Abstract
Enfuvirtide was the first fusion inhibitor approved by the Food and Drug Administration (FDA) in 2003 for HIV-1 infection in treatment-experienced patient. It is the first approved antiviral agent to attack the HIV life cycle in its early stages. For HIV fusion to occur, the HR1 and HR2 domains in the gp41 region need to interact. Enfuvirtide is a synthetic peptide that corresponds to 36 amino acids of the HR2, which competitively binds to HR1 inhibiting the interaction with the HR2 domain thus preventing fusogenic conformation and inhibiting viral entry into host cells. Resistance to enfuvirtide is conferred by mutations occurring in the HR1 region involving residues 36–45. Mozambique, a sub-Saharan country, with an HIV prevalence of 11.5%, provides first line and second line antiretroviral therapy (ART)-based treatment. In poor resource settings such as Mozambique the lack of adequate infrastructures, the high costs of viral load tests, and the availability of salvage treatment have hindered the intended objective of monitoring HIV treatment, suggesting an important concern regarding the development of drug resistance. The general aim of this study was to evaluate naturally occurring polymorphisms and resistance-associated mutations in the gp41 region of HIV-1 isolates from Mozambique. The study included 78 patients naive to ARV treatment and 28 patients failing first line regimen recruited from Centro de Saúde Alto-Maé situated in Maputo. The gp41 gene from 103 patients was sequenced and resistance-associated mutations for enfuvirtide were screened. Subtype analysis revealed that 96% of the sequences were classified as subtype C, 2% as subtype G, 1% as subtype A1, and the other 1% as a mosaic form composed of A1/C. No enfuvirtide resistance-associated mutations in HR1 of gp41 were detected. The major polymorphisms in the HR1 were N42S, L54M, A67T, and V72I. This study suggests that this new class of antiviral drug may be effective as a salvage therapy in patients failing first line regimens in Mozambique. However, further phenotypic studies are required to determine the clinical relevance of the polymorphisms detected in this study.
Since the beginning of the HIV pandemic approximately 60 million people have died of AIDS. Globally the World Health Organization (WHO) estimated that by the end of 2011 33.4 million people were living with HIV. To date sub-Saharan Africa still remains the most affected region accounting 69% of HIV infections worldwide in which 1 in every 20 adults (49%) is infected.1 Mozambique, a sub-Saharan country, faces serious HIV epidemics with a national prevalence of 11.5% of adults aged 15–49 years; the overall prevalence rate is 17.8% in the southern region, 12.5% in the central region, and 5.6% in the northern region.2 Thus, according to the Ministry of Health in 2010 the number of people receiving antiretroviral treatment (ART) was approximately 211,000 adults and 16,800 children and the ART coverage for people in need of treatment was 40% for adults and 26% for children.3 As recommended by the WHO, the first line ART regimen in Mozambique consists of two nucleoside reverse transcriptase inhibitors (NRTIs) and one nonnucleoside reverse transcriptase inhibitor (NNRTI) and the second line ART regimen consists of a boosted protease inhibitor (PIs) and two NRTIs for patients failing the first line regimen. However, due to the rapid expansion of ART in Mozambique the emergence of drug resistance viruses is an important public health problem.4 During the past decade, the intensive use of highly active antiretroviral treatment (HAART) has substantial reduced mortality and morbidity. Despite the success of HAART, treatment failure associated with the lack of adherence, drug toxicity, and the emergence of drug-resistant viruses is an important issue for the effectiveness of antiretroviral (ARV) programs. Thus, the use of new drug classes to increase drug combinations is necessary to suppress drug-resistant virus replication.5,6
The HIV envelope glycoproteins encoded by the env gene is composed of two subunits, gp120 (surface) and gp41 (transmembrane), that play an important role during viral entry into the host cell.7 On the surface of the target cell, gp120 interacts with the CD4 receptors and coreceptors (CCR5 or CXR4) and gp41 mediates the fusion between the viral and cellular membrane. As a consequence of this interaction, conformational changes in gp120 and gp41 occur, causing the membranes to fuse followed by the release of the viral genetic material into the cytoplasm of the cell.8 To retain the conformational changes specific regions within gp41 need to interact making it a good target for drug development.
