Abstract
Generation of induced pluripotent stem cells (iPSCs) via the ectopic expression of reprogramming factors is a simple, advanced, yet often perplexing technology due to low efficiency, slow kinetics, and the use of numerous distinct systems for factor delivery. Scientists have used almost all available approaches for the delivery of reprogramming factors. Even the well-established retroviral vectors confuse some scientists due to different tropisms in use. The canonical virus-based reprogramming poses many problems, including insertional mutagenesis, residual expression and re-activation of reprogramming factors, uncontrolled silencing of transgenes, apoptosis, cell senescence, and strong immunogenicity. To eliminate or alleviate these problems, scientists have tried various other approaches for factor delivery and transgene removal. These include transient transfection, nonintegrating viral vectors, Cre-loxP excision of transgenes, excisable transposon, protein transduction, RNA transfection, microRNA transfection, RNA virion, RNA replicon, nonintegrating replicating episomal plasmids, minicircles, polycistron, and preintegration of inducible reprogramming factors. These alternative approaches have their own limitations. Even iPSCs generated with RNA approaches should be screened for possible transgene insertions mediated by active endogenous retroviruses in the human genome. Even experienced researchers may encounter difficulty in selecting and using these different technologies. This survey presents overviews of iPSC technologies with the intention to provide a quick yet comprehensive reference for both new and experienced reprogrammers.
Introduction
Generation of induced pluripotent stem cells (iPSCs) is a long process with a low efficiency. Conventional iPSC technology (or factor reprogramming) is based on integrating vectors [1,2], which have problems of cell death, residual expression and re-activation of reprogramming factors [3], immunogenicity [4], uncontrolled silencing of transgenes, and insertional mutagenesis. To address these issues, numerous alternative approaches have been developed. Figure 1 summarizes systems and strategies for such efforts. Approaches to factor reprogramming broadly fall into two categories: chemical and transgene reprogramming. Many small molecules are reported to promote reprogramming when used with the canonical reprogramming factors [5–8]. Recently, mouse iPSCs were generated exclusively with a combination of seven small-molecule compounds without resort to any transgene [9]. There are many forms of transgene reprogramming, and these are classified into three groups: direct cell transduction of gene products (protein transduction), RNA- and DNA-based reprogramming. RNA reprogramming can be achieved through transfection/transduction of synthetic mRNAs, microRNAs (miRNAs), RNA viruses, or synthetic RNA replicons. DNA-based technologies are the most widely used, and they also take three major forms: virus particles, transposons, and plasmids. Viruses can be retroviruses or DNA viruses. Retroviral vectors are included in this category, because these vectors have a DNA phase. Retroviral reprogramming is the founding method, and it includes gamma retroviral vectors and human immunodifficiency virus 1 (HIV1)-derived lentiviral vectors (LV). The DNA adenovirus was later used to avoid integration of transgenes into the reprogrammed genomes. PiggyBac transposons were used to integrate the reprogramming factors for lasting ectopic expression, and subsequent excision after the completion of reprogramming. Reprogramming plasmids can be circular, or they can be linearized for enhanced integration into the genome to achieve lasting expression for efficient reprogramming. Circular reprogramming plasmids are used to avoid integration, and they include conventional and episomal plasmids. Minicircle DNA is grouped into plasmids in Fig. 1, as this circular, supercoiled DNA molecule resembles a plasmid.
FIG. 1.
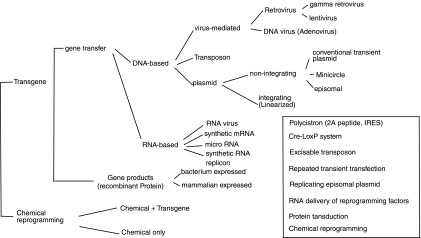
Systems and strategies for factor-mediated pluripotent reprogramming. IRES, internal ribosome entry site.
The first batch of mouse and human iPSC lines were generated using virus-based genome-integrating systems due to the need for lasting transgene expression required for succesful reprogramming. However, any insertion of foreign DNA into the reprogrammed genome will pose a risk of insertional mutagenesis. A second major concern with integrating systems is reactivation and residual expression of the integrated reprogramming factors. All of the reprogramming factors are tumorigenic to some extent with c-Myc as the most notorious oncogene [10]. Actually, c-Myc was found to be responsible for the tumors found in iPSC chimeric mice [3,11]. Therefore, integrating vectors are not the choice for reprogramming when safety becomes a concern. Reduction or complete elimination of transgene integrations has been one of the major goals for all of the improvements cited earlier. The inset of Fig. 1 depicts the strategies for such efforts, which include (i) use of a polycistron to reduce the number of integrations; (ii) use of the Cre-LoxP system to excise the transgenes from the reprogrammed genome; (iii) direct delivery of reprogramming RNA (synthetic mRNA, RNA virus, RNA replicon, or miRNA) to avoid integration of the transgene sequences; (iv) transposon transposition of transgenes into the reprogramming genomes, and subsequent excision of transgenes from the reprogrammed genomes; (v) repeated transfections of cells with nonreplicating plasmids; (vi) protein transduction; (vii) use of reprogramming-promoting small molecules; and (viii) use of nonintegrating and replicating episomal plasmids.
Given the fact that almost all systems of gene delivery have been employed in factor reprogramming, even experienced reprogrammers encounter difficulty and frustration in choosing which approaches to use for a specific reprogramming project. For example, the most widely used retroviral vectors include three different systems representing various tropisms: ecotropism, amphotropism, and pantropism. Which system fits a specific reprogramming experiment will depend on cell types to be reprogrammed, end use of the iPSCs, and the nature of a reprogramming project. Wise selection of reprogramming vehicles is the key to success, and it requires knowledge of the technologies of various factor delivery systems. Therefore, a survey of different iPSC reprogramming technologies is provided. A supporting companion review on vectorology and factor delivery systems used in factor reprogramming is available [12]. Chemical reprogramming is not covered here, because it does not involve a special technology other than a simple inclusion of reprogramming chemicals in the reprogramming medium. Several reviews on chemical reprogramming are available [13–17].
Reprogramming with Retroviral Vectors
Retroviral vectors [12] are the most widely used vehicles for gene therapy and transgene delivery in cultured cells and animal models [18]. They also played a seminal role in the historical development of iPSC technology due to their ability to efficiently transduce murine cells for a lasting expression of transgenes. After screening 24 murine genes using ecotropic retroviral vectors, Yamanaka and Takahashi found that the quadruplet of Oct4 (also known as Pou5f1), Sox2, c-Myc, and Klf4 can convert murine somatic cells into embryonic stem cell (ESC)-like cells (iPSC) [1]. The early and many ensuing murine reprogramming experiments employed retroviral vectors [1,3,19–21]. However, ecotropic retroviral vectors do not transduce human cells, yet human iPSCs were generated with an ecotropic retrosystem after the human fibroblasts were sensitized by a murine viral receptor, mCAT1, delivered by LV [19,22,23]. This extra step could have been avoided if the authors had used pantropic retroviral vectors or LV for the delivery of reprogramming genes (see below). A second pseudotype of retroviral vectors, amphotropic retroparticles, transduces human cells at a low efficiency (16% for fibroblasts) that is generally insufficient for reprogramming although some human cells, including human mesenchymal stem cells [24] and neonatal dermal fibroblasts [25], are reported to be reprogrammed by amphotropic vectors. However, amphotropic retroparticles appear to transduce human keratinocytes efficiently with two rounds of spinoculations, and reprogrammed human keratinocytes into iPSCs [26,27]. Although transduction efficiency of human CD133+ cells with amphotropic retroviral vectors is as low as 28%, CD133+ cells were still reprogrammed by three rounds of viral transductions [28] due to the high reprogrammability of hematopoietic stem cells [28,29]. Amphotropic retroviral vectors can reprogram nonhuman primate fibroblasts [30,31]. A third type of retroviral vectors, pantropic retroviral vectors pseudotyped with vesicular stomatitis virus G (VSV-G) protein, can transduce human cells more efficiently. VSV-G-pseudotyped retroviral vectors reprogrammed activated human T cells [32], human umbilical vein endothelial cells [33], human adipose stem cells (hASCs) [34,35], fibroblasts [36,37], human neural stem cells (hNSCs) [38], and diseased human fibroblasts [39]. Pantropic retroviral vectors also enable reprogramming of nonhuman primate cells [40,41]. Recently, human cells from urine samples were reprogrammed surprisingly with ecotropic retroviral vectors [42,43]. This success indicates that ecotropic retroviral vectors may transduce some types of human cells. The efficiency of many human reprogramming experiments using amphotropic or ecotropic vectors could have been significanly increased if the authors had used pantropic retroviral vectors. Retroviral vectors transduce only dividing cells, and, therefore, nondividing cells and slow-growing cells are more difficult to be reprogrammed with retroviral vectors. For example, due to low transduction efficiency with retroviral vectors, researchers used preintegrated reprogramming factors to reprogram mouse B cells, for which the starting B cells were isolated from transgenic mice that harbored inducible reprogramming factors [44]. Such a requirement of cell division for transduction may explain the frequent use of repeated transductions in many retroviral reprogramming protocols. Since retroviral vectors transduce only cells that divide shortly after infection, consecutive transductions increase the population of infected cells. Reprogramming with retroviral vectors might be compromised by premature silencing of reprogramming factors [45], by de novo methytransferase Dnmt3a and Dnmt3b [46], and by the retroviral silencing factor TRIM28 [45,47]. Retroviral vector silencing starts as early as day 4 of reprogramming [45]. Viral integration is not required for successul reprogramming, as many nonintegration approaches have proved to be capable of reprogramming [9,31,48–53]. However, viral transduction per se surprisingly promotes reprogramming due to favorable epigenetic changes provoked by viral activation of the TLR3 innate immune response pathway [54]. The titers of ecotropic and amphotropic vectors decrease dramatically after concentration and/or freezing, and, therefore, fresh preparations of retroviral vectors are usually used in retroviral reprogramming protocols [55]. This is less convenient than the standard LV, which can be concentrated and frozen without significant loss of titer. Nevertheless, pantropic RV pseudotyped with VSV-G can be concentrated and frozen for later use in reprogramming [56]. In addition to many other drawbacks of retroviral reprogramming (insertional mutagenesis, transgene reactivation, and residual expression), retroviral vector-generated iPSCs are much more immunogenic than those induced with nonviral vectors [4], which compromises the value of such cells in clinical applications.
