Abstract
Using bioinformatic methods, we identified a total of 221 and 199 tRNA genes in the nuclear genomes of Nasonia vitripennis and honey bee (Apis mellifera), respectively. We performed comparative analyses of Nasonia tRNA genes with honey bee and other selected insects to understand genomic distribution, sequence evolution and relationship of tRNA copy number with codon usage patterns. Many tRNA genes are located physically close to each other in the form of small clusters in the Nasonia genome. However, the number of clusters and the tRNA genes that form such clusters vary from species to species. In particular, the Ala-, Pro-, Tyr- and His-tRNA genes tend to accumulate in clusters in Nasonia but not in honey bee, whereas the bee contains a long cluster of 15 tRNA genes (of which 13 are Gln-tRNAs) that is absent in Nasonia. Though tRNA genes are highly conserved, contrasting patterns of nucleotide diversity are observed among the arm and loop regions of tRNAs between Nasonia and honey bee. Also, the sequence convergence between the reconstructed ancestral tRNAs and the present day tRNAs suggests a common ancestral origin of Nasonia and honey bee tRNAs. Furthermore, we also present evidence that the copy number of isoacceptor tRNAs (those having a different anticodon but charge the same amino acid) is correlated with codon usage patterns of highly expressed genes in Nasonia.
Keywords: isoacceptor, codon usage, gene cluster, sequence divergence, anticodon
Introduction
Transfer RNA (tRNA) genes are ubiquitous in all organisms and have been attractive gene models for evolutionary studies (Cedergren et al. 1980; Hani and Feldmann 1998; Björk et al. 2001; Sun and Caetano-Anollés 2008). They transcribe 75–95 nt non-coding RNA molecules that are required at the ribosome to transfer specific amino acids to a growing polypeptide chain. Each tRNA is conservatively folded into an L-shaped tertiary structure required for base-pairing of its anticodon triplet to the corresponding codon on the mRNA. It contains a charge-site at the opposite end with CCA nucleotides where an amino acid cognate to the tRNA is attached. The genes coding for different tRNA isotypes (those that transfer different amino acids) are selected differentially at the sequence level in different species. Mutations in the anticodon site generate new isoacceptor families comprising tRNAs with different anticodons but still charging the same amino acid. The isoacceptor genes also show polyphyletic grouping suggesting that these tRNAs accumulate mutations at other parts of gene than the anticodon (Saks and Sampson, 1995).
The recent sequencing of genomes for different insect species has provided opportunities to analyze the arthropod non-coding RNA genes. Hymenoptera, comprising the sawflies, wasps, bees, ants, is one of the larger orders of insects and many of these that have important economical and ecological impacts. The completion of genome sequencing projects for honey bee and now for Nasonia presents an important step toward better understanding the general and molecular biology of these two important hymenopteran insects. In this regard, the tRNA genes are important for evolutionary investigations as they are key components of the translational machinery.
Identification of tRNA genes provides avenues to evaluate their functional relevancies on codon degeneracy (Ikemura 1985; Percudani et al. 1997; Hani and Feldmann 1998). The correlation between codon usage bias and tRNA gene abundance is known in prokaryotes as well as some eukaryotes (Ikemura 1985; Kanaya et al. 1999, Higgs and Ran 2008). The link between tRNA genes and codon bias is an evolutionary process that is dependent upon the translational efficiencies of species (Rocha 2004; Higgs and Ran 2008).
In the present study, we identify the tRNA genes in the N. vitripennis genome to understand 1) gene number and genomic distribution pattern of these genes, 2) relationship between tRNA gene copies and codon usage 3) patterns of sequence evolution, and 4) intron evolution in a comparative manner with A. mellifera. Our results indicate that although tRNA genes are highly conserved, these genes show contrasting patterns of genomic distribution and copy number as well as sequence evolution between the two species. We also performed limited comparative analysis of tRNAs of Nasonia and honey bee with that of other arthropods in which tRNA genes have been annotated.
