Abstract
A novel HIV-1 circulating recombinant form (CRF) designated CRF65_cpx was recently characterized from three epidemiologically unlinked individuals infected through heterosexual contact in western Yunnan province of China. This is the first complex mosaic HIV-1 CRF, consisting of contributions from three or more different subtypes, identified in China. An additional full-length genome sequence with identical recombinant breakpoints was found among a previously reported recombinant strain from a man who had sex with a man in Anhui province of East Central China. The breakpoint analysis of the recombinants showed a complex genome organization composed of parental subtypes B′ (Thailand variant of subtype B), C, and CRF01_AE, with 13 recombination breakpoints observed in almost all structure genes of HIV-1. The generation of complex recombinant forms is likely due to cocirculation of multiple lineages of HIV-1 strains in high-risk populations in western Yunnan.
The global HIV/AIDS pandemic is dominated by the circulation of highly diversified HIV-1 group M (main) strains that consist of nine subtypes (A to D, F to H, J, and K), subsubtypes, and, at present, more than 60 circulating recombinant forms (CRFs).1 In addition to mutation processes inherent to viral replication, genetic recombination between HIV-1 strains is a significant mechanism that contributes to the genetic diversity and complexity of HIV-1 genomes, resulting in various HIV-1 recombinant strains in the world.2,3 Unique recombinant forms (URFs) are frequently found in many regions, where multiple subtypes and CRFs cocirculate, in so-called “recombination hotspots.”1 URFs may give rise to CRFs if transmission becomes widespread and sustained in the population.1,4
As the epicenter of HIV-1 transmission in China, the southwestern province of Yunnan is considered to be one such recombination hotspot. Over the past 20 years, many HIV-1 strains with different genotypes (B, C, CRF07_BC, CRF08_BC, CRF01_AE, and diverse patterns of URFs) were found in Yunnan. In 1989, HIV-1 subtype B′ (the Thai variant of subtype B) was first found among injection drug users (IDUs) in the southwestern region of Yunnan province.5,6 In the mid-1990s, subtype C strains presumably introduced from India were also identified in IDUs from this region.7,8 Cocirculation of subtypes B' and C in the same places and risk populations enables the generation of recombinants. Many B′C recombinants were identified in the following years.9–14 Two closely related and well-known CRFs (CRF07_BC and CRF08_BC) circulating in China are believed to have come from these recombinants.15,16 In recent years, infections by CRF01_AE have been increasing, and mosaic strains containing CRF01_AE fragments have also frequently arisen in this area,11,12,17 which complicates the HIV-1 epidemic in Yunnan. In this study, we detected a novel HIV-1 circulating recombinant form, designated CRF65_cpx, by near full-length genome (NFLG) analysis, from four individuals with obvious epidemiological linkages.
Three NFLG sequences (YNFL01, YNFL02, and YNFL05) were amplified and sequenced with the methods reported previously.18 The patients were diagnosed as HIV-1 positive between 2009 and 2010 and recruited from a national HIV-1 drug resistance survey in Yunnan. The study was approved by the institutional review boards of the National Center for AIDS/STD Control and Prevention. Extensive searching of the Los Alamos HIV database (www.hiv.lanl.gov) revealed that one CRF01_AE/BC recombinant strain, HF104,19 sampled in 2011 from a man who had sex with men in Anhui province, displayed a high level of similarity with the strains form Yunnan. The epidemiological information of the four patients is listed in Table 1.
Table 1.
Demographic Characteristics of Study Subjects Infected with CRF65_cpx
| Strain name | Sampling year | Sampling region | Sex | Age | Ethnic group | Risk history | Accession number |
|---|---|---|---|---|---|---|---|
| 10CN.YNFL1 | 2010 | Dehong, Yunnan | Female | 23 | Han | Heterosexual | KC870027 |
| 10CN.YNFL2 | 2010 | Dehong, Yunnan | Female | 22 | Dai | Heterosexual | KC870028 |
| 09CN.YNFL5 | 2009 | Dehong, Yunnan | Male | 18 | Han | Heterosexual | KC870030 |
| 11CN.HF104 | 2011 | Hefei, Anhui | Male | NA | Han | MSM | KC183778 |
NA, not available; MSM, men who have sex with men.