Reprogramming with LV
LV [12] are another widely used vehicle for the delivery of transgenes in reprogramming. Due to the broadened tropism with VSV-G pseudotyping, LV are ubiquitously used for reprogramming of human cells [2,57–59], murine cells [45,58,60], rat cells [61], and pig cells [62]. One advantage of LV is their abilities to transduce nondividing cells. Due to this trait, LV were used to successfully reprogram murine terminal β cells, which divide only one time in culture [60]. Although LV tranduce nondividing cells, repeated transductions were still employed for reprogramming of mouse and human melanocytes [58]. LV are reported to be more efficient in reprogramming than retroviral vectors because of slower silencing of reprogramming factors [45]. Inducible LV are useful tools in the study of reprogramming [44,45,58,63–65]. The use of inducible LV enables secondary reprogramming to be performed with cells differentiated from iPSCs previously generated with such vectors [44,58]. Such secondary cells avoid inefficient viral transduction of multiple reprogramming factors, and they are more homogeneous in copy number and insertion position of reprogramming factors [64]. Therefore, inducible LV furnish a valuable tool for the study of reprogramming mechanisms owing to the homogeneity and controlled expression of transgenes [45,64]. For example, inducible LV allow for the demonstration that the reprogramming of mouse fibroblasts requires at least 8 days of expression of exogenous reprogramming factors, and that transgene expression is still required for a period of time beyond the point at which Oct4 or Sox2 is activated [45]. Different reprogramming genes have a great impact on viral titer. For example, in our experience, LV encoding human KLF4 or LIN28 give titers of 2–10 times or 10–50 times lower, respectively, than those of LV encoding the OCT4 and c-MYC reprograming factors (our unpublished observations).
Reprogramming by Transient Transfection
Although transient transfection was not the choice of methods in the pioneering screening reprogramming, it turns out that transient transfection can generate both mouse and human iPSCs, albeit at a very low efficiency [51,66,67]. Yamanaka's group reported the successful generation of mouse iPSCs free of transgene sequences from mouse embryonic fibroblast (MEF) using conventional plasmids [51]. However, integrations were found in some of these iPSC lines; they had to transfect the cells at least four times and the reprogramming efficiency was at least one order lower than that of the virus-based methods. Interestingly, mouse iPSCs were generated by just one or two transfections of transient plasmids when nucleofection was employed. Nucleofection delivers plasmids directly into nuclei, and it gives a strong and early expression of transgenes. However, the majority of the iPSC lines generated by nucleofection harbored random integrations [66]. It is more difficult to generate human iPSCs with transient tranfections. In combination with valproic acid (VPA), Si-Tayeb et al. generated a single human iPSC line with regular plasmids [67]. Another limitation with reprogramming by transient transfection is that most types of normal cells are difficult to transfect with current transfection technologies [68]. For the generation of patient-specific iPSC lines, primary cells should be used as the starting cells. Electroporation can be used to transfect the hard-to-transfect primary cells, but it is not practical to use electroporation if multiple rounds of transfections are required, because electroporation requires detachment of cells (adherent cells). In addition, electroporation usually results in significant cell death. As mentioned earlier, electroporation also promotes transgene integration and this voids the point of using this approach to eliminate integration. A new class of biodegradable, low toxic transfection reagents, poly-β-amino esters, has enhanced transfection efficiency for human cells [69], and facilitates the generation of human iPSCs with transient plasmids [70]. However, as with nucleofection, the enhanced reprogramming may be attributed to the increased integration of the reprogramming factors, as all tested iPSC lines harbored integrated copies of transgenes.
Another factor impairing reprogramming with transient plasmids is the rapid silencing of transgenes because of the bacterial backbone sequences. Minicircles containing a eukaryotic expression cassette without a bacterial backbone provide longer and stronger expression. Such a system is reported to reprogram hASCs into iPSCs, but at a very low efficiency of 0.005% even with three consecutive transfections due to the transient nature of minicircle DNA [71,72]. Literally, six canonical reprogramming factors were acting in their reprogramming, as hASCs express higher levels of KLF4 and c-MYC than ES cells, and the authors used the four reprogramming factors OCT4, SOX2, NANOG, and LIN28. The same set of reprogramming factors in minicircles has not been sucessful in reprogramming human adult dermal fibroblasts although neonatal fibroblasts (IMR90) were reprogrammed into iPSCs at an efficiency 10 times lower than hASC. It is not clear whether a minicircle is really more efficient in reprogramming than regular plasmids, as no side-by-side comparison has ever been done.
Protein Reprogramming
Any DNA-based reprogramming approach will present the possibility of random integration. The chance of integration increases for reprogramming with transient plasmids because of repeated transfections. Nucleofection appears to promote the integration of reprogramming plasmids [51,66]. One ideal method for the generation of transgene-free iPSCs is, therefore, protein transduction [12]. Zhou et al. reported the generation of mouse iPSCs using four rounds of protein transduction of the reprogramming factors tagged with poly-arginine in the presence of VPA [48]. Their reprogramming proteins were produced in Escherichia coli. Protein purification and refolding are, therefore, necessary. Human iPSC lines have also been established with protein transductions without VPA [73]. In contrast to producing the mouse reprogramming proteins in E. coli, the human reprogramming employed reprogramming proteins produced in HEK293 cells. Reprogramming proteins produced in mammalian cells might be superior to those from bacteria, as bacteria lack eukaryotic posttranslational modifications. In addition, the bacterial recombinant proteins may have been mis-folded during the artificial renaturation process, and this might explain why proteins from HEK293 do not require VPA for complete reprogramming. However, this method is extremely inefficient (0.001%), and slow (8 weeks). This low efficacy might partially be a result of the fact that the investigators used whole cell lysates from the expressing cells; purification of the reprogramming factors might improve the efficiency. It was recently shown that activation of the TLR3 pathway due to the innate immune response to viral infection benefits reprogramming, and, indeed, the efficiency of protein reprogramming was increasesed by treatment of the reprogramming cells with the TLR3 agonist poly(I:C) [54]. Interestingly, mouse iPSC lines were successfully established from primary fibroblasts with only one single treatment with cell-free protein extracts from mouse ESCs [74]; while previous reprogramming of mouse NIH/3T3 fibroblasts and human 293T cells with mouse ESC extracts and extracts of human pluripotent carcinoma cells, respectively, generated only partially reprogrammed pluripotent cells [75,76]. Since the acting factors in the reprogramming with pluripotent cell extracts are native proteins without any protein transduction domain, these experiments had to use streptolysin O to reversibly permeate the cell membrane for protein entrance [77].