Results
Identification of tRNA genes
A combinatorial procedure that incorporated BlastN (Altschul et al. 1990), ARAGORN (Laslett and Canback, 2004) and tRNAscanSE (Lowe and Eddy, 1997) was used to systematically identify tRNA genes in the N. vitripennis and A. mellifera genomes. The D. melanogaster tRNA sequences were used as query to identify putative tRNA genes and these were then analyzed by ARAGORN and tRNAscanSE. Sequences validated as tRNA genes by both ARAGORN and tRNAscanSE were used in this analysis. They have been assigned numerical temporary IDs (and will be eventually assigned official identifiers) and the complete list (sequence and genome positions) is available in Supplementary Table 1. A total of 221 tRNA genes were identified in N. vitripennis, compared to 199 tRNAs in A. mellifera. The number of tRNAs genes cognate to each amino acid varies (Table 1). It ranges from 4 (Trp) to 18 (Leu and Lys) in Nasonia and from 3 (Cys) to 18 (Gln) in honey bee. The number of tRNA genes cognate to each of the 20 standard amino acids shows a significant correlation (t-test p < 0.05) between Nasonia and honey bee as well as among other insects such as silk worm and different Drosophila species. But the total number of tRNA genes varies among different arthropods. The silk worm genome contains 496 tRNA genes (http://silkworm.genomics.org.cn/), the Aedes aegypti and Anopheles gambie mosquito genomes contain 906 and 414 tRNA genes (www.vectorbase.org), whereas Drosophila species contain from 200–400 tRNA genes (www.flybase.org). On the other hand, the branchiopod Daphnia pulex (water flea) genome contains an excessively large number (n = 3778) of tRNA genes (http://wfleabase.org/release1/current_release/trna-ncrna/). This suggests that genomic abundance of tRNA gene copies varies within arthropods. Among vertebrates also, specific species such as zebrafish genome (12752 tRNA genes) and cow genome (4112 tRNA genes) have excessive number of tRNA genes compared to that of other sequenced vertebrate genomes (that ranges within 200 to 900 only; see http://gtrnadb.ucsc.edu/). Thus, genomic abundance of tRNA copies doesn’t seem to have correlation with phylogenetic relationships but probably vary in species specific manner.
Table 1.
Number of tRNA genes in Nasonia and honey bee in comparison with that of silk worm and fruit fly.
| tRNA_Gene | Nvit | Amel | Bmor | Dmel |
|---|---|---|---|---|
| Ala | 16 | 14 | 54 | 17 |
| Arg | 10 | 13 | 26 | 26 |
| Asn | 8 | 8 | 29 | 10 |
| Asp | 10 | 9 | 34 | 14 |
| Cys | 5 | 3 | 11 | 7 |
| Gln | 9 | 18 | 17 | 12 |
| Glu | 14 | 11 | 28 | 20 |
| Gly | 17 | 14 | 49 | 20 |
| His | 8 | 7 | 15 | 5 |
| Ile | 12 | 8 | 23 | 11 |
| Leu | 18 | 11 | 27 | 22 |
| Lys | 18 | 13 | 21 | 19 |
| Met | 7 | 7 | 26 | 6 |
| Phe | 7 | 6 | 12 | 8 |
| Pro | 14 | 12 | 20 | 17 |
| Ser | 12 | 15 | 40 | 21 |
| Thr | 10 | 10 | 19 | 17 |
| Trp | 4 | 4 | 10 | 8 |
| Tyr | 9 | 5 | 12 | 10 |
| Val | 13 | 11 | 23 | 15 |
| TOTAL | 221 | 199 | 496 | 285 |
Patterns of genomic distribution of tRNA genes