Enfuvirtide (ENF, T20, or FUZEON, Hoffmann-La Roche, Inc.) is a 36-amino acid synthetic peptide that mimics the HR2 domain of the gp41 at residues 127–162. It was approved by the Food and Drug Administration (FDA) in 2003 to be used in patients undergoing ART.9,10 ENF competitively binds with HR2 and blocks the conformational changes required for the fusion process.1,8 Resistance to ENF is associated with mutations occurring at residues 35–45, which decreases the susceptibility to ENF.11–13 Therefore to predict the effectiveness of ENF in Mozambique as an alternative ART, this study aimed to evaluate the genetic diversity, describe the natural occurring polymorphisms, and examine the presence of ENF mutations associated with resistance in patients naive to ART and failing the first line regimen in the HIV-1 isolates circulating in Maputo city.
The study included 103 patients attending the Alto-Maé Health Centre in Maputo, Mozambique from December 2009 to August 2010. Seventy-eight of the patients were naive to ART and 33 were failing the first line regimen. The study protocol was approved by the national ethics review committee of Mozambique (number 307/CNBS). Every potential participant was given a written informed consent and a subjects consent form was signed. Genomic DNA was extracted from buffy coat samples using the QIAamp DNA blood mini kit according to the manufacturer's instructions. The full-length gp41 of the approximately 697-bp region within the env gene was amplified by nested polymerase chain reaction (PCR). The amplified PCR products were purified using Millipore Motagem (Life Science kits). Purified amplified products were sequenced with the ABI Big Dye terminator system (v3.1) according to the manufacturer's instructions on the ABI prism 3100 (Applied Biosystems, EUA).
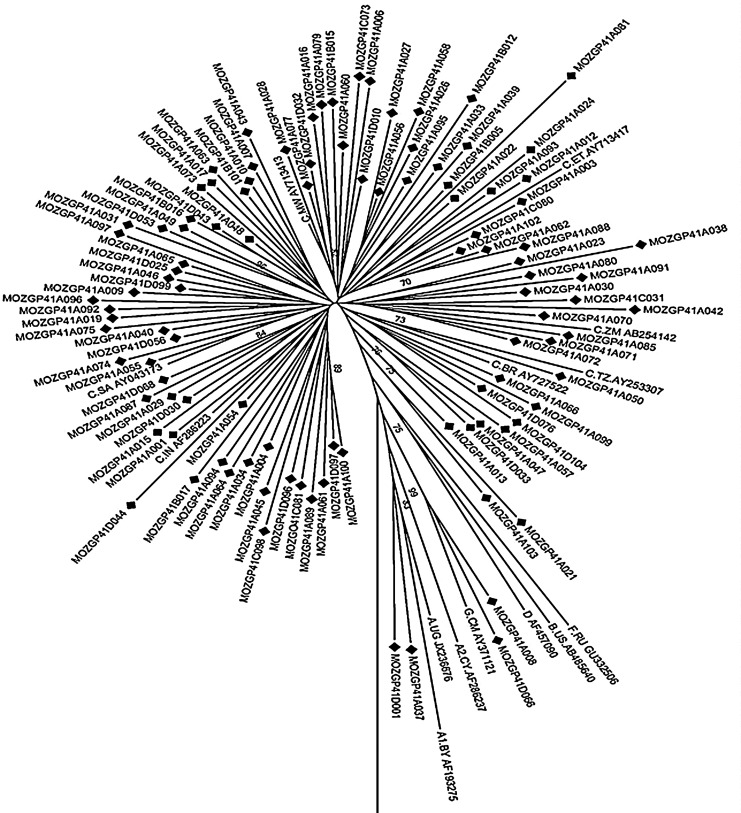
The sequences were edited using the DNASATR (SeqMan) program and HIV-1 subtype B (HXB2) was used as a reference sequence. For subtyping analysis the sequences were submitted to the REGA (www.bioafrica.net/rega-genotype/html/subtypinghiv.html) algorithm. Subsequently for recombinant analysis the sequences were submitted to the jumping profile hidden Markov model (jpHMM) (http//jphmm.gobics.de/) algorithm. The sequences were aligned using the MEGA program (version 5.0) and used for phylogenetic and molecular analysis. A phylogenetic tree was performed by neighbor-joining (NJ) using the Kimura two-parameter method. The stability of the nodes was assessed by bootstrap analysis (1,000 replicates) and bootstrap values >70% were considered significant.
