, Osame M, et al. : Quantitative in situ PCR assay of HTLV-1 infected cells in peripheral blood lymphocytes of patients with ATL, HAM/TSP and asymptomatic carriers. J Neurol Sci 1998;159(1):67–72 [DOI] [PubMed] [Google Scholar]
- 35.Kubota R, Kawanishi T, Matsubara H, et al. : Demonstration of human T lymphotropic virus type I (HTLV-I) tax-specific CD8+ lymphocytes directly in peripheral blood of HTLV-I-associated myelopathy/tropical spastic paraparesis patients by intracellular cytokine detection. J Immunol 1998;161(1):482–488 [PubMed] [Google Scholar]
- 36.Nagai M, Usuku K, Matsumoto W, et al. : Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: High proviral load strongly predisposes to HAM/TSP. J Neurovirol 1998;4(6):586–593 [DOI] [PubMed] [Google Scholar]
- 37.Vine AM, Heaps AG, Kaftantzi L, et al. : The role of CTLs in persistent viral infection: Cytolytic gene expression in CD8+ lymphocytes distinguishes between individuals with a high or low proviral load of human T cell lymphotropic virus type 1. J Immunol 2004;173(8):5121–5129 [DOI] [PubMed] [Google Scholar]
- 38.Oliere S, Hernandez E, Lezin A, et al. : HTLV-1 evades type I interferon antiviral signaling by inducing the suppressor of cytokine signaling 1 (SOCS1). PLoS Pathog 2010;6(11):e1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandis AZ, Prasad A, Band H, et al. : Regulation of CXCR4-mediated chemotaxis and chemoinvasion of breast cancer cells. Oncogene 2004;23(1):157–167 [DOI] [PubMed] [Google Scholar]
- 40.Hartmann TN, Burger JA, Glodek A, et al. : CXCR4 chemokine receptor and integrin signaling co-operate in mediating adhesion and chemoresistance in small cell lung cancer (SCLC) cells. Oncogene 2005;24(27):4462–4471 [DOI] [PubMed] [Google Scholar]
- 41.Luker KE. and Luker GD: Functions of CXCL12 and CXCR4 in breast cancer. Cancer Lett 2006;238(1):30–41 [DOI] [PubMed] [Google Scholar]
- 42.Ganju RK, Brubaker SA, Meyer J, et al. : The alpha-chemokine, stromal cell-derived factor-1alpha, binds to the transmembrane G-protein-coupled CXCR-4 receptor and activates multiple signal transduction pathways. J Biol Chem 1998;273(36):23169–23175 [DOI] [PubMed] [Google Scholar]
- 43.Bustin SA: Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J Mol Endocrinol 2002;29(1):23–39 [DOI] [PubMed] [Google Scholar]
- 44.Chuaqui RF, Bonner RF, Best CJ, et al. : Post-analysis follow-up and validation of microarray experiments. Nat Genet 2002;32(Suppl):509–514 [DOI] [PubMed] [Google Scholar]
- 45.Freeman WM, Walker SJ, and Vrana KE: Quantitative RT-PCR: Pitfalls and potential. Biotechniques 1999;26(1):112–122, 124–125 [DOI] [PubMed] [Google Scholar]
- 46.Wurmbach E, Yuen T, and Sealfon SC: Focused microarray analysis. Methods 2003;31(4):306–316 [DOI] [PubMed] [Google Scholar]
- 47.Yang YH, Dudoit S, Luu P, et al. : Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 2002;30(4):e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan PK, Downey TJ, Spitznagel EL Jr, et al. : Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res 2003;31(19):5676–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yauk CL, Berndt ML, Williams A, et al. : Comprehensive comparison of six microarray technologies. Nucleic Acids Res 2004;32(15):e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu B, Ping G, Shinohara Y, et al. : Comparison of gene expression measurements from cDNA and 60-mer oligonucleotide microarrays. Genomics 2005;85(6):657–665 [DOI] [PubMed] [Google Scholar]
- 51.Jazin EE, Soderstrom S, Ebendal T, et al. : Embryonic expression of the mRNA for the rat homologue of the fusin/CXCR-4 HIV-1 co-receptor. J Neuroimmunol 1997;79(2):148–154 [DOI] [PubMed] [Google Scholar]
- 52.Moepps B, Frodl R, Rodewald HR, et al. : Two murine homologues of the human chemokine receptor CXCR4 mediating stromal cell-derived factor 1alpha activation of Gi2 are differentially expressed in vivo. Eur J Immunol 1997;27(8):2102–2112 [DOI] [PubMed] [Google Scholar]
- 53.Ideker T, Thorsson V, Ranish JA, et al. : Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 2001;292(5518):929–934 [DOI] [PubMed] [Google Scholar]