Many tRNA genes are localized physically close to each other in insect genomes, including Nasonia and honey bee (Supplementary Table 2). In N. vitripennis, about 53% of the tRNA genes (n = 119) are located in small clusters. There are 40 tRNA-clusters that are distributed across 28 scaffolds in Nasonia. The number of tRNAs in these clusters varies from 2 to 6. In the A. mellifera genome, about 49% of the tRNA genes (n = 99) are also present in small physical clusters. A total of 30 tRNA-clusters are identified in the honey bee. They are localized in 23 linkage groups. The tRNA gene clusters contain either copies of a specific tRNA gene or different tRNA genes (Figure 1). The individual tRNA genes for which duplicate copies tend to form clusters vary among insect species (Table 2). Ala- and Pro-tRNA gene copies tend to accumulate in physical clusters in Nasonia but not in honey bee. Conversely, the bee genome contains a long cluster of 15 tRNA genes (of which 13 are Gln-tRNAs) that is absent in the Nasonia genome. Moreover, it seems that the number of clusters containing copies of a specific tRNA gene is generally lower in Nasonia and honey bee compared to that observed in different fruit fly species (Table 3). Although the biological significance of such clustering patterns of tRNAs in insects is not known, computational predictions (RNAfold) of the primary transcript of clustered tRNAs shows fold-back secondary structures in each (data not shown). It is plausible that clustering of tRNA copies may be related to the expression and biogenesis of these specific tRNA genes. Some of these clusters may be generated by tandem duplications of tRNA genes as evident from nearly equal intergenic distances between tRNA genes. The tRNA-Gln cluster of honey bee contains copies of this gene that are separated by similar distance (85–91 bp). However, we did not observe such patterns in the tRNA clusters of Nasonia.
Figure 1.
Clusters of tRNA genes (clusters with > 5 genes are shown) in Nasonia (filled) and honey bee (empty). Letters on top represent the amino acid the gene is cognate to, and numbers on the bottom show the intergenic distance in kb (rounded to one decimal).
Table 2.
tRNA genes that form clusters in genomes of different insect species.
| Species | tRNA Genes* |
|---|---|
| Nvit | Ala,Pro |
| Amel | Gln |
| Bmor | Tyr,Asp,Gly,Met,Asn,Ala |
| Dpul | Asp,Gly,Leu,Pro, |
| Dsim | Glu |
| Dsec | Asp,Glu |
| Dyak | Arg,Asn,Glu |
| Dmel | Glu |
| Dwil | Pro |
| Dvir | Asp |
| Dmoj | Gly |
| Dgri | None |
| Dere | Glu,Gly |
| Dana | Asn, Glu, Leu |
| Dpse | Leu |
| Dper | Leu |
A common criterion was set to compare only those clusters that contain more than 5 copies of the same tRNA gene in these genomes.
Table 3.
Number of clusters* of tRNA genes in genomes of Nasonia, honey bee, silk worm and different fruit flies.
| Species | Nt | Ns | Percentage |
|---|---|---|---|
| Nvit | 40 | 11 | 27.50 |
| Amel | 30 | 8 | 26.67 |
| Bmor | 34 | 15 | 44.12 |
| Dsim | 55 | 34 | 61.82 |
| Dsec | 73 | 49 | 67.12 |
| Dyak | 78 | 59 | 75.64 |
| Dmel | 69 | 52 | 75.36 |
| Dwil | 69 | 56 | 81.16 |
| Dvir | 52 | 36 | 69.23 |
| Dmoj | 48 | 35 | 72.92 |
| Dgri | 47 | 35 | 74.47 |
| Dere | 66 | 46 | 69.70 |
| Dana | 97 | 78 | 80.41 |
| Dpse | 68 | 52 | 76.47 |
| Dper | 64 | 48 | 75.00 |
Nt = Total number of clusters; Ns = No. of clusters that contain copies of the same tRNA gene. Percentage: percentage of Ns to Nt.
Each of these clusters contains at least 3 tRNA genes.