Recombination breakpoints were determined using RIP and jpHMM (www.hiv.lanl.gov). Bootscanning and similarity plot analyses were performed to define their recombinant breakpoints using SimPlot 3.2. The breakpoint confirmation and origin of each segment were then analyzed by subregion phylogenetic trees. The reference sequence alignments including all of HIV-1 group M, group P, Thai subtype B (M041, M081, M145), CRF01_AE, CRF07_BC, and CRF08_BC sequences were downloaded from the Los Alamos National Laboratory HIV Database (www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html#ref). An initial alignment was performed using Clustal W, and then adjusted manually using BioEdit.20 Phylogenetic tree analyses were implemented in MEGA 5.0 using the neighbor-joining method with the Kimura two-parameter model and 1,000 bootstrap replications test. Branches with bootstrap values higher than 0.7 were considered as phylogenetic clusters.
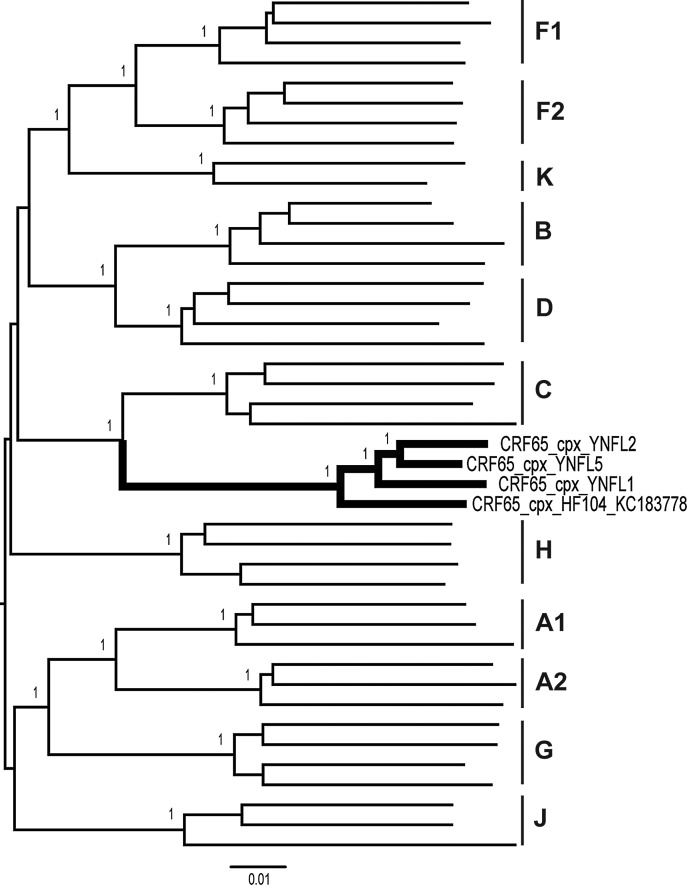
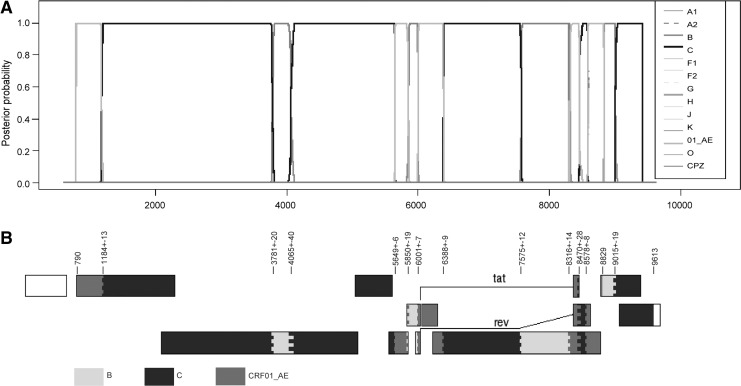
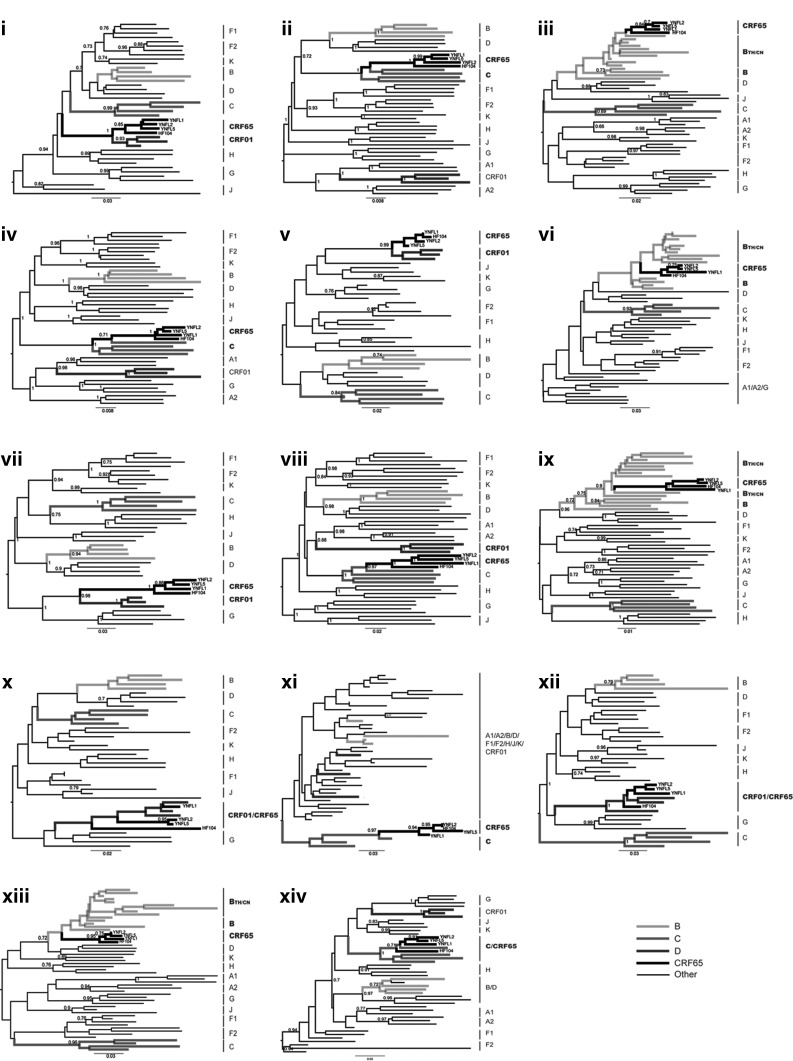
As shown in Fig. 1, the four strains formed a distinct and well-supported (bootstrap value=100%) monophyletic cluster, distantly related to all known HIV-1 subtype/CRFs. Recombination analysis shows that the sequences were composed of subtypes B, C, and CRF01_AE (Fig. 2). The breakpoints were defined by HIV Sequence Locator (www.hiv.lanl.gov/content/sequence/LOCATE/locate.html) relative to the HXB2 coordinates. The mosaic structure of the recombinant genome is described as follows: ICRF01_AE (790–1,170 nt), IIC (1,197–3,760 nt), IIIB (3,802–4,024 nt), IVC (4,105–5,643 nt), VCRF01_AE (5,655–5,830 