Reprogramming with RNA Viral Vectors
Sendai virus (SeV) is a nonsegmented negative-strand RNA virus that has an exclusively cytoplasmic replication cycle without any DNA phase. To establish transgene-free iPSCs, cytoplasmic SeV-based vectors (SeVV) [12] have been used to reprogram mouse and human cells into iPSCs [52,78–82]. F-deficient SeV vector (SeVV/ΔF) can efficiently reprogram human fibroblasts [78] and human activated T cells from peripheral blood [52,79]. A portion of the reprogrammed cells lost all viral genomes due to dilution over cell proliferation, and transgene-free iPSCs were subsequently established by removing virus-containing cells through negative selection by treatment of the reprogrammed cells with an antibody against the viral surface protein HN residing on the surface of the reprogrammed cells. The clearance of viral genomes from the established iPSCs can be accelerated by using temperature-sensitive SeVV rendered by point mutations in the viral polymerase genes P and L [81]. iPSCs generated using ts-SeVV lose viral genomes faster than the conventional SeVV/ΔF, and a short temperature shift can virtually remove all viral genomes from iPSCs. The earlier methods use multiple vectors for delivery of the four reprogramming factors, and coordinated expression is compromised due to homologous viral interference when coinfected [80,83]. An SeVV/ΔMΔFΔHN vector was, therefore, developed to deliver all four reprogramming factors in a single vector, in which all structure genes are deleted from the vector and replaced with the four reprogramming factors. Reprogramming with single SeVV/ΔMΔFΔHN was more efficient than with multiple SeVVs, and it might be the most efficient system as it is 100 times more efficient than the standard retroviral reprogramming [80]. Such an SeVV/ΔMΔFΔHN devoid of all structure genes also has the additional merits of enhanced safety and reduced immune response to the infected cells. Without the HN surface protein on the established iPSCs, virus genomes can still be efficiently removed by transfection of the generated iPSCs with siRNAs against the L gene, which encodes the enzyme essential for viral genome replication [80]. Genes delivered by SeVVs cannot be silenced by epigenetic modification, as there is no DNA phase. While this is an advantage in terms of persistent gene expression, it is not sure whether this will be a problem after the endogenous pluripotent genes are activated considering that the expression levels of pluripotent genes should be strictly controlled. Although SeV is not pathogenic to human, it naturally infects the airway epithelium [84], and, therefore, extreme care should be exercised in handing packaged virions when oncogenes such as c-Myc and Klf4 are installed in this vector. SeV is strongly fusogenic [85]. It is not known whether any tetraploid iPSCs have ever been generated in previous research due to cell fusion during reprogramming.
Reprogramming with Synthetic mRNA and Synthetic RNA Replicon
A second RNA strategy for the generation of iPSCs free of transgene integration is to deliver synthetic mRNA encoding reprogramming factors directly into the cells to be reprogrammed. Indeed, Warren et al. [50] converted the human ESC-derived fibroblasts into iPSCs using synthetic mRNA of the Yamanaka factors [OCT4, SOX2, KLF4, and c-MYC (OSKM)]. The addition of LIN28 mRNA to the Yamanaka combination (OSKML) can reprogram more refractory human fibroblasts (D551, MRC-5, BJ, and skin biopsy from an adult cystic fibrosis patient). Due to the transient nature of RNA transfection, daily transfection for appoximately 17 days is required in their protocol. Foreign single-stranded RNA induces innate immune response from the transfected cells through the interferon/NFκB-dependent pathway [50,86]. To make reprogramming successful, modifications of mRNAs are, therefore, necessary to reduce the innate immune response. In addition to removal of the immunogenic 5′ triphosphate from the residual uncapped mRNAs by phosphatase treatment, the reprogramming mRNAs should be modified by both 5-methyl-cytosine and pseudouridine for efficient reprogramming. Medium supplementation with interferon inhibitors is also advantageous. Interestingly, the iPSCs generated by mRNA reprogramming were more closely related to ESCs than those reprogrammed using viral vectors, and the authors suggested that the lack of residual expression of the reprogramming factors in mRNA reprogramming may contribute to this difference. The same mRNA cocktail can also reprogram human cells in a feeder-free system [87]. The key to successful reprogramming with synthetic mRNAs is to avoid the cell death due to a strong innate immune response to long synthetic mRNAs, and this explains why several other groups achieved only partial reprogramming with synthetic unmodified reprogramming mRNAs [86,88,89].
mRNA reprogramming requires repeated tranfections for approximately 17 times, which needs careful optimization of the transfection reagent, media, and procedure. Repeated tranfections exclude the use of electroporation. The need for multiple transfections also limits the use of mRNA reprogramming in hard-to-transfect cells such as blood cells, which are one of the most convenient sources of cells for reprogramming [29,53]. To overcome this, Dowdy's group recently used synthetic RNA replicon to deliver reprogramming factors into human reprogramming cells [90]. Their RNA replicon is based on the Venezuelan equine encephalitis (VEE) virus. VEE virus belongs to alphaviruses that are enveloped, single-stranded, postive-sense RNA viruses without any DNA phase in their life. An alphavirus replicates exclusively in the cytoplasm using the virus-encoded RNA-dependent RNA polymerase (RdRP). A noninfectious, but self-replicating alphaviral vector (RNA replicon) can be constructed by replacing the structure genes with genes of interest. RNA replicons can be delivered into cells in the form of packaged virions or as synthetic RNA genomes (RNA replicons). The latter can be achieved by in vitro transcription using SP6 or T7 promoter systems. On delivery into cells, a synthetic alphaviral RNA genome can self-replicate, as it retains RdRP. RNA replicons need only one transfection for successful reprogramming. However, alphavirus is extremely cytopathic [91,92], and reprogramming cannot be achieved without the suppression of interferon with the B18R protein. A caveat to the use of RNA replicons (SeVV and alphaviral vectors) is that RNA or RNA viruses can be converted into cDNA by possible reverse transcriptases in the human genome, and the resulting cDNA can still integrate [93,94], as demonstrated by the prevailing presence of processed pseudogenes in the human genome [95], and recent experimental evidence for the genomic integration of nonretroviral cDNAs [96]. Such reverse transcriptase activity can emanate from the active members of the human endogenous retroviruses, HERV-K [97,98]. Therefore, even iPSC lines generated by RNA approaches should be carefully examined for possible integrations.
Episomal Reprogramming
Reprogramming with conventional plasmids requires multiple rounds of transfections because of their incapability to replicate in mamallian cells. To overcome this, Yu et al. used Epstein Barr virus (EBV)-based self-replicating episomal vectors to successfully reprogram human fibroblasts [49]. EBV episomal plasmids need only one transfection for successful reprogramming due to their ability to replicate and partition in mammalian cells. However, the efficiency is so low that the four canonical reprogramming factors are not sufficient to drive reprogramming to completion, and a total of five to seven factors are required for the generation of iPSCs [31,49,99–102]. The efficiency of episomal reprogramming can be increased by using blood progenitors as starting cells, but additional factors are still used [29,31,53,100]. The reprogramming efficiency can also be greatly increased using a xenofree culture system developed recently, but seven reprogramming factors were still used for reprogramming human fibroblasts [99]. Using a similar EBV episomal system, OCT4 and NANOG are sufficient to reprogram the hNSC line, ReNcell-VM [103]. This is not surprising, as neural stem cells represent the most reprogrammable cells [103–105] possibly due to their expression of the reprogramming factors SOX2, MYC, and KLF4 [38,104,105]. Integration-free iPSC lines can be established owing to the instability of EBV plasmids in primate cells [106,107]. The episomal iPSCs may have higher quality, as such iPSCs displayed much lower immunogenicity than those generated by viral vectors [4]. The relatively lower efficiency of episomal reprogramming seems intrinsic to the vector itself given the following three observations. First, transgenes in the EBV vector are silenced quickly by human cells due to quick and extensive methylation of the vector sequence [107,108]. Second, although EBV plasmids can replicate efficiently, the establishment of a stable state is an infrequent event, and 30% of the transfected cells lose plasmids with each cell division cycle during the first 2 weeks post-transfection [107]. Third, the “established” EBV plasmids in human cells are still not stable on withdrawal of the selection pressure, and are reported to be lost in 2% to 5% of the cells in each cell division cycle [106,107]. The expression level of Epstein-Barr virus nuclear antigen 1 (EBNA1) in current constructs seems to be a limiting factor, as reprogramming efficiency can be enhanced by either inclusion of a separate EBNA1-expressing plasmid [109], or introduction of the EBNA1-coding mRNA [99]. Recently, it has been reported that an improved EBV vector can reprogram human cord blood CD34+ cells into iPSCs with only two factors (OCT4 and SOX2) [110]. The EBV plasmids cannot replicate in murine cells. Fortunately, polyoma-based episomal vectors can be stably maintained in mouse cells [111,112], and such vectors were used to reprogram murine cells into iPSCs [4].
Polycistronic Reprogramming
To achieve complete reprogramming, four factors are required. Early reprogramming constructs contained a single individual reprogramming gene on each plasmid, and, therefore, at least four integrations were necessary for successful reprogramming due to the use of four separate vectors [1,2]. In fact, it was found that each reprogramming factor had multiple integrations [3,19,20] with as many as 8 copies per reprogrammed genome for a single reprogramming factor [3]. To reduce the number of integrations, single-vector reprogramming systems were developed [66,113–115]. In such systems, the four genes were linked by 2A sequences. This single-vector system expresses the four reprogramming factors as a single transcript, but results in four individual proteins through mechanisms of self-cleavage or ribosomal skip mediated by the 2A peptides [116,117]. The single-vector approach can reduce the number of integrations to one, but integration still exists.