All the aligned sequences were converted into amino acids and exported into excel format. For extensive analysis the frequency of each amino acid position was determined. For the consensus sequences all the 99 gp41 sequences for the Mozambique subtype C gp41 sequences obtained in the study were used to generate the Mozambique C consensus sequence. For the Africa C and Global C consensus sequences, 15 gp41 subtype C sequences from Africa and another 15 gp41 subtype C sequences from non-African countries were retrieved LASD (www.hiv/lanl.gov/content/sequence/HIV/refer.html). All the three consensus sequences (Mozambique C, Africa C, and Global C) were generated individually by aligning the sequences from the study and those retrieved from LASD using the BioEdit Sequence Alignment Editor (version 5.0, Carlsbad, CA). Once the sequences were aligned a consensus sequence for the Mozambique C, Africa C, and Global C was generated. For molecular characterization comparison all Mozambique C, Africa C, and Global C sequences were aligned with subtype B (HXB2) using BioEdit.
Subtyping analysis using Rega revealed that 99 of the sequences were subtype C, two sequences were subtype G, and one was subtype A1. This is in accordance with previous studies showing that subtype C is the most frequent in Mozambique14,15 and is consistent with previous studies showing the existence of this subtype in neighboring countries such as South Africa, with the exception to Tanzania where subtype A1 predominates.16,17 Similarly, in Mozambique subtype G has previously been found in drug-naive patients enrolled from 2002 to 2004 and subtype A1 was recently introduced from patients enrolled in 2010.15,18
Furthermore, recombinant analysis using the jpHMM program showed that one (Sequence ID A037) of the sequences was a mosaic form composed of subtypes C/A1. The C/A1 mosaic form identified in this study could represent a new CRF or URF, but full genomic analysis and further studies in other regions of Mozambique are required to confirm this finding.
Based on the phylogenetic analysis, 99 of the sequences clustered with subtype C, two sequences with subtype G (Sequence ID: D066 and A008), and the other two sequences with subtype A1 (Sequence ID: A037 and D001) (Fig. 1). Additionally, seven clusters within the phylogeny tree were observed, and subtype C sequences segregated into phylogenetic subclusters representing viruses originally from different regions. These results demonstrate that the Mozambican sequences formed a country-specific cluster and sequences spread throughout the tree. This therefore suggests an epidemiological link among the Mozambican sequences and different subtype C sequence homology with neighboring countries and others. The high genetic variation observed in this study suggests multiple introductions of subtype C viruses in Mozambique, thus confirming the multiple subtype C clade sublineages circulating in southern Africa as previously described.17
FIG. 1.
Phylogenetic relationship of newly characterized full-length gp41 sequences (n=103) circulating in Maputo, Mozambique with HIV-1 subtype references sequences and the outgroup (group N from Cameroon, AJ006022) from the LASB. The phylogenetic tree was constructed using neighbor joining and the Kimura two-parameter distance method. The reliability of the branching order was estimated from 1,000 bootstrap replicates, and only bootstraps values of 70% or higher are shown.