Patterns of sequence evolution
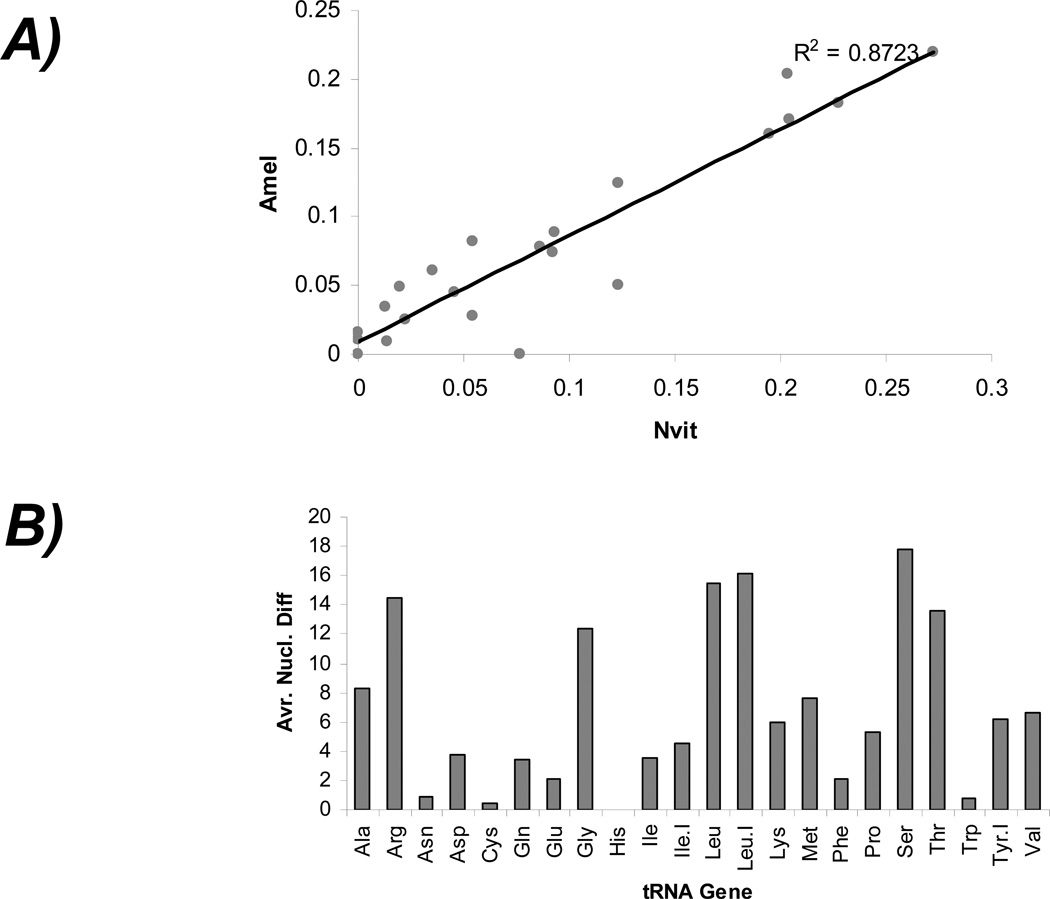
The tRNA genes show characteristic patterns of nucleotide sequence differences in Nasonia and honey bee. The nucleotide diversity (π values) within each tRNA gene family between Nasonia and honey bee is highly correlated (p = 0.05) suggesting high sequence conservation of tRNAs genes. However, the π values of Ala-, Arg-, Gly-, Leu-, Ser- and Thr- tRNA families are relatively higher compared to that of Asn, Cys and Trp tRNA genes between the two species (Figure 2). There is a high diversity of the tRNA-Met genes in spite of the fact that there is only one isoacceptor family in these tRNAs. Probably this is because separate initiator and elongator tRNAs are retained in the genome. In initiator tRNAs, the anticodon sequence CAT is preceded by C instead of T. We observed just one initiator tRNA-Met gene in each genome, all others were elongator tRNA-Met genes. We re-checked the sequence diversity of tRNA-Met genes after removing the initiator tRNA genes. Although the overall nucleotide diversity among the genes was reduced, we still found nucleotide variations in other parts of the genes (data not shown). We also compared sequence diversity of tRNA genes among other selected arthropods (Drosophila, silk worm, beetle, body louse and Daphnia) where genome sequences are available. The Leu-tRNAs and Glu-tRNAs show significant correlation (p = 0.0013) in nucleotide diversity in these species, whereas others like the Leu-tRNAs and Val-tRNAs do not. Similarly, the π values of Gly-tRNAs of these species show correlation with that of Ala-tRNA genes (p = 0.02) but not with that of Ser-tRNA genes (p = 0.286). These results suggest differential selection pressures on different tRNA gene families in these insects.