A second advantage by installing all of the four reprogramming genes on one vector is that every infected cell receives a full set of reprogramming genes. It is known that an increased number of different types of viral particles in a mixed viral sample (different genes) decreases the co-infection of an individual cell [118]. However, placing four genes in one vector significantly compromises the packaging efficiency of viral particles due to the increased size of the viral genome. Internal ribosome entry site was also used for co-expression of reprogramming factors in the generation of human iPSCs in combination with multiple promoters [49].
Excisable Reprogramming Constructs
The Cre-Lox system was used to excise integrated transgenes from the reprogrammed genomes. Kaji et al. [119] constructed a single plasmid containing the four reprogramming factors linked by 2A sequences. The polycistron is flanked by Lox sequences. Linearization of the plasmids promotes the integration of the reprogramming cassette into the mouse genome, and therefore permits the reprogramming with a single transfection. The resulting iPSCs were then transfected with Cre recombinase to excise the transgenes. The overall reprogramming efficiency is actually low. This method is not yet demonstrated for the reprogramming of human cells. This approach can remove the risk of residual expression and re-activation of reprogramming factors, but the risk of insertional mutagenesis remains, because any sequence beyond the Lox sequences cannot be excised out. Such insertional mutagenesis by the residual inserted sequence can be reduced or eliminated by selecting a reprogrammed genome that harbors a single integration in an intron of a gene [120]. The same excisable system can be incorperated into LV [113].
A more advanced excision-after-reprogramming approach is the use of PiggyBac (PB) transposon. The PB vector was successfully used to reprogram MEF, and, subsequently, the introduced exogenous sequences were completely excised seamlessly by transient tranfection of the resulting iPSCs with the PB transposase to successfully generate several mouse iPSC lines without any genomic alteration of the reprogrammed genomes [121]. Four individual donor plasmids appear to give rise to too many integrations in the reprogrammed genomes (an average of nine integrations per reprogrammed genome), and the subsequent excision becomes a daunting task due to the low efficiency of transposon excision, and secondary transposition. To facilitate transposon removal, the authors then designed a single-plasmid donor vector containing the four reprogramming genes linked by 2A so as to reduce the number of transposon integrations. The authors actually obtained two mouse iPSC lines with a single integration per line out of 48 iPSC lines, and the integrated sequences were cleanly excised at a frequency of 2% by transient expression of transposase. The authors also reported the generation of human iPSCs by PB transposition, but no human transgene-free line was mentioned in the report. The excision process can be facilitated by reducing the amount of transposon plasmids, but this can decrease reprogramming efficiency too as at least two sets of integrated reprogramming factors are needed for efficient reprogramming [122]. The screening process can be simplified by inclusion of the negative selection gene, HSV-tk, in the transposon cassette [122]. PiggyBac systems provide an avenue to generate iPSC lines without genome alteration. It is also simpler and less immunogenic than the viral systems [123]. There are several concerns though. First, it is still an integration-based method, and therefore requires an extra excision step. Second, in the excision process, the transposase not only excises the transposon, but also transposes transposons into other locations of the genome. This secondary transposition complicates the excision step. Third, due to efficient integration and inefficient excision of this system, an extra screening step for iPSC lines with only a single integration using Southern hybridization is highly preferred before the excision process. It is reported that single integration is suboptimal for reprogramming and represents the rare populations of reprogrammed cells, and thus Yusa et al. had to use iPSC lines with two integrations for the subsequent transposon excision [122]. Fourth, although the transposase cuts the transposons precisely, imperfect excisions occur. Fifth, due to the incomplete removal, imperfect excision (microdeletion and small duplication), and re-integration of the excised copy, a high-resolution screening after the excision is required to ensure the integrity of the reprogrammed genomes. Sixth, during transposition, the transposase cuts the donor plasmid into two parts, and the linearized nontransposon half might have a higher chance to undergo random integrations. The current protocols ignore the possible integrations of nontransposon elements of the donor plasmids. Finally, the human genome harbors many PB-like transposons [124,125] and this raises two concerns. First, it is not known whether these PB-like human “transposases” transpose the introduced PB transposons into any cryptic locations that can no longer be excised by the “canonical” transposase. The second concern is the possible mobilization of human PB-like elements by the introduced active transposase. While the human PB-transposases might be degenerate and inactive, the latter possibility could be highly real given that the cis-elements of PiggyBac transposon are very short. Evidence showed that terminal repeats only are sufficient in transposition although the additional sequences increase the transposition efficiency [126,127]. This low stringency might increase the chance for some homologous sequences in the human genome to be targets of the introduced transposase given that there are around 2,000 PB-like elements dispersed in the human genome [125]. One last particular concern is that the transposases should be used twice (one for reprogramming transfection, and the other for excision). Therefore, double uses of transposase plasmids also increase the risk of integrations of the transposase plasmids. Unfortunately, current screening protocols ignore the possible integrations of this helper plasmid.
Secondary Reprogramming with Preintegrated Inducible Reprogramming Factors
Reprogramming is a slow, inefficient, stochastic process with a long latency. Little is known about this complex process, and it is difficult to study due to its complex nature. Inefficient cell transfection with multiple reprogramming factors is a limiting factor in the study of the reprogramming process. To develop controllable and efficient reprogramming approaches for the study of the reprogramming process, iPSCs were generated using inducible lentiviral or PiggyBac constructs [128,129]. Such iPSCs were then used to generate transgenic mice. Somatic cells harboring the four inducible reprogramming factors were then isolated from these mice for the study of the reprogramming process. Unlike the cells from de novo transduction, these transgenic cells are genetically homogeneous in terms of location and number of transgene integrations. Many types of cells isolated from such iPSC-derived mice can be efficiently reprogrammed into secondary iPSCs by doxycycline induction, and therefore prove that this system is helpful in the study of reprogramming kinetics, drug screening, and screening for additional reprogramming factors and for reprogramming roadblocks [129]. This strategy helped identify C/EBP as a promoter of, and Pax5 as an inhibitor of reprogramming the hard-to-transfect terminally differentiated B cells [44]. Using the same system, Hanna et al. established that reprogramming is a stochastic process, and disproved the elite model of reprogramming [63]. A similar system was also used in human cells [64,130]. Human fibroblasts and keratinocytes have been reprogrammed into iPSCs using inducible lentiviral reprogramming constructs. The resulting human iPSCs were then differentiated into secondary fibroblasts, and theses iPSC-derived fibroblasts were used for secondary reprogramming. Kinetics of human reprogramming was examined using such secondary inducible reprogramming by timed withdrawal of the inducer (doxycycline) [64,130]. A limitation of this system is that some preintegrated factors become noninducible due to silencing (position effect) [130]. Cells from different reprogrammable mouse strains also vary in reprogrammability due to varied integration numbers of reprogramming cassette introduced randomly by LV or PiggyBac transposon [129]. This variability might be overcome by targeting a single inducible reprogramming cassette to a defined locus, such as Col1a1, using Flp recombinase [131].
Adenoviral Reprogramming
Stadtfeld et al. reported the generation of transgene-free mouse iPSCs from hepatocytes using adenoviral vectors [132]. However, they failed to produce iPSCs from mouse fibroblasts with this system. This might be because hepatocytes need lower expression of the reprogramming factors for the generation of iPSCs than fibroblasts, and because adenoviral vectors transduce hepatocytes more efficiently [133]. Even with hepatocytes, reprogramming efficiency is very low compared with retrovirus-mediated reprogramming. This is reflected by the fact that Yamanaka's group failed to generate mouse iPSCs from hepatocytes with adenoviral vectors [51]. The low reprogramming efficiency with adenoviral vectors may be attributed to transgene dilution over time due to cell division. Human iPSC lines were established later from fibroblasts using adenoviral vectors, but extremely high viral titers had to be used (>200 pfu/cell for adenovirus vs 5 cfu/cell for integrating lentiviral or retroviral vectors) [134]. One caveat with adenoviral reprogramming is that adenoviral vectors have an extremely high level of integration compared with naked plasmid DNA [135] even though adenoviral vectors are generally considered nonintegrating vehicles. At much lower multiplicity of infection/transduction (MOI, 10 pfu per cell), they have an integration efficiency of 10−3 to 10−5 per cell, and can reach approximately 1% for some types of cells [135]. As mentioned earlier, reprogramming with adenoviral vectors requires a much higher MOI, and this implies an increased chance of transgene integration. Furthermore, integrated adenoviral vectors undergo rearrangement [135], and this makes it difficult to identify iPSC lines free of foreign sequences.