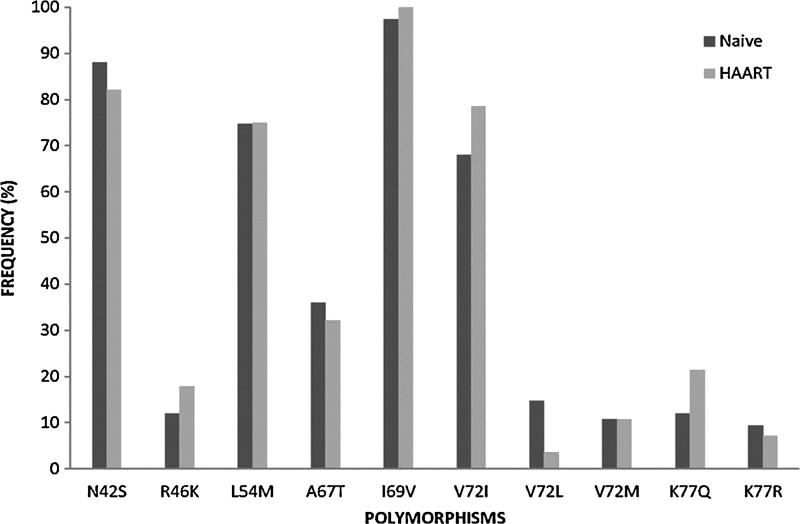
For HIV-1 fusion to occur, the HR1 and HR2 domains in the gp41 region need to interact. ENF is a synthetic peptide that corresponds to 36 amino acids of the HR2, which competitively binds to HR1 inhibiting the interaction with the HR2 domain and thus preventing the virus from fusing. Resistance to ENF is conferred by mutations occurring in the HR1 region involving residues 36, 37, and 38 (GIV motif). In addition, mutations between residues 36 and 45 (G36D/E, I37V, V38A/M/E, Q40H, N42T, and N43D) may also confer resistance to ENF.21,22 In a total of 103 sequences analyzed no mutations associated with resistance mutation at residues 3–38 were detected, but polymorphisms in the HR1 domain of the gp41 region were observed. The N42S polymorphisms previously associated with increased susceptibility to ENF19 were detected in 90% (n=89) of the sequences (Fig. 2).
FIG. 2.
Frequency of polymorphism above 3% within the gp41 HR1 (residues 29–82) region in patients naive to antiretroviral therapy (ART) (n=75) and patients failing first line regimen (n=28).
Similar to previous studies polymorphism L54M was detected at a high frequency of 74% (n=76) (Fig. 2). Thus, it was demonstrated that L54M induces structural changes in the gp41 complex affecting the accessibility of the HR1 region to ENF and thereby reducing sensitivity to inhibition. Although natural occurring sequence variation within the gp41 HR1 may reduce sensitivity of HIV-1 inhibition by ENF, further studies will be necessary to elucidate if this observed polymorphism may affect the efficacy of treatment with ENF in Mozambique.
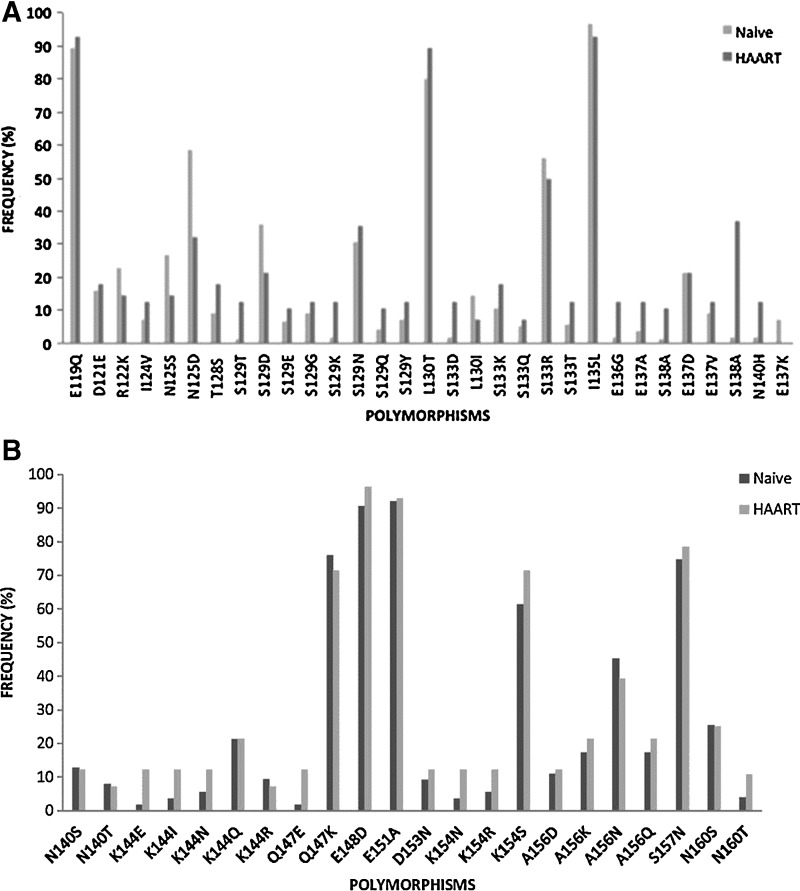
The HR1 domain across group M subtypes is highly conserved in comparison to HR2, which contains greater sequence variability.20 Similar to Araújo and collaborators,19 several polymorphisms within the HR2 domain were observed: polymorphism L130I in 13% (n=13) of the sequences, S129N in 31% (n=32) of the sequences, E137K in 6% (n=6) of the sequences, and S138A in 4% (n=4) of the sequences (Fig. 3); these recently detected sequences were related to decreased susceptibility to ENF.12
FIG. 3.
