Editor: HIV-1 is classically classified into three main groups: M (major), N (non-M, non-O), and O (outlier),1 with Group P being identified most recently.2 Evolutionary studies suggest each group represents a distinct cross species transmission of SIVcpz or SIVgor into the human population.2,3 HIV-1, Group O has mostly remained endemic to Cameroon and inaccurate diagnosis of Group O infection is a major barrier to treatment in this country.4 A unique subset of individuals, termed long-term nonprogressors (LTNPs), is able to maintain stable CD4+ T cell counts for greater than 5 years in the absence of antiretroviral therapy. Here we report a patient infected with a Group O HIV-1 isolate who has maintained long-term control of HIV replication in the absence of antiretroviral treatment (ART).
The patient is a 38-year-old treatment-naive female originally from Cameroon. She was diagnosed with HIV-1 infection in 1998 in Cameroon, and first presented to the United States in 2004 with an initial CD4 count of 576 and a viral load of <50 copies/ml as measured by the Roche AMPLICOR HIV-1 MONITOR 1.5 ultrasensitive assay. She maintained stable CD4+ T cell counts and viral loads of <50 copies/ml in the absence of ART (Fig. 1A) over the next 6 years. The rate of CD4+ T cell decline was calculated as described previously.5 We found that there was no significant decline in CD4+ T cells between 2005 and 2010 (p=0.0615). HLA typing revealed that she was HLA-A*02/66 and HLA-B*42/53 positive.
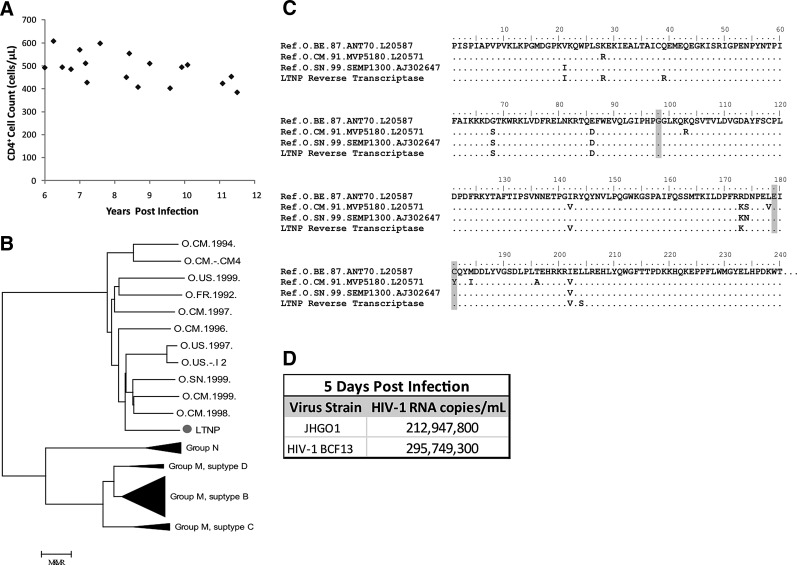
FIG. 1.
(A) Patient's CD4 count over time. (B) Phylogenetic analysis demonstrating that the env sequence from the patient's isolate (gray dot) clusters with the env sequence from Group O isolates. (C) Reverse transcriptase sequence from the patient and two other Group O isolates showing the presence of nonnucleoside reverse transcriptase inhibitor mutations (gray). (D) HIV RNA copies/ml in culture supernatant at day 5 postinfection with the patient's isolate (JHG01) or a laboratory isolate.