Reprogramming with miRNA Mimics
miRNAs are noncoding, single-stranded, short regulatory RNAs with around 22 nts. Their main function is the suppression of gene expression by either breakdown of target mRNAs or translation supression [136,137]. It is now well known that miRNAs play major roles in reprogramming [138,139]. Knockdown of Ago2, Drosha, or Dicer, components for miRNA biogenesis, severely impaired reprogramming [140]. Some miRNAs enriched in or specific to ESCs were demonstrated to promote reprogramming in either the form of synthetic mature miRNA or virus-delivered transgenes [140–143]. Unexpectedly, miRNAs can reprogram both human and mouse cells into pluripotent cells without the help of any other reprogramming factors [144–147]. Therefore, reprogramming with synthetic miRNAs may represent an ideal modality for the generation of human iPSCs for clinical application considering several advantages of miRNA mimics [148]. First, they have no risk of genome integration. Second, miRNAs are small, and can be readily synthesized and delivered into cells. In addition, on incorporation into ribonucleoprotein complexes, they remain stable for several days, and, therefore, they function longer in transfected cells than the coding RNAs, which is a basic requirement for the reprogramming process. However, transfection of the reprogramming cells with miRNA mimics still provides only a transient action, and multiple transfections are required for complete reprogramming. Finally, feasibility of miRNA-mimic reprogramming awaits confirmation and/or improvement from more laboratories.
Concluding Remarks
Factor reprogramming to pluripotency is an inefficient, slow, stochastic process requiring the overexpression of multiple genes. Canonical and efficient reprogramming is mediated by integrating viral vectors. Retroviral reprogramming is still a prefered method in research reprogramming, but vector tropisms should be considered. HIV1-based, VSV-G-pseudotyped LV should be a better choice than retroviral vectors due to their broadened tropism, ability to transduce nondividing cells, ease of handling, reprogramming efficiency, and availability of inducible systems. One major goal of reprogramming research since the advent of iPSC technology is to eliminate or reduce transgene integrations so as to avoid insertional mutagenesis, and residual expression and re-activation of reprogramming factors. To achieve this, many alternative approaches have been introduced. One common limitation for most of these improvements is the lower efficiency of reprogramming compared with the founding LV and retroviral vectors. SeVV might be the only method that has a higher reprogramming efficiency, while factor integrations are eliminated. The EBV-based episomal system is the simplest nonintegrating approach with reasonable efficiency, because it needs only one transfection with standard Maxiprep DNA. EBV-episomal reprogramming might be a good choice for clinical reprogramming if immunogenicity of SeVV reprogramming is considered. iPSCs generated with RNA virus/replicon still has the possibility of transgene insertion given the existence of active endogenous retroviruses in human genome. With this said, reprogramming with protein, chemicals, and miRNA mimics might be the most reliable approach to avoid genome alterations. One focus of future studies will be the enhancement of reprogramming efficiency. The four canonical reprogramming factors play different roles and function at different time points of reprogramming, and sequential introductions of the reprogramming factors are reported to be beneficial to reprogramming [149]. The stoichiometry of reprogramming factors affects both reprogramming efficiency and iPSC quality [150,151]. Protocol optimization should consider these facts.
Glossary
Spinoculation: also known as centrifugal infection, a viral transduction/infection process that resorts to centrifugation to enhance deposition of viral particles onto cell surface for increased transduction/infection.
Pseudotype: pseudotype is a virus in which its original native envelope protein is replaced with that of another virus. Pseudotyping is used to alter the tropism (usually expand) of viral vector and enhance the stability of vector particles.
Nucleofection: a proprietary technology for cell transfection through electric shock and cell-type specific buffers, which can efficiently deliver nucleic acid directly into the nucleus for quick and strong expression.
Acknowledgments
The author's research is supported by the UAB new faculty development fund. The author thanks Dr. Kevin Pawlik for his critical reading of this article. He is also grateful to the two anonymous reviewers for their critical review. Their inputs make Table 1 and other improvements possible.
Table 1.
Comparison of Different Reprogramming Methods
| Delivery methods | Efficiency | Cell types | Advantage | Disadvantage | References | ||
|---|---|---|---|---|---|---|---|
| Retroviral vector | Gamma | Ecotropic | High | MEF, mouse gingival fibroblasts, sensitized human fibroblasts | Lasting expression, packaging cell lines available | Insertional mutagenesis, residual expression of reprogramming factors, reactivation of reprogramming factor in iPSC-derived cells, titer loss during viral concentration and storage, infect rodents only, do not infect nondividing cells, do not infect human cells without mCAT1 sensitization, premature silencing, more immunogenic | 1,19,22,23 |
| Amphotropic | High | Human mesenchymal stromal cells, human dermal fibroblasts, human keratinocytes, human CD133+ cells, nonhuman primate fibroblasts | Lasting expressoin, packaging cell lines available | Insertional mutagenesis, residual expression of reprogramming factors, reactivation of reprogramming factors in iPSC-derivaved cells, titer loss during viral concentration and storage, limited tropism, do not infect nondividing cells, premature silencing, more immunogenic, multiple transduction required for human cells | 24–28,30 | ||
| Pantropic | High | Human T cells, HUVEC, human adipose stem cells, human fibroblasts, human NSC, diseased human fibroblasts, monkey fibroblasts and liver cells | Infect wide range of cells, lasting expression | Insertional mutagenesis, residual expression of reprogramming factors, reactivation of reprogramming factors in iPSC-derived cells, do not infect nondividing cells, no efficient packaging cell lines available due to the toxicity of VSV-G, premature silencing, more immunogenic | 32–41 | ||
| Lentiviral | High | Human cells, murine terminal β cells, rat cells, pig cells | Lasting expression of transgenes; infect wide range of cells; easy concentration and storage of virus. Infect nondividing cells, slower silencing, available inducible systems | Insertional mutagenesis, residual expression of reprogramming factors, reactivation of reprogramming factors in iPSC-derived cells, No efficient packaging cell lines avaible due to toxicity of VSV-G | 2,58–62 | ||
| Sendai viral vector | Very high | Human T cells, BJ, HDF, MEF, Human CD34+ cells | Nonintegrating, no premature silencing, broad tropism | Immunogenic, fusogenic, multiple vectors required, expensive, virus curing process needed, screening for integrations may be required, virus preparation | 52,78–82 | ||
| Transient transfection | Very low | MEF, human foreskin fibroblasts | Nonintegrating, inexpensive, no virus preparation | Low efficiency, multiple rounds of transfection, occational integration, difficulty in transfection of primary cells, excludes the use of nucleofection for adherent cells due to multiple transfection, vector silencing | 51,66,67,70,110 | ||
| EBV episomal plasmids | Low | Human CD34+ cells, cord blood MNC, bone marrow MNC, human fibroblast, human NSC | Nonintegrating, inexpensive, no virus preparation, single transfection, transfect primary cells using nucleofection, reasonable efficiency when additional factor used, low immunogenicity | Need additional reprogrammming factors, occasional genomic integration, inefficient, cell death when nucleofection used, does not work with rodent cells, vector silencing | 31,49,53,99–103 | ||
| Protein | Very low, long kinetics | MEF, Human newborn fibroblasts, BJ, Mouse fibroblasts | Abosolute nongenome integration, controllable factor stimulation (timing, concentration and temporal sequence) | Inefficient, multiple transductions, affected by quality of recombinant proteins | 48,54,73,74 | ||
| Synthetic mRNA | Low | ESC-derived fibroblasts, BJ, D551, MRC-5, patient fibroblasts | Nonintegrating, controllable factor stimulation (timing, concentration, and temporal sequence) | Intense innate immune response, multiple transfections, limited cell types, expensive, labor-intensive, challenging transfection, RNA instability, low reprogramming efficiency, additional factor (LIN28 and/or NANOG) required | 50,87 | ||
| Replicating synthetic RNA | Low | BJ, primary HFF | Nonintegrating, single transfection | Strong innate immune response, cytopathic, expensive, inefficient | 90 | ||
| Adenoviral vector | Very low | Hepatocytes, human fibroblasts | Nonintegrating | High spontaneous integration, inefficient, virus-based, limited cell types, high virus titer required, transient expression | 132,134 | ||
| PiggyBac | Low | MEF | Excisible integration, no foot-print, less immunogenic | Insertional mutagenesis, extra excision step, inefficient excision, inefficient reprogramming, integration of helper plasmids | 121,122 | ||
| microRNA mimic | Low | Mouse adipose stromal cells, human adipose stromal cells, HDF | Nonintegrating, easy synthesis, controllable administration (time and concentration) | Inefficient, transient expression, multiple transfections | 146 | ||
| Preintegrated, inducilbe system | Very high | All reprogrammable murine cells from transgenic mice including cells difficult to culture and cells refractory to direct transduction, iPSC-derived human fibroblasts, | Temporal control of expression, reprogram terminally differentiated cells, reprogram hard-to-transfect cells, genetically homogenous cells, good for research reprogramming (genetic and chemical screening, mechanistic study), no direct virus transduction, available transgenic mice | Position effect, insertional mutagenesis, limited power for human cells, not suitable for clinical reprogramming, transgenic cells from current transgenic mice may not be optimal for efficient reprogramming, varied efficiency between available transgenic mice. | 64,128,129 | ||
| Minicircle | Very low | Human adipose stem cells | Nonintegrating, inexpensive, low immunogenic | Inefficient, occasional integration, additional factors used, limited cell types, multiple transfections | 71,72 |
Cell types: listed are generally the types of reprogramming cells used in the references. Efficiency in column 2 is the reprogramming efficiency rated by the author. A percentage is not listed given that such a number is not accurate and may be misleading, because it is affected by cell types, criteria used, and other factors. For example, reprogramming efficiencies were evaluated in different references by AP+ colonies, Tra-1-60+ colonies, SSEA1+ colonies, colony morphology, Oct4-GFP+ colonies, Nanog+ colonies, and number of iPSC lines established. In addition, flowcytometry percentage of marker positive cells in the reprogramming populations was frequently used in the literature. However, examination time has a great impact on this number due to the rapid proliferation of the reprogrammed cells.