- 54.Greenbaum D, Colangelo C, Williams K, et al. : Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 2003;4(9):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gygi SP, Rochon Y, Franza BR, et al. : Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 1999;19(3):1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwanhausser B, Busse D, Li N, et al. : Global quantification of mammalian gene expression control. Nature 2011;473(7347):337–342 [DOI] [PubMed] [Google Scholar]
- 57.Ghazalpour A, Bennett B, Petyuk VA, et al. : Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet 2011;7(6):e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogel C. and Marcotte EM: Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012;13(4):227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arai M, Ohashi T, Tsukahara T, et al. : Human T-cell leukemia virus type 1 Tax protein induces the expression of lymphocyte chemoattractant SDF-1/PBSF. Virology 1998;241(2):298–303 [DOI] [PubMed] [Google Scholar]
- 60.Kawaguchi A, Orba Y, Kimura T, et al. : Inhibition of the SDF-1alpha-CXCR4 axis by the CXCR4 antagonist AMD3100 suppresses the migration of cultured cells from ATL patients and murine lymphoblastoid cells from HTLV-I Tax transgenic mice. Blood 2009;114(14):2961–2968 [DOI] [PubMed] [Google Scholar]
- 61.Pflanz S, Timans JC, Cheung J, et al. : IL-27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4(+) T cells. Immunity 2002;16(6):779–790 [DOI] [PubMed] [Google Scholar]
- 62.Niedbala W, Cai B, Wei X, et al. : Interleukin 27 attenuates collagen-induced arthritis. Ann Rheum Dis 2008;67(10):1474–1479 [DOI] [PubMed] [Google Scholar]
- 63.Batten M, Li J, Yi S, et al. : Interleukin 27 limits autoimmune encephalomyelitis by suppressing the development of interleukin 17-producing T cells. Nat Immunol 2006;7(9):929–936 [DOI] [PubMed] [Google Scholar]
- 64.Hamano S, Himeno K, Miyazaki Y, et al. : WSX-1 is required for resistance to Trypanosoma cruzi infection by regulation of proinflammatory cytokine production. Immunity 2003;19(5):657–667 [DOI] [PubMed] [Google Scholar]
- 65.Stumhofer JS, Laurence A, Wilson EH, et al. : Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nat Immunol 2006;7(9):937–945 [DOI] [PubMed] [Google Scholar]
- 66.Yoshimura T, Takeda A, Hamano S, et al. : Two-sided roles of IL-27: Induction of Th1 differentiation on naive CD4+ T cells versus suppression of proinflammatory cytokine production including IL-23-induced IL-17 on activated CD4+ T cells partially through STAT3-dependent mechanism. J Immunol 2006;177(8):5377–5385 [DOI] [PubMed] [Google Scholar]
- 67.Fakruddin JM, Lempicki RA, Gorelick RJ, et al. : Noninfectious papilloma virus-like particles inhibit HIV-1 replication: Implications for immune control of HIV-1 infection by IL-27. Blood 2007;109(5):1841–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frank AC, Zhang X, Katsounas A, et al. : Interleukin-27, an anti-HIV-1 cytokine, inhibits replication of hepatitis C virus. J Interferon Cytokine Res 2010;30(6):427–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greenwell-Wild T, Vazquez N, Jin W, et al. : Interleukin-27 inhibition of HIV-1 involves an intermediate induction of type I interferon. Blood 2009;114(9):1864–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Imamichi T, Yang J, Huang DW, et al. : IL-27, a novel anti-HIV cytokine, activates multiple interferon-inducible genes in macrophages. AIDS 2008;22(1):39–45 [DOI] [PubMed] [Google Scholar]
- 71.Larousserie F, Bardel E, Pflanz S, et al. : Analysis of interleukin-27 (EBI3/p28) expression in Epstein-Barr virus- and human T-cell leukemia virus type 1-associated lymphomas: Heterogeneous expression of EBI3 subunit by tumoral cells. Am J Pathol 2005;166(4):1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Wang G, Sun B, et al. : Interleukin-27 suppresses experimental autoimmune encephalomyelitis during bone marrow stromal cell treatment. J Autoimmun 2008;30(4):222–229 [DOI] [PubMed] [Google Scholar]
- 73.Amadi-Obi A, Yu CR, Liu X, et al. : TH17 cells contribute to uveitis and scleritis and are expanded by IL-2 and inhibited by IL-27/STAT1. Nat Med 2007;13(6):711–718 [DOI] [PubMed] [Google Scholar]
- 74.Miyazaki Y, Shimanoe Y, Wang S, et al. : Amelioration of delayed-type hypersensitivity responses by IL-27 administration. Biochem Biophys Res Commun 2008;373(3):397–402 [DOI] [PubMed] [Google Scholar]