nt), VIB (5,870–5,593 nt), VIICRF01_AE (6,009–6,378 nt), VIIIC (6,398–7,562 nt), IXB (7,588–8,301 nt), XCRF01_AE (8,331–8,441 nt), XIC (8,499–8,569 nt), XIICRF01_AE (8,587–8,828 nt), XIIIB (8,829–8,995 nt), and XIVC (9,035–9,613 nt). Subregion phylogenetic analyses were used to verify the breakpoints, which were well confirmed by bootstrap values greater than 0.7 except for VI, XI, and XIII. The main reason for reduced confidence on these regions is likely to be their short lengths (125 bp, 72 bp, and 167 bp, respectively). Further analysis of the three regions was performed using the HIV Genotyping Tool from NCBI (www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi), and results supported our findings above (data not shown). Subregion neighbor-joining trees (Fig. 3 III, VI, IX, and XIII) also show that clusters of the newly identified CRF are much closer to subtype B′ (a variant of subtype B prevalent in Thailand and China, defined as BTH/CN) than other subtype B clades; hence, the parental origin of the B segments in the CRF is most likely from the Thai B lineage. The recombinant structure of the new CRF is distinct from any known CRFs. Since the CRF consists of contributions from three different subtypes, it is designated CRF65_cpx (complex),4 and is the first complex CRF identified in China.
FIG. 1.
Phylogenetic tree analysis of the near full-length genome (NFLG) sequences of CRF65_cpx. All HIV-1 group M reference sequences were used to construct the neighbor-joining phylogenetic tree with the Kimura two-parameter model and 1,000 bootstrap replications test. The tree branches of CRF65_cpx are displayed as a bold line.
FIG. 2.