MNC, mononulcear cells; HDF, human dermal fibroblasts; NSC, neural stem cells; HFF, human foreskin fibroblasts; HUVEC, human umbilical vein endothelial cells; EBV, Epstein Barr virus; ESC, embryonic stem cell; iPSC, induced pluripotent stem cell; MEF, mouse embryonic fibroblast; VSV-G, vesicular stomatitis virus G protein.
Author Disclosure Statement
The author declares no competing financial interests.
References
- 1.Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920 [DOI] [PubMed] [Google Scholar]
- 3.Okita K, Ichisaka T. and Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448:313–317 [DOI] [PubMed] [Google Scholar]
- 4.Zhao T, Zhang ZN, Rong Z. and Xu Y. (2011). Immunogenicity of induced pluripotent stem cells. Nature 474:212–215 [DOI] [PubMed] [Google Scholar]
- 5.Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H. and Ding S. (2009). Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell 4:16–19 [DOI] [PubMed] [Google Scholar]
- 6.Liang G, Taranova O, Xia K. and Zhang Y. (2010). Butyrate promotes induced pluripotent stem cell generation. J Biol Chem 285:25516–25521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, et al. (2009). A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 5:491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, et al. (2009). Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci U S A 106:8912–8917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, et al. (2013). Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341:651–654 [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa M, Takizawa N, Narita M, Ichisaka T. and Yamanaka S. (2010). Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci U S A 107:14152–14157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N. and Yamanaka S. (2008). Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 26:101–106 [DOI] [PubMed] [Google Scholar]
- 12.Hu K. (2014). Vectorology and factor delivery in induced pluripotent stem cell reprogramming. Stem Cells Dev [Epub ahead of print]; DOI: 10.1089/scd.2013.0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anastasia L, Pelissero G, Venerando B. and Tettamanti G. (2010). Cell reprogramming: expectations and challenges for chemistry in stem cell biology and regenerative medicine. Cell Death Differ 17:1230–1237 [DOI] [PubMed] [Google Scholar]
- 14.Feng B, Ng JH, Heng JCD. and Ng HH. (2009). Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 4:301–312 [DOI] [PubMed] [Google Scholar]
- 15.Xu Y, Shi Y. and Ding S. (2008). A chemical approach to stem-cell biology and regenerative medicine. Nature 453:338–344 [DOI] [PubMed] [Google Scholar]
- 16.Li WL. and Ding S. (2010). Small molecules that modulate embryonic stem cell fate and somatic cell reprogramming. Trends Pharmacol Sci 31:36–45 [DOI] [PubMed] [Google Scholar]
- 17.Li W, Li K, Wei W. and Ding S. (2013). Chemical approaches to stem cell biology and therapeutics. Cell Stem Cell 13:270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cepko CW. and Pear (1996). Overview of the retrovirus transduction system. In: Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA. and Struhl K, eds. John Wiley & Sons, Inc., New York, pp. 9.9.1–9.9.16 [DOI] [PubMed] [Google Scholar]
- 19.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872 [DOI] [PubMed] [Google Scholar]
- 20.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE. and Jaenisch R. (2007). In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448:318–324 [DOI] [PubMed] [Google Scholar]
- 21.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, et al. (2007). Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1:55–70 [DOI] [PubMed] [Google Scholar]
- 22.Egusa H, Okita K, Kayashima H, Yu GN, Fukuyasu S, Saeki M, Matsumoto T, Yamanaka S. and Yatani H. (2010). Gingival fibroblasts as a promising source of induced pluripotent stem cells. PLoS One 5:e12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohta S, Imaizumi Y, Okada Y, Akamatsu W, Kuwahara R, Ohyama M, Amagai M, Matsuzaki Y, Yamanaka S, Okano H. and Kawakami Y. (2011). Generation of human melanocytes from induced pluripotent stem cells. PLoS One 6:e16182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oda Y, Yoshimura Y, Ohnishi H, Tadokoro M, Katsube Y, Sasao M, Kubo Y, Hattori K, Saito S, et al. (2010). Induction of pluripotent stem cells from human third molar mesenchymal stromal cells. J Biol Chem 285:29270–29278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT. and Plath K. (2008). Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A 105:2883–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aasen T. and Izpisua Belmonte JC. (2010). Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nat Protoc 5:371–382 [DOI] [PubMed] [Google Scholar]
- 27.Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, et al. (2008). Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol 26:1276–1284 [DOI] [PubMed] [Google Scholar]
- 28.Giorgetti A, Montserrat N, Aasen T, Gonzalez F, Rodriguez-Piza I, Vassena R, Raya A, Boue S, Barrero MJ, et al. (2009). Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell 5:353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu K. and Slukvin I. (2013). Induction of pluripotent stem cells from umbilical cord blood. In Stem Cells: From Biology to Therapy. Meyers RA, ed. Wiley-Blackwell, Singapore, pp. 345–370 [Google Scholar]
- 30.Wu Y, Zhang Y, Mishra A, Tardif SD. and Hornsby PJ. (2010). Generation of induced pluripotent stem cells from newborn marmoset skin fibroblasts. Stem Cell Res 4:180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu K, Yu J, Suknuntha K, Tian S, Montgomery K, Choi KD, Stewart R, Thomson JA. and Slukvin II. (2011). Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood 117:e109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown ME, Rondon E, Rajesh D, Mack A, Lewis R, Feng X, Zitur LJ, Learish RD. and Nuwaysir EF. (2010). Derivation of induced pluripotent stem cells from human peripheral blood T lymphocytes. PLoS One 5:e11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panopoulos AD, Ruiz S, Yi F, Herrerias A, Batchelder EM. and Izpisua Belmonte JC. (2011). Rapid and highly efficient generation of induced pluripotent stem cells from human umbilical vein endothelial cells. PLoS One 6:e19743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugii S, Kida Y, Berggren WT. and Evans RM. (2011). Feeder-dependent and feeder-independent iPS cell derivation from human and mouse adipose stem cells. Nat Protoc 6:346–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugii S, Kida Y, Kawamura T, Suzuki J, Vassena R, Yin YQ, Lutz MK, Berggren WT, Izpisua Belmonte JC. and Evans RM. (2010). Human and mouse adipose-derived cells support feeder-independent induction of pluripotent stem cells. Proc Natl Acad Sci U S A 107:3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Piza I, Richaud-Patin Y, Vassena R, Gonzalez F, Barrero MJ, Veiga A, Raya A. and Belmonte JCI. (2010). Reprogramming of human fibroblasts to induced pluripotent stem cells under xeno-free conditions. Stem Cells 28:36–44 [DOI] [PubMed] [Google Scholar]
- 37.Park IH, Lerou PH, Zhao R, Huo HG. and Daley GQ. (2008). Generation of human-induced pluripotent stem cells. Nat Protoc 3:1180–1186 [DOI] [PubMed] [Google Scholar]
- 38.Hester ME, Song S, Miranda CJ, Eagle A, Schwartz PH. and Kaspar BK. (2009). Two factor reprogramming of human neural stem cells into pluripotency. PLoS One 4:e7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, et al. (2008). Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321:1218–1221 [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, et al. (2008). Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell 3:587–590 [DOI] [PubMed] [Google Scholar]
- 41.Tomioka I, Maeda T, Shimada H, Kawai K, Okada Y, Igarashi H, Oiwa R, Iwasaki T, Aoki M, et al. (2010). Generating induced pluripotent stem cells from common marmoset (Callithrix jacchus) fetal liver cells using defined factors, including Lin28. Genes Cells 15:959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou T, Benda C, Dunzinger S, Huang Y, Ho JC, Yang J, Wang Y, Zhang Y, Zhuang Q, et al. (2012). Generation of human induced pluripotent stem cells from urine samples. Nat Protoc 7:2080–2089 [DOI] [PubMed] [Google Scholar]
- 43.Zhou T, Benda C, Duzinger S, Huang Y, Li X, Li Y, Guo X, Cao G, Chen S, et al. (2011). Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol 22:1221–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, et al. (2008). Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 133:250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stadtfeld M, Maherali N, Breault DT. and Hochedlinger K. (2008). Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2:230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okano M, Bell DW, Haber DA. and Li E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257 [DOI] [PubMed] [Google Scholar]
- 47.Wolf D. and Goff SP. (2007). TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46–57 [DOI] [PubMed] [Google Scholar]
- 48.Zhou HY, Wu SL, Joo JY, Zhu SY, Han DW, Lin TX, Trauger S, Bien G, Yao S, et al. (2009). Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4:381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II. and Thomson JA. (2009). Human induced pluripotent stem cells free of vector and transgene sequences. Science 324:797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, et al. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7:618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okita K, Nakagawa M, Hong HJ, Ichisaka T. and Yamanaka S. (2008). Generation of mouse induced pluripotent stem cells without viral vectors. Science 322:949–953 [DOI] [PubMed] [Google Scholar]