Virus was cultured from the patient's latent reservoir as previously described6 in 2010 and designated JHGO1. Phylogenetic analysis was performed on full length env sequences of the patient's virus compared to Group O, N, and M isolates. Classical, maximum likelihood, and Bayesian phylogenetic analysis was performed. The patient's virus was most closely related to HIV-1, Group O (Fig. 1B). Group O nested-PCR primers were developed utilizing full genome Group O sequences from the Los Alamos HIV Sequence database and full genome sequencing was performed (GenBank accession number JN571034). No gross sequence defects were seen compared to other Group O viral sequences. Evolutionary differences in Group O reverse transcriptase (RT) have resulted in resistance to nonnucleoside reverse transcriptase inhibitors (NNRTIs) in many treatment-naive patients.7 The patient's virus had mutations A98G, V179E, and Y181C in RT (Fig. 1C), all of which have been reported to confer resistance to NNRTIs.7 The virus isolated from the patient's latent reservoir was found to be as replication-competent as a Group O HIV-1 laboratory strain (HIV-1 BCF18) in a viral fitness assay where activated CD4+ T cells from seronegative donors were infected with 200 ng/ml p24 of either the patient's isolate or the laboratory strain as previously described6 and HIV-1 RNA in culture supernatant was quantified on day 5 postinfection with the Abbott m2000 RealTime HIV Assay (Fig. 1D).9
The patient's plasma viral load was subsequently determined to be 2,682 RNA copies/ml by the Abbott m2000 RealTime HIV Assay. The Roche AMPLICOR HIV-1 MONITOR test remained <50 copies/ml consistent with studies showing that the latter test does not routinely detect Group O HIV-1 isolates.9
Taken together, although this patient was initially misclassified as an elite controller, her stable CD4+ T cell counts and low viral load indicate that she is a long-term nonprogressor (LTNP). LTNPs have previously been identified in Group M infection, but to our knowledge, long-term nonprogression in patients infected with a Group O virus has not been previously reported. Infection with a defective virus accounts for some cases of long-term nonprogression,10 but multiple studies have also shown that replication-competent virus can be isolated from some LTNPs and elite controllers.11 It is clear that host factors, such as protective HLA types and HIV-specific CD8+ T cells, play a large role in the control of HIV-1 infection.12 This patient possessed no previously reported protective HLA alleles, but a substantial proportion of LTNPs and elite controllers (EC) do not have these alleles.13 Our fitness assay results suggest that this patient is not infected with a defective virus. While we were not able to study HIV-specific CD8+ T cell function in this patient, it appears that host factors were responsible for control of viral replication. This case suggests that some Group O-infected individuals can partially control fully pathogenic virus.
Acknowledgments
The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1BCF13 from Drs. Sentob Saragosti, Françoise Brun-Vézinet, and François Simon.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Yamaguchi J, Bodelle P, Vallari AS, et al. : HIV infections in northwestern Cameroon: identification of HIV type 1 group O and dual HIV type 1 group M and group O infections. AIDS Res Hum Retroviruses 2004;20:944–957 [DOI] [PubMed] [Google Scholar]
- 2.Plantier JC, Leoz M, Dickerson JE, et al. : A new human immunodeficiency virus derived from gorillas. Nat Med 2009;15:871–872 [DOI] [PubMed] [Google Scholar]
- 3.Gao F, Bailes E, Robertson DL, et al. : Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 1999;397:436–441 [DOI] [PubMed] [Google Scholar]
- 4.Aghokeng AF, Mpoudi-Ngole E, Dimodi H, et al. : Inaccurate diagnosis of HIV-1 group M and O is a key challenge for ongoing universal access to antiretroviral treatment and HIV prevention in Cameroon. PloS One 2009;4:e7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sedaghat AR, Rastegar DA, O'Connell KA, Dinoso JB, Wilke CO, and Blankson JN: T cell dynamics and the response to HAART in a cohort of HIV-1-infected elite suppressors. Clin Infect Dis 2009;49:1763–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey JR, Zhang H, Wegweiser BW, et al. : Evolution of HIV-1 in an HLA-B*57-positive patient during virologic escape. J Infect Dis 2007;196:50–55 [DOI] [PubMed] [Google Scholar]
- 7.Tebit DM, Lobritz M, Lalonde M, et al. : Divergent evolution in reverse transcriptase (RT) of HIV-1 group O and M lineages: Impact on structure, fitness, and sensitivity to nonnucleoside RT inhibitors. J Virol 2010;84:9817–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Descamps D, Collin G, Letourneur F, et al. : Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: In vitro phenotypic and genotypic analyses. J Virol 1997;71:8893–8898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyne MT, Konnick EQ, Phansalkar A, and Hillyard DR: Evaluation of the Abbott investigational use only realtime HIV-1 assay and comparison to the Roche Amplicor HIV-1 monitor test, version 1.5. J Mol Diagnost: JMD 2009;11:347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deacon NJ, Tsykin A, Solomon A, et al. : Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 1995;270:988–991 [DOI] [PubMed] [Google Scholar]
- 11.Buckheit RW, 3rd, Salgado M, Martins KO, and Blankson JN: The implications of viral reservoirs on the elite control of HIV-1 infection. Cell Mol Life Sci: CMLS 2013;70:1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hersperger AR, Migueles SA, Betts MR, and Connors M: Qualitative features of the HIV-specific CD8+ T-cell response associated with immunologic control. Curr Opin HIV AIDS 2011;6:169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emu B, Sinclair E, Hatano H, et al. : HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J Virol 2008;82:5398–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]