Recombinant analysis of the novel identified CRF65_cpx. (A) Posterior probabilities analysis of the recombinants calculated by jpHMM. Inset: the different symbols represent different references. (B) Mosaic structure of CRF65_cpx, which was generated by using the Recombinant HIV-1 Drawing Tool (www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html). The±refers to the ambiguous region in which the recombinant may occur and cannot be defined by the subtype to which it belongs.
FIG. 3.
Subregion tree analysis of the novel identified CRF65_cpx. The phylogenetic trees of the 14 mosaic segments defined by bootscanning plot analysis were constructed with MEGA 5.0 using the neighbor-joining method. The stability of the nodes was assessed by bootstrap analysis with 1,000 replications, and bootstrap values of 0.7 and higher are shown. The subtypes of sequences are listed on the right side of the trees, and the sequences of subtype B, C, CRF01_AE and CRF65_cpx are bold marked, respectively. The scale bars are shown at the bottom of each tree.
Due to Yunnan's unique geographic position, history, and the social behavior patterns of local ethnicities, the region plays a very important role in the introduction and spread of HIV in China.5,7,21–24 From the earliest epidemic of subtype B in 1989, subtype C, CRF07_BC, and CRF08_BC appeared here one after another.11,12 In more recent decades, CRF01_AE has seen widespread transmission nationally,25 and recombinants containing CRF01_AE segments have been reported in different regions and populations.9,19,26,27 In this study, we characterized a novel HIV-1 circulating recombinant form from three epidemiologically unlinked individuals infected through the heterosexual route in the western Yunnan province of China. Of note, we also identified another HIV-1 isolate from previous studies that shares the same breakpoint structure with new identified strains from a man who had sex with men in Anhui province approximately 2,000 km away. Further molecular epidemiological investigation is needed to identify the influence of CRF65_cpx on the HIV epidemic in China. The emergence of CRF65_cpx indicates the increasing complexity of HIV-1 genomes in China, which may complicate disease prevention and treatment efforts that depend on genotype-specific mechanisms such as vaccine development.
Sequence Data
The nucleotide sequence of the three isolates YNFL01, YNFL02, and YNFL05 has been submitted to GenBank with the accession numbers KC870027, KC870028, and KC870030, respectively.
Acknowledgments
This work was supported by the National Major Projects for Infectious Diseases Control and Prevention Grants (2012ZX10001-008 and 2012ZX10001-002), the National Natural Science Foundation of China Grants (81020108030 and 81261120393), the State Key Laboratory for Infectious Disease Development Grant (2012SKLID103), and the International Development Research Center of Canada Grant (104519-010). We would like to thank Brian Foley from the Los Alamos National Laboratory for advice on CRF nomenclature.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Hemelaar J: The origin and diversity of the HIV-1 pandemic. Trends Mol Med 2012;18(3):182–192 [DOI] [PubMed] [Google Scholar]
- 2.McCutchan FE: Understanding the genetic diversity of HIV-1. AIDS 2000;14(Suppl 3):S31–S44 [PubMed] [Google Scholar]
- 3.Robertson DL, Sharp PM, McCutchan FE, and Hahn BH: Recombination in HIV-1. Nature 1995;374(6518):124–126 [DOI] [PubMed] [Google Scholar]
- 4.Robertson DL, Anderson JP, Bradac JA, et al. : HIV-1 nomenclature proposal. Science 2000;288(5463):55–56 [DOI] [PubMed] [Google Scholar]
- 5.Shao Y, Chen Z, Wang B, Zeng Y, Zhao S, and Zhang Z: Isolation of viruses from HIV infected individuals in Yunnan. Chin J Epidemiol 1991;12:129 [Google Scholar]
- 6.Zheng X, Tian C, Choi KH, et al. : Injecting drug use and HIV infection in southwest China. AIDS 1994;8(8):1141–1147 [DOI] [PubMed] [Google Scholar]
- 7.Luo CC, Tian C, Hu DJ, Kai M, Dondero T, and Zheng X: HIV-1 subtype C in China. Lancet 1995;345(8956):1051–1052 [DOI] [PubMed] [Google Scholar]
- 8.Shao Y, Guan Y, Zhao Q, et al. : Genetic variations and molecular epidemiology of the Ruili HIB-1 strains of Yunnan in 1995. Chin J Virol 1996;12(1):9–17 [Google Scholar]