- 52.Seki T, Yuasa S, Oda M, Egashira T, Yae K, Kusumoto D, Nakata H, Tohyama S, Hashimoto H, et al. (2010). Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell 7:11–14 [DOI] [PubMed] [Google Scholar]
- 53.Hu K. and Slukvin I. (2013). Generation of transgene-free iPSC lines from human normal and neoplastic blood cells using episomal vectors. Methods Mol Biol 997:163–176 [DOI] [PubMed] [Google Scholar]
- 54.Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E. and Cooke JP. (2012). Activation of innate immunity is required for efficient nuclear reprogramming. Cell 151:547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takahashi K, Okita K, Nakagawa M. and Yamanaka S. (2007). Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2:3081–3089 [DOI] [PubMed] [Google Scholar]
- 56.Srinivasakumar N, Zaboikin M, Tidball AM, Aboud AA, Neely MD, Ess KC, Bowman AB. and Schuening FG. (2013). Gammaretroviral vector encoding a fluorescent marker to facilitate detection of reprogrammed human fibroblasts during iPSC generation. PeerJ 1:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K. and Ding S. (2010). Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7:651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Utikal J, Maherali N, Kulalert W. and Hochedlinger K. (2009). Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci 122:3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, et al. (2008). Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell 3:475–479 [DOI] [PubMed] [Google Scholar]
- 60.Stadtfeld M, Brennand K. and Hochedlinger K. (2008). Reprogramming of pancreatic beta cells into induced pluripotent stem cells. Curr Biol 18:890–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao J, Cui C, Chen S, Ren J, Chen J, Gao Y, Li H, Jia N, Cheng L, Xiao H. and Xiao L. (2009). Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell 4:11–15 [DOI] [PubMed] [Google Scholar]
- 62.Wu Z, Chen J, Ren J, Bao L, Liao J, Cui C, Rao L, Li H, Gu Y, et al. (2009). Generation of pig induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol 1:46–54 [DOI] [PubMed] [Google Scholar]
- 63.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A. and Jaenisch R. (2009). Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462:595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C. and Hochedlinger K. (2008). A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 3:340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eminli S, Foudi A, Stadtfeld M, Maherali N, Ahfeldt T, Mostoslavsky G, Hock H. and Hochedlinger K. (2009). Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet 41:968–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonzalez F, Monasterio MB, Tiscornia G, Pulido NM, Vassena R, Morera LB, Piza IR. and Belmonte JCI. (2009). Generation of mouse-induced pluripotent stem cells by transient expression of a single nonviral polycistronic vector. Proc Natl Acad Sci U S A 106:8918–8922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Si-Tayeb K, Noto FK, Sepac A, Sedlic F, Bosnjak ZJ, Lough JW. and Duncan SA. (2010). Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Dev Biol 10:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dickens S, Van den Berge S, Hendrickx B, Verdonck K, Luttun A. and Vranckx JJ. (2010). Nonviral transfection strategies for keratinocytes, fibroblasts, and endothelial progenitor cells for ex vivo gene transfer to skin wounds. Tissue Eng Part C Methods 16:1601–1608 [DOI] [PubMed] [Google Scholar]
- 69.Yang F, Green JJ, Dinio T, Keung L, Cho SW, Park H, Langer R. and Anderson DG. (2009). Gene delivery to human adult and embryonic cell-derived stem cells using biodegradable nanoparticulate polymeric vectors. Gene Ther 16:533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Montserrat N, Garreta E, Gonzalez F, Gutierrez J, Eguizabal C, Ramos V, Borros S. and Belmonte JCI. (2011). Simple generation of human induced pluripotent stem cells using poly-beta-amino esters as the non-viral gene delivery system. J Biol Chem 286:12417–12428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, et al. (2010). A nonviral minicircle vector for deriving human iPS cells. Nat Methods 7:197–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Narsinh KH, Jia FJ, Robbins RC, Kay MA, Longaker MT. and Wu JC. (2011). Generation of adult human induced pluripotent stem cells using nonviral minicircle DNA vectors. Nat Protoc 6:78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R. and Kim KS. (2009). Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4:472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cho HJ, Lee CS, Kwon YW, Paek JS, Lee SH, Hur J, Lee EJ, Roh TY, Chu IS, et al. (2010). Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood 116:386–395 [DOI] [PubMed] [Google Scholar]
- 75.Taranger CK, Noer A, Sorensen AL, Hakelien AM, Boquest AC. and Collas P. (2005). Induction of dedifferentiation, genomewide transcriptional programming, and epigenetic reprogramming by extracts of carcinoma and embryonic stem cells. Mol Biol Cell 16:5719–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rajasingh J, Lambers E, Hamada H, Bord E, Thorne T, Goukassian I, Krishnamurthy P, Rosen KM, Ahluwalia D, et al. (2008). Cell-free embryonic stem cell extract-mediated derivation of multipotent stem cells from NIH3T3 fibroblasts for functional and anatomical ischemic tissue repair. Circ Res 102:e107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walev I, Bhakdi SC, Hofmann F, Djonder N, Valeva A, Aktories K. and Bhakdi S. (2001). Delivery of proteins into living cells by reversible membrane permeabilization with streptolysin-O. Proc Natl Acad Sci U S A 98:3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fusaki N, Ban H, Nishiyama A, Saeki K. and Hasegawa M. (2009). Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad B Phys 85:348–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seki T, Yuasa S. and Fukuda K. (2012). Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus. Nat Protoc 7:718–728 [DOI] [PubMed] [Google Scholar]
- 80.Nishimura K, Sano M, Ohtaka M, Furuta B, Umemura Y, Nakajima Y, Ikehara Y, Kobayashi T, Segawa H, et al. (2011). Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. J Biol Chem 286:4760–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, Takada N, Inoue M, Hasegawa M, et al. (2011). Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A 108:14234–14239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Macarthur CC, Fontes A, Ravinder N, Kuninger D, Kaur J, Bailey M, Taliana A, Vemuri MC. and Lieu PT. (2012). Generation of human-induced pluripotent stem cells by a nonintegrating RNA sendai virus vector in feeder-free or xeno-free conditions. Stem Cells Int 2012:564612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shimazu Y, Takao S, Irie T, Kiyotani K, Yoshida T. and Sakaguchi T. (2008). Contribution of the leader sequence to homologous viral interference among Sendai virus strains. Virology 372:64–71 [DOI] [PubMed] [Google Scholar]
- 84.Yonemitsu Y, Kitson C, Ferrari S, Farley R, Griesenbach U, Judd D, Steel R, Scheid P, Zhu J, et al. (2000). Efficient gene transfer to airway epithelium using recombinant Sendai virus. Nat Biotechnol 18:970–973 [DOI] [PubMed] [Google Scholar]
- 85.Okada Y. (1993). Sendai virus-induced cell-fusion. Method Enzymol 221:18–41 [DOI] [PubMed] [Google Scholar]
- 86.Angel M. and Yanik MF. (2010). Innate immune suppression enables frequent transfection with RNA encoding reprogramming proteins. PLoS One 5:e11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Warren L, Ni Y, Wang J. and Guo X. (2012). Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Sci Rep 2:657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yakubov E, Rechavi G, Rozenblatt S. and Givol D. (2010). Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem Biophys Res Commun 394:189–193 [DOI] [PubMed] [Google Scholar]
- 89.Plews JR, Li J, Jones M, Moore HD, Mason C, Andrews PW. and Na J. (2010). Activation of pluripotency genes in human fibroblast cells by a novel mRNA based approach. PLoS One 5:e14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yoshioka N, Gros E, Li HR, Kumar S, Deacon DC, Maron C, Muotri AR, Chi NC, Fu XD, Yu BD. and Dowdy SF. (2013). Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell 13:246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Petrakova O, Volkova E, Gorchakov R, Paessler S, Kinney RM. and Frolov I. (2005). Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in Mammalian cells. J Virol 79:7597–7608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gorchakov R, Frolova E, Williams BR, Rice CM. and Frolov I. (2004). PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J Virol 78:8455–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu K. (2006). Intron exclusion and the mystery of intron loss. FEBS Lett 580:6361–6365 [DOI] [PubMed] [Google Scholar]
- 94.Hu K. (2008). Homologous recombination and innocuous intron elimination In: Genetic Recombination Research and Progress. Schulz JH, ed. Nova Biomedical Books, New York, pp. 323–333 [Google Scholar]
- 95.Torrents D, Suyama M, Zdobnov E. and Bork P. (2003). A genome-wide survey of human pseudogenes. Genome Res 13:2559–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Geuking MB, Weber J, Dewannieux M, Gorelik E, Heidmann T, Hengartner H, Zinkernagel RM. and Hangartner L. (2009). Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science 323:393–396 [DOI] [PubMed] [Google Scholar]