Frequency of polymorphisms above 3% in the gp41 HR2 region in patient naive to HAART (n=75) and patients failing first line regimen (n=28) (A) from residues 116–137 and (B) from residues 140–161.
The structural basis of the heptad repeats for membrane fusion by the enveloped viruses consists of an α-helical coiled-coil conformation labeled a-b-c-d-e-f-g that interacts with hydrophobic residues at positions a and d. Hence between α-helices, serine at position 138 (a position) of the C helix interacts with leucine at position 45 (e position) of the N helix (HR1) in an antiparallel manner. In addition, serine-138 interacts with glutamine at position 40 (g position) of the N helix (HR1) and at close proximity to asparagine at position 43 (c position) in the HR1. However, polymorphism S138A at motif a may have an impact on the interaction between HR1 and HR2, subsequently influencing the fusion property and the inhibitory property of ENF. It was demonstrated that S138A accompanied by a mutation in HR1 at residues 42 and 43 increased the level of resistance by 3-fold, suggesting that this substitution might be a secondary mutation when it coexists with a mutation at residues 43.23 Hence, polymorphism S138A, mostly prevalent in subtypes C (n=3) and A1 (n=1), but in the absence of a mutation in the HR1 was detected. Although the S138A polymorphism did not occur accompanied by mutations in residue 43, as suggested by Xu and collaborators, there is still a possibility that this polymorphism might contribute to drug resistance.24
Other polymorphisms such as E148D, E151D, E119Q, Q147K, and A156N previously reported in patients naive to ENF and patients on ENF treatment were also observed at a high frequency. This suggests that certain polymorphic sites are due to the natural variability of the HR2 domain; however, in vitro studies are necessary to evaluate the impact of these polymorphisms on ENF susceptibility (Fig. 3).
ENF is a 36-amino acid peptide that was designed based on the HR2 domain of the HIV-1 subtype B gp41 envelope glycoprotein. The genetic differences among HIV variants may influence the emergence of drug resistance, since the viral nucleotide composition is different. However, since subtype C is responsible for the local epidemic in Mozambique, the genetic variability and molecular signature of subtype C from Mozambique gp41 sequences were determined. In total, genetic variability representing 15% (29/199) was observed for the gp41 region within the env gene (Fig. 4).
FIG. 4.
Alignment of HIV-1 gp41 consensus sequence LASDB, Mozambique C, Africa C, and Global C with respect to HXB2 subtype B gp41. The FP (residues 8–28) is highlighted in the first box, the HR1 domain (residues 29–82) is highlighted in the second box, and the HR2 domain (residues 116–161) is highlighted in the third box. The mutations associated with resistance to ENF are indicated between the two arrows at positions 36–45.