Figure 2.
Sequence evolution patterns of Nasonia and honey bee tRNA genes. A) Correlation in the nucleotide diversity (π value) of tRNA genes between Nasonia and honey bee. B) The average nucleotide differences (y-axis) vary among the tRNA genes of Nasonia and honey bee. Some tRNAs show the least number of nucleotide differences (highly conserved) whereas others show elevated numbers of nucleotide differences (relatively less conserved).
We also measured nucleotide differences between Nasonia and honey bee in specific critical regions that are important for tRNA structure and function. Based on nucleotide variation (π values) in the tRNA loop, arm or internal promoter regions, three distinct patterns of sequence evolution emerge: 1) perfectly conserved tRNA sequences ((π value = 0), 2) moderate level of nucleotide diversity (π value ranges within 0 and 0.2) and 3) least conserved tRNA sequences (π value > 0.2) (Table 4). Our data shows that the acceptor arm and the anti-codon loop are relatively more prone to nucleotide variation than the D-arm, T-arm or the internal promoter regions of the tRNA genes. The acceptor stem region is the 7-bp stem of a tRNA molecule that is formed by base pairing of the 5'-terminal nucleotides with the 3'-terminal nucleotides of the tRNA. This is used to attach the amino acid (using the 3’-CCA terminal motif) during the aminoacylation reaction. The anticodon loop is a 5-bp region that contains the anticodon triplet required for baseparing with the codon in the mRNA. The D arm is a 4 bp stem ending in a loop that often contains dihydrouridine. The T arm is a 5 bp stem containing the sequence TΨC where Ψ is a pseudouridine. The relatively higher diversity of acceptor arm and anticodon loop sequences compared to that of the D- and T-arm box sequences suggests stronger natural selection in these critical regions of tRNAs in these insects. The A-box (position: 8–19) and B-box (position: 52–62) represent the internal promoter sites for transcriptional regulation of tRNA genes. The B-box sequences show more variation than A-box sequences in these insects. Similar results are evident in human tRNA sequences (Goodenbour and Pan 2006). The greater conservation of the A-box sequence across species suggests a possible universal role in tRNA expression, whereas variation in B-box sequences may be involved in gene or species specific expression of tRNAs.
Table 4.
Nucleotide diversity (π) in different regions of tRNAs between Nasonia and honey bee.