- 97.Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A. and Tristem M. (2005). Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J Virol 79:12507–12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bannert N. and Kurth R. (2004). Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A 101Suppl 2:14572–14579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, et al. (2011). Chemically defined conditions for human iPSC derivation and culture. Nat Methods 8:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chou BK, Mali P, Huang X, Ye Z, Dowey SN, Resar LM, Zou C, Zhang YA, Tong J. and Cheng L. (2011). Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res 21:518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu J, Chau KF, Vodyanik MA, Jiang J. and Jiang Y. (2011). Efficient feeder-free episomal reprogramming with small molecules. PLoS One 6:e17557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, et al. (2011). A more efficient method to generate integration-free human iPS cells. Nat Methods 8:409–412 [DOI] [PubMed] [Google Scholar]
- 103.Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH. and Muotri AR. (2009). Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One 4:e7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, et al. (2009). Oct4-induced pluripotency in adult neural stem cells. Cell 136:411–419 [DOI] [PubMed] [Google Scholar]
- 105.Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M. and Scholer HR. (2008). Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454:646–650 [DOI] [PubMed] [Google Scholar]
- 106.Kirchmaier AL. and Sugden B. (1995). Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J Virol 69:1280–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leight ER. and Sugden B. (2001). Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol Cell Biol 21:4149–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kameda T, Smuga-Otto K. and Thomson JA. (2006). A severe de novo methylation of episomal vectors by human ES cells. Biochem Biophys Res Commun 349:1269–1277 [DOI] [PubMed] [Google Scholar]
- 109.Okita K, Yamakawa T, Matsumura Y, Sato Y, Amano N, Watanabe A, Goshima N. and Yamanaka S. (2012). An efficient non-viral method to generate integration-free human iPS cells from cord blood and peripheral blood cells. Stem Cells 31:458–466 [DOI] [PubMed] [Google Scholar]
- 110.Meng X, Neises A, Su RJ, Payne KJ, Ritter L, Gridley DS, Wang J, Sheng M, Lau KH, Baylink DJ. and Zhang XB. (2012). Efficient reprogramming of human cord blood CD34+ cells into induced pluripotent stem cells with OCT4 and SOX2 alone. Mol Ther 20:408–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Camenisch G, Gruber M, Donoho G, Van Sloun P, Wenger RH. and Gassmann M. (1996). A polyoma-based episomal vector efficiently expresses exogenous genes in mouse embryonic stem cells. Nucleic Acids Res 24:3707–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gassmann M, Donoho G. and Berg P. (1995). Maintenance of an extrachromosomal plasmid vector in mouse embryonic stem cells. Proc Natl Acad Sci U S A 92:1292–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chang CW, Lai YS, Pawlik KM, Liu K, Sun CW, Li C, Schoeb TR. and Townes TM. (2009). Polycistronic lentiviral vector for “Hit and Run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells 27:1042–1049 [DOI] [PubMed] [Google Scholar]
- 114.Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN. and Mostoslavsky G. (2009). Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 27:543–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shao L, Feng W, Sun Y, Bai H, Liu J, Currie C, Kim J, Gama R, Wang Z, et al. (2009). Generation of iPS cells using defined factors linked via the self-cleaving 2A sequences in a single open reading frame. Cell Res 19:296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D. and Ryan MD. (2001). Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol 82:1013–1025 [DOI] [PubMed] [Google Scholar]
- 117.Ryan MD. and Drew J. (1994). Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J 13:928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu Y, Melton DW, Zhang Y. and Hornsby PJ. (2009). Improved coinfection with amphotropic pseudotyped retroviral vectors. J Biomed Biotechnol 2009:901079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P. and Woltjen K. (2009). Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 458:771–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Awe JP, Lee PC, Ramathal C, Vega-Crespo A, Durruthy-Durruthy J, Cooper A, Karumbayaram S, Lowry WE, Clark AT, et al. (2013). Generation and characterization of transgene-free human induced pluripotent stem cells and conversion to putative clinical-grade status. Stem Cell Res Ther 4:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, et al. (2009). piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458:766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yusa K, Rad R, Takeda J. and Bradley A. (2009). Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods 6:363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.VandenDriessche T, Ivics Z, Izsvak Z. and Chuah MK. (2009). Emerging potential of transposons for gene therapy and generation of induced pluripotent stem cells. Blood 114:1461–1468 [DOI] [PubMed] [Google Scholar]
- 124.Sarkar A, Sim C, Hong YS, Hogan JR, Fraser MJ, Robertson HM. and Collins FH. (2003). Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol Genet Genomics 270:173–180 [DOI] [PubMed] [Google Scholar]
- 125.Feschotte C. (2006). The piggyBac transposon holds promise for human gene therapy. Proc Natl Acad Sci U S A 103:14981–14982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mitra R, Fain-Thornton J. and Craig NL. (2008). piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J 27:1097–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li X, Harrell RA, Handler AM, Beam T, Hennessy K. and Fraser MJ., Jr. (2005). piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect Mol Biol 14:17–30 [DOI] [PubMed] [Google Scholar]
- 128.O'Malley J, Skylaki S, Iwabuchi KA, Chantzoura E, Ruetz T, Johnsson A, Tomlinson SR, Linnarsson S. and Kaji K. (2013). High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature 499:88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, Staerk J, Markoulaki S. and Jaenisch R. (2008). A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol 26:916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M. and Jaenisch R. (2008). A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3:346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stadtfeld M, Maherali N, Borkent M. and Hochedlinger K. (2010). A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods 7:53–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stadtfeld M, Nagaya M, Utikal J, Weir G. and Hochedlinger K. (2008). Induced pluripotent stem cells generated without viral integration. Science 322:945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Alba R, Bosch A. and Chillon M. (2005). Gutless adenovirus: last-generation adenovirus for gene therapy. Gene Ther 12Suppl 1:S18–S27 [DOI] [PubMed] [Google Scholar]
- 134.Zhou WB. and Freed CR. (2009). Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells 27:2667–2674 [DOI] [PubMed] [Google Scholar]
- 135.Harui A, Suzuki S, Kochanek S. and Mitani K. (1999). Frequency and stability of chromosomal integration of adenovirus vectors. J Virol 73:6141–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Filipowicz W, Bhattacharyya SN. and Sonenberg N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114 [DOI] [PubMed] [Google Scholar]
- 137.Bartel DP. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 138.Anokye-Danso F, Snitow M. and Morrisey EE. (2012). How microRNAs facilitate reprogramming to pluripotency. J Cell Sci 125:4179–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Leonardo TR, Schultheisz HL, Loring JF. and Laurent LC. (2012). The functions of microRNAs in pluripotency and reprogramming. Nat Cell Biol 14:1114–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li ZH, Yang CS, Nakashima K. and Rana TM. (2011). Small RNA-mediated regulation of iPS cell generation. EMBO J 30:823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Judson RL, Babiarz JE, Venere M. and Blelloch R. (2009). Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27:459–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R. and Blelloch R. (2011). Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 29:443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liao B, Bao X, Liu L, Feng S, Zovoilis A, Liu W, Xue Y, Cai J, Guo X, et al. (2011). MicroRNA cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem 286:17359–17364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DTS, Chen DT. and Ying SY. (2008). Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 14:2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang YZ, Yang WL, Gruber PJ, Epstein JA. and Morrisey EE. (2011). Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8:376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, et al. (2011). Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8:633–638 [DOI] [PubMed] [Google Scholar]
- 147.Lin SL, Chang DC, Lin CH, Ying SY, Leu D. and Wu DT. (2011). Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res 39:1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chang HM. and Gregory RI. (2011). MicroRNAs and reprogramming. Nat Biotechnol 29:499–500 [DOI] [PubMed] [Google Scholar]
- 149.Liu X, Sun H, Qi J, Wang L, He S, Liu J, Feng C, Chen C, Li W, et al. (2013). Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat Cell Biol 15:829–838 [DOI] [PubMed] [Google Scholar]
- 150.Carey BW, Markoulaki S, Hanna JH, Faddah DA, Buganim Y, Kim J, Ganz K, Steine EJ, Cassady JP, et al. (2011). Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell 9:588–598 [DOI] [PubMed] [Google Scholar]
- 151.Papapetrou EP, Tomishima MJ, Chambers SM, Mica Y, Reed E, Menon J, Tabar V, Mo Q, Studer L. and Sadelain M. (2009). Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci U S A 106:12759–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]