To identify the molecular signatures within the subtype C Mozambique consensus sequence, an alignment of the gp41 consensus sequence generated for Africa C, Global C was performed. When the Mozambique C, Africa C, and Global subtype C consensus sequences were compared to subtype B HXB2, 20 polymorphic codons at positions 7, 24, 42, 54, 67, 69, 72, 101,108 110, 113, 114, 115, 119, 129, 130,132, 135, 173, and 182 were observed. Thus, seven extra polymorphic codons at positions 125, 133, 147, 148, 151, 154, and 187 were observed within the Mozambique C and Africa C consensus sequences confirming these codons as Africa subtype C molecular signatures for the gp41 region within the env gene. In addition, two polymorphic codons for which the frequency of asparagine (N) was 46% at position 96 and 53% at position 157 were observed only within the Mozambique subtype C consensus sequence suggesting that these codons were molecular signatures. In addition, high homology among the Mozambique C and Africa C consensus sequence, showing only seven differences at codons 125, 133, 147, 148, 151, 154, and 187 when compared to Global C and HXB2 subtype B (Fig. 4), were observed. This suggests an epidemiological linkage among gp41 sequences from these regions. Notably, further studies are required to clarify whether the genetic background of HIV subtypes affects the efficiency of treatment with fusion inhibitors, especially in Mozambique where subtype C is most frequent.19
These results demonstrate that the HIV-1 epidemic in Mozambique is quite homogeneous with regard to the subtype mostly composed of C clade viruses influenced by foreign and neighboring countries. In addition, the recent introduction of other subtypes and mosaic forms suggests that the genetic complexity in Mozambique is evolving rapidly. Therefore, it is important to continually monitor the impact and the potential of HIV-1 genetic diversity relevant for diagnosis, primary drug resistance, and vaccination. In conclusion, the absence of primary resistance mutations that confer resistance to ENF suggests that these new ARV drugs can be used as salvage treatment in Mozambique. However, whether the presence of the polymorphisms described in this study would have an impact on the response of fusion inhibitors and trigger faster than unusual mutations associated with resistance to ENF still remains to be addressed in future in vitro and in vivo studies. Therefore, these results show that in vitro studies are necessary to understand the relevance of the above-mentioned signatures observed for HIV-1-specific subtype C.
Acknowledgments
We are thankful to the hospital staff of the Centro de Saúde do Alto Maé. Special thanks to the staff of the Animal Molecular Virology of Rio de Janeiro Federal University. In addition, we also wish to thank the staff of the Molecular Virology Laboratory of the National Institute of Health of Mozambique for their collaboration on this project. The study was supported by the Brazilian National Research Council, CNPq (grant 471299/2009-0) and CAPES, National AIDS Program/Ministério da Saúde do Brasil and UFRJ.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.UNAIDS: Report on the Global AIDS Epidemic “UNAIDS/JC2417E,”. 2012.
- 2.Instituto Nacional de Saúde (INS), Instituto Nacional de Estatística (INE), e ICF Macro: Inquérito Nacional de Prevalência, Riscos Comportamentais e Informação sobre o HIV e SIDA em Moçambique 2009. Calverton, MD, EUA: INS, INE e ICF Macro, 2010 [Google Scholar]
- 3.Ministério da Saúde (MISAU): Moçambique MdS. Estatisticas do Programa Nacional de Controlo do HIV/SIDA, 2010
- 4.World Health Organization (WHO): Antiretroviral therapy for HIV infection in adults and adolescents: Recommendations for a public health approach. World Health Organization, Department of HIV/AIDS, Geneva, Switzerland, 2010 [PubMed] [Google Scholar]
- 5.Bennett DE, Bertagnolio S, Sutherland D, and Gilks CF: The World Health Organization's global strategy for prevention and assessment of HIV drug resistance. Antivir Ther 2008;13(Suppl 2):1–13 [PubMed] [Google Scholar]
- 6.Gupta RK. and Pillay D: HIV resistance and the developing world. Int J Antimicrob Agents 2007;29(5):510–517 [DOI] [PubMed] [Google Scholar]
- 7.Chan DC, Fass D, Berger J M, and Kim PS: Core structure of gp41 from the HIV envelope glycoprotein. Cell 1997;89(2):263–273 [DOI] [PubMed] [Google Scholar]