| tRNA_location | Conserved+/+ | Conserved+/− | Moderately_Conserved& | Least_Conserved# |
|---|---|---|---|---|
| Acceptor_Arm | H,F | N,C,W,E,I | V,D,Q,K,P,M,A,T | R,G,L,S |
| D_Arm | N,C,E,Q,H,I,L,K,F,W | D,M,P,A,V | R,S,T | G |
| T_Arm | D,C,CQ,H,P,W | N,G,E,F,I,L,S | A,V,M,K,R | T |
| Anticodon_Loop | N,D,C,H,I,F,W | none | M,Q,K,E,G,T,P,A | R,V,S,L |
| A-box | C,E,H,W | P,N,D,Q,F,A,G | I,V,K,M,R,L,T,S | none |
| B-box | C,E,H,W | P,N,D,Q | F,I,V,K,A,M,R,L,T | S |
Note: +/+ means that each of these are perfectly conserved in both Nvit and Amel; +/− means that these genes are conserved in one species but show variation (π values within 0 to 0.2) in the other; the symbol ‘&’ means that genes show moderate levels of sequence variation (π values within 0 to 0.2); the symbol ‘#’ means that these genes show higher sequence variation (π > 0.2) suggesting that these are least conserved among tRNA genes of these two species. The entries with single letter abbreviations represent the standard amino acids to which the tRNA gene is cognate.
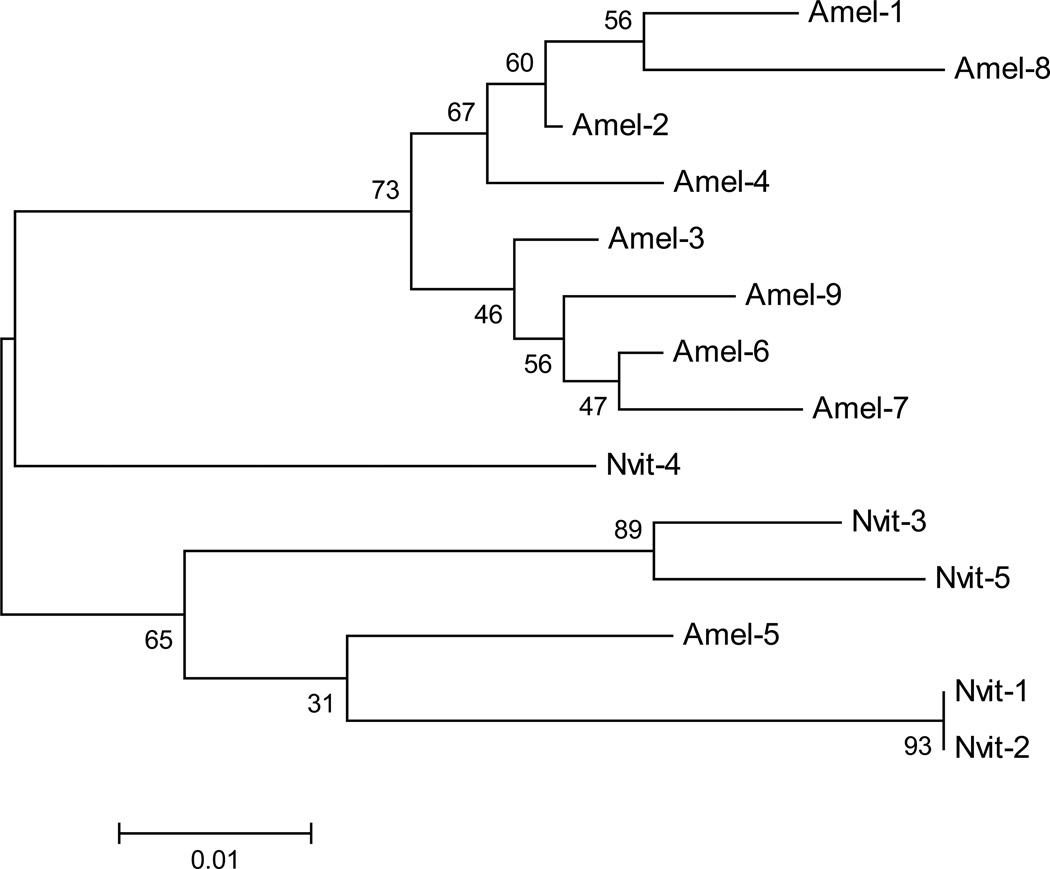
We also analyzed intron sequences of tRNAs. Only Tyr-, Ile- and Leu- tRNA copies contain introns in all the insects we analyzed. But, in water flea (Dpahnia pulex), copies of the Thr-, His-, Arg- and Gly- tRNA genes also contain intron. The copies of tRNAs genes other than Tyr-, Ile- and Leu- tRNAs are also known to contain introns in different non-insect species (Genomic tRNA database; http://gtrnadb.ucsc.edu/). In Nasonia and honey bee, each of the Tyr-tRNA gene copies contains an intron. On the other hand, the tRNA-Ile genes and tRNA-Leu genes contain intron only in specific copies (Table 5). Invariably, the introns are inserted at nucleotide position 38 in all Tyr-tRNA genes and at position 39 in the Ile- and Leu-tRNA genes in both Nasonia and honey bee. The intron sequences of Ile-tRNAs are highly conserved unlike those of the Tyr- and Leu-tRNAs in these species. Moreover, the Ile-, Leu- and Tyr- tRNA copies (intron-containing copies) seem to have originated from independent ancestral sequences. This is evident from convergence of present-day sequences of these genes with that of the reconstructed ancestral genes (Table 6). The ancestral DNAs were reconstructed independently for Ile-, Leu- and Tyr- tRNA genes of Nasonia and honey bee using maximum likelihood methodology (Yang 2007) and the convergence was determined as suggested in Cedergren et al. (1980). A phylogenetic analysis based on minimum evolution of Tyr-tRNA genes (Figure 3) of Nasonia and honey bee further supports that these sequences may have a common ancestor.