- 9.Yang R, Xia X, Kusagawa S, Zhang C, Ben K, and Takebe Y: On-going generation of multiple forms of HIV-1 intersubtype recombinants in the Yunnan Province of China. AIDS 2002;16(10):1401–1407 [DOI] [PubMed] [Google Scholar]
- 10.Qiu Z, Xing H, Wei M, et al. : Characterization of five nearly full-length genomes of early HIV type 1 strains in Ruili city: Implications for the genesis of CRF07_BC and CRF08_BC circulating in China. AIDS Res Hum Retroviruses 2005;21(12):1051–1056 [DOI] [PubMed] [Google Scholar]
- 11.Pang W, Zhang C, Duo L, et al. : Extensive and complex HIV-1 recombination between B', C and CRF01_AE among IDUs in south-east Asia. AIDS 2012;26(9):1121–1129 [DOI] [PubMed] [Google Scholar]
- 12.Li L, Chen L, Yang S, et al. : Recombination form and epidemiology of HIV-1 unique recombinant strains identified in Yunnan, China. PLoS One 2012;7(10):e46777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Chen L, Yang S, et al. : Near full-length genomic characterization of a novel HIV type 1 subtype B/C recombinant strain from Yunnan, China. AIDS Res Hum Retroviruses 2010;26(6):711–716 [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Li L, Yang S, et al. : Identification and characterization of two new HIV type 1 unique (B/C) recombinant forms in China. AIDS Res Hum Retroviruses 2011;27(4):445–451 [DOI] [PubMed] [Google Scholar]
- 15.Takebe Y, Liao H, Hase S, et al. : Reconstructing the epidemic history of HIV-1 circulating recombinant forms CRF07_BC and CRF08_BC in East Asia: The relevance of genetic diversity and phylodynamics for vaccine strategies. Vaccine 2010;28(Suppl 2):B39–B44 [DOI] [PubMed] [Google Scholar]
- 16.Tee KK, Pybus OG, Li XJ, et al. : Temporal and spatial dynamics of human immunodeficiency virus type 1 circulating recombinant forms 08_BC and 07_BC in Asia. J Virol 2008;82(18):9206–9215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei H, Su L, Feng Y, et al. : Near full-length genomic characterization of a novel HIV type 1 CRF07_ BC/01_AE recombinant in MSM from Sichuan, China. AIDS Res Hum Retroviruses 2013; 29(8):1173–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, He X, Wang Z, et al. : Tracing the origin and history of HIV-1 subtype B' epidemic by near full-length genome analyses. AIDS 2012;26(7):877–884 [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Meng Z, Xu J, et al. : New emerging recombinant HIV-1 strains and close transmission linkage of HIV-1 strains in the Chinese MSM population indicate a new epidemic risk. PLoS One 2013;8(1):e54322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall TA: BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Paper presented at the Nucleic Acids Symposium Series 1999 [Google Scholar]
- 21.Cheng H, Zhang J, Capizzi J, Young NL, and Mastro TD: HIV-1 subtype E in Yunnan, China. Lancet 1994;344(8927):953–954 [DOI] [PubMed] [Google Scholar]
- 22.Shao Y, Zhao F, Yang W, Zhang Y, and Gong X: The identification of recombinant HIV-1 strains in IDUs in southwest and northwest China. Chin J Exp Clin Virol 1999;13:109. [PubMed] [Google Scholar]
- 23.Su L, Graf M, Zhang Y, et al. : Characterization of a virtually full-length human immunodeficiency virus type 1 genome of a prevalent intersubtype (C/B') recombinant strain in China. J Virol 2000;74(23):11367–11376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beyrer C, Razak MH, Lisam K, Chen J, Lui W, and Yu XF: Overland heroin trafficking routes and HIV-1 spread in south and south-east Asia. AIDS 2000;14(1):75–83 [DOI] [PubMed] [Google Scholar]
- 25.Feng Y, He X, Hsi JH, et al. : The rapidly expanding CRF01_AE epidemic in China is driven by multiple lineages of HIV-1 viruses introduced in the 1990s. AIDS 2013;27(11):1793–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Li L, Bao Z, et al. : Identification of a novel HIV type 1 circulating recombinant form (CRF52_01B) in Southeast Asia. AIDS Res Hum Retroviruses 2012;28(10):1357–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Chen L, Yang S, et al. : Recombination form and epidemiology of HIV-1 unique recombinant strains identified in Yunnan, China. PLoS One 2012;7(10):e46777. [DOI] [PMC free article] [PubMed] [Google Scholar]