- 8.Sattentau QJ, Moore JP, Vignaux F, Traincard F, and Poignard P: Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol 1993;67(12):7383–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melby T, Sista P, DeMasi R, Kirkland T, Roberts N, Salgo M, Heilek-Snyder G, Cammack N, Matthews TJ, and Greenberg ML: Characterization of envelope glycoprotein gp41 genotype and phenotypic susceptibility to enfuvirtide at baseline and on treatment in the phase III clinical trials TORO-1 and TORO-2. AIDS Res Hum Retroviruses 2006;22(5):375–385 [DOI] [PubMed] [Google Scholar]
- 10.Fung HB. and Guo Y: Enfuvirtide: A fusion inhibitor for the treatment of HIV infection. Clin Ther 2004;26(3):352–378 [DOI] [PubMed] [Google Scholar]
- 11.Rimsky LT, Shugars DC, and Matthews TJ: Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J Virol 2008;72(2):986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Alvarez L, Carmona R, Ocampo A, Asorey A, Miralles C, Perez de Castro S, Pinilla M, Contreras G, Taboada JA, and Najera R: Long-term monitoring of genotypic and phenotypic resistance to T20 in treated patients infected with HIV-1. J Med Virol 2006;78(2):141–147 [DOI] [PubMed] [Google Scholar]
- 13.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, and Kappes JC: Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 2002;46(6):1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abreu CM, Brindeiro PA, Martins AN, Arruda MB, Bule E, Stakteas S, Tanuri A, and Moraes BR: Genotypic and phenotypic characterization of human immunodeficiency virus type 1 isolates circulating in pregnant women from Mozambique. Arch Virol 2008;153(11):2013–2017 [DOI] [PubMed] [Google Scholar]
- 15.Bartolo I, Casanovas J, Bastos R, Rocha C, Abecasis AB, Folgosa E, Mondlane J, Manuel R, and Taveira N: HIV-1 genetic diversity and transmitted drug resistance in health care settings in Maputo, Mozambique. J Acquir Immune Defic Syndr 2009;51(3):323–331 [DOI] [PubMed] [Google Scholar]
- 16.Bartolo I, Epalanga M, Bartolomeu J, Fonseca M, Mendes A, Gama A, and Taveira N: High genetic diversity of human immunodeficiency virus type 1 in Angola. AIDS Res Hum Retroviruses 2005;21(4):306–310 [DOI] [PubMed] [Google Scholar]
- 17.Gordon M, De Oliveira T, Bishop K, Coovadia HM, Madurai L, Engelbrecht S, Janse van Rensburg E, Mosam A, Smith A, and Cassol S: Molecular characteristics of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: Implications for vaccine and antiretroviral control strategies. J Virol 2003;77(4):2587–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira MF, Ramalho DB, Abreu CM, Vubil A, Mabunda N, Ismael N, Francisco C, Jani IV, and Tanuri A: Genetic diversity and naturally polymorphisms in HIV type 1 integrase isolates from Maputo, Mozambique: Implications for integrase inhibitors. AIDS Res Hum Retroviruses 2012;28(12):1788–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Araujo LA, Junqueira DM, de Medeiros RM, Matte MC, and Almeida SE: Naturally occurring resistance mutations to HIV-1 entry inhibitors in subtypes B, C, and CRF31_BC. J Clin Virol 2012;54(1):6–10 [DOI] [PubMed] [Google Scholar]
- 20.Dorn J, Masciotra S, Yang C, Downing R, Biryahwaho B, Mastro TD, Nkengasong J, Pieniazek D, Rayfield MA, Hu DJ, and Lal RB: Analysis of genetic variability within the immunodominant epitopes of envelope gp41 from human immunodeficiency virus type 1 (HIV-1) group M and its impact on HIV-1 antibody detection. J Clin Microbiol 2000;38(2):773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabrera C, Marfil S, Garcia E, Martinez-Picado J, Bonjoch A, Bofill M, Moreno S, Ribera E, Domingo P, Clotet B, and Ruiz L: Genetic evolution of gp41 reveals a highly exclusive relationship between codons 36, 38 and 43 in gp41 under long-term enfuvirtide-containing salvage regimen. AIDS 2006;20(16):2075–2080 [DOI] [PubMed] [Google Scholar]
- 22.Shafer RW, and Schapiro JM: HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev 2008;10(2):67–84 [PMC free article] [PubMed] [Google Scholar]
- 23.Chinnadurai R, Munch J, and Kirchhoff F: Effect of naturally-occurring gp41 HR1 variations on susceptibility of HIV-1 to fusion inhibitors. AIDS 2005;19(13):1401–1405 [DOI] [PubMed] [Google Scholar]
- 24.Oliveira AC, Martins AN, Pires AF, Arruda MB, Tanuri A, Pereira HS, and Brindeiro R. M:Enfuvirtide (T-20) resistance-related mutations in HIV type 1 subtypes B, C, and F isolates from Brazilian patients failing HAART. AIDS Res Hum Retroviruses 2009;25(2):193–198 [DOI] [PubMed] [Google Scholar]