- 75.Grossman WJ, Verbsky JW, Barchet W, et al. : Human T regulatory cells can use the perforin pathway to cause autologous target cell death. Immunity 2004;21(4):589–601 [DOI] [PubMed] [Google Scholar]
- 76.Zhao DM, Thornton AM, DiPaolo RJ, et al. : Activated CD4+CD25+ T cells selectively kill B lymphocytes. Blood 2006;107(10):3925–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hori S, Nomura T, and Sakaguchi S: Control of regulatory T cell development by the transcription factor Foxp3. Science 2003;299(5609):1057–1061 [DOI] [PubMed] [Google Scholar]
- 78.Fontenot JD. and Rudensky AY: A well adapted regulatory contrivance: Regulatory T cell development and the forkhead family transcription factor Foxp3. Nat Immunol 2005;6(4):331–337 [DOI] [PubMed] [Google Scholar]
- 79.Roncador G, Garcia JF, Maestre L, et al. : FOXP3, a selective marker for a subset of adult T-cell leukaemia/lymphoma. Leukemia 2005;19(12):2247–2253 [DOI] [PubMed] [Google Scholar]
- 80.Oh U, Grant C, Griffith C, Fugo K, et al. : Reduced Foxp3 protein expression is associated with inflammatory disease during human T lymphotropic virus type 1 infection. J Infect Dis 2006;193(11):1557–1566 [DOI] [PubMed] [Google Scholar]
- 81.Hayashi D, Kubota R, Takenouchi N, et al. : Reduced Foxp3 expression with increased cytomegalovirus-specific CTL in HTLV-I-associated myelopathy. J Neuroimmunol 2008;200(1–2):115–124 [DOI] [PubMed] [Google Scholar]
- 82.Toulza F, Heaps A, Tanaka Y, et al. : High frequency of CD4+FoxP3+ cells in HTLV-1 infection: Inverse correlation with HTLV-1-specific CTL response. Blood 2008;111(10):5047–5053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Best I, Lopez G, Verdonck K, et al. : IFN-gamma production in response to Tax 161-233, and frequency of CD4+Foxp3+ and Lin HLA-DRhigh CD123+ cells, discriminate HAM/TSP patients from asymptomatic HTLV-1-carriers in a Peruvian population. Immunology 2009;128(1 Suppl):e777–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malec M, van der Velden VH, Bjorklund E, et al. : Analysis of minimal residual disease in childhood acute lymphoblastic leukemia: Comparison between RQ-PCR analysis of Ig/TcR gene rearrangements and multicolor flow cytometric immunophenotyping. Leukemia 2004;18(10):1630–1636 [DOI] [PubMed] [Google Scholar]
- 85.Corti B, Altimari A, Gabusi E, et al. : Potential of real-time PCR assessment of granzyme B and perforin up-regulation for rejection monitoring in intestinal transplant recipients. Transplant Proc 2005;37(10):4467–4471 [DOI] [PubMed] [Google Scholar]
- 86.Sarwal MM, Jani A, Chang S, et al. : Granulysin expression is a marker for acute rejection and steroid resistance in human renal transplantation. Hum Immunol 2001;62(1):21–31 [DOI] [PubMed] [Google Scholar]
- 87.Shin GT, Kim SJ, Lee TS, et al. : Gene expression of perforin by peripheral blood lymphocytes as a marker of acute rejection. Nephron Clin Pract 2005;100(3):c63–70 [DOI] [PubMed] [Google Scholar]
- 88.Vlad G, Ho EK, Vasilescu ER, et al. : Anti-CD25 treatment and FOXP3-positive regulatory T cells in heart transplantation. Transpl Immunol 2007;18(1):13–21 [DOI] [PubMed] [Google Scholar]
- 89.Shi R, Yang J, Jaramillo A, et al. : Correlation between interleukin-15 and granzyme B expression and acute lung allograft rejection. Transpl Immunol 2004;12(2):103–108 [DOI] [PubMed] [Google Scholar]
- 90.Vukmanovic-Stejic M, Zhang Y, Cook JE, et al. : Human CD4+ CD25hi Foxp3+ regulatory T cells are derived by rapid turnover of memory populations in vivo. J Clin Invest 2006;116(9):2423–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Satou Y, Utsunomiya A, Tanabe J, et al. : HTLV-1 modulates the frequency and phenotype of FoxP3+CD4+ T cells in virus-infected individuals. Retrovirology 2012;9:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tachibana N, Okayama A, Ishizaki J, et al. : Suppression of tuberculin skin reaction in healthy HTLV-I carriers from Japan. Int J Cancer 1988;42(6):829–831 [DOI] [PubMed] [Google Scholar]
- 93.Malta TM, Silva IT, Pinheiro DG, et al. : Altered expression of degranulation-related genes in CD8(+) T cells in human T lymphotropic virus Type I infection. AIDS Res Hum Retroviruses 2013;28:826–836 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.