Table 5.
Number of intron containing and intronless Ile-, Leu- and Tyr- tRNA genes in Nasonia and honey bee.
| tRNA_Gene | Nvit | Amel |
|---|---|---|
| Tyr (I+) | 9 | 5 |
| Tyr (I−) | 0 | 0 |
| Ile (I+) | 3 | 2 |
| Ile (I−) | 9 | 6 |
| Leu (I+) | 3 | 3 |
| Leu (I−) | 15 | 8 |
I+ : intron-containing genes, I− : intron-less genes
Table 6.
Average nucleotide differences in the present-day tRNA genes of Nasonia and honey bee in comparison to that of the reconstructed ancestral sequences.
| tRNA Genes | Nvit vs. Amel Present-day |
Nvit vs. Amel Ancestral |
common or independent ancestor |
|---|---|---|---|
| Ile_intronless | 3.276 | 1.205 | common ancestor |
| Leu_intronless | 12.67 | 11.283 | common ancestor |
| Leu_intron | 10.467 | 5.333 | common ancestor |
| Tyr_intron | 8.736 | 4 | common ancestor |
Figure 3.
A minimum evolution phylogenetic tree of Tyr-tRNA genes of Nasonia and honey bee. The genetic distance scale is shown at the bottom.
Relationship of tRNA gene abundance and codon usage bias in Nasonia
The tRNA genes show differential copy numbers among the isoacceptor families between Nasonia and honey bee. Isoaceptors are tRNAs that are cognate to the same amino acid but recognize different codons (for that amino acid) in the mRNA. The variation of genomic abundance of these isoacceptor genes was estimated from the relative abundance of isoacceptor tRNAs (henceforth ‘RAIT’). RAIT is the ratio of observed number of tRNA gene copies to the expected number of isoacceptor copies in the tRNA gene family. The expected number of tRNA isoacceptors represents the average number of tRNAs with the same anticodon in a particular isotype tRNA gene family. This expectation should be valid if there were no bias in the anticodon abundance. However, the observed data suggests that the isoacceptor tRNA genes are not equally abundant. The Pro(CGG), Leu (CAA), Leu (TAA), Leu (CAG), Ser (AGA), Leu (AAG), Ser (GCT) and Ala (AGC) tRNA genes showed relatively higher RAIT variation than that of Ile(TAT), Met(CAT), Glu(TTC), Gly(TTC) tRNA genes between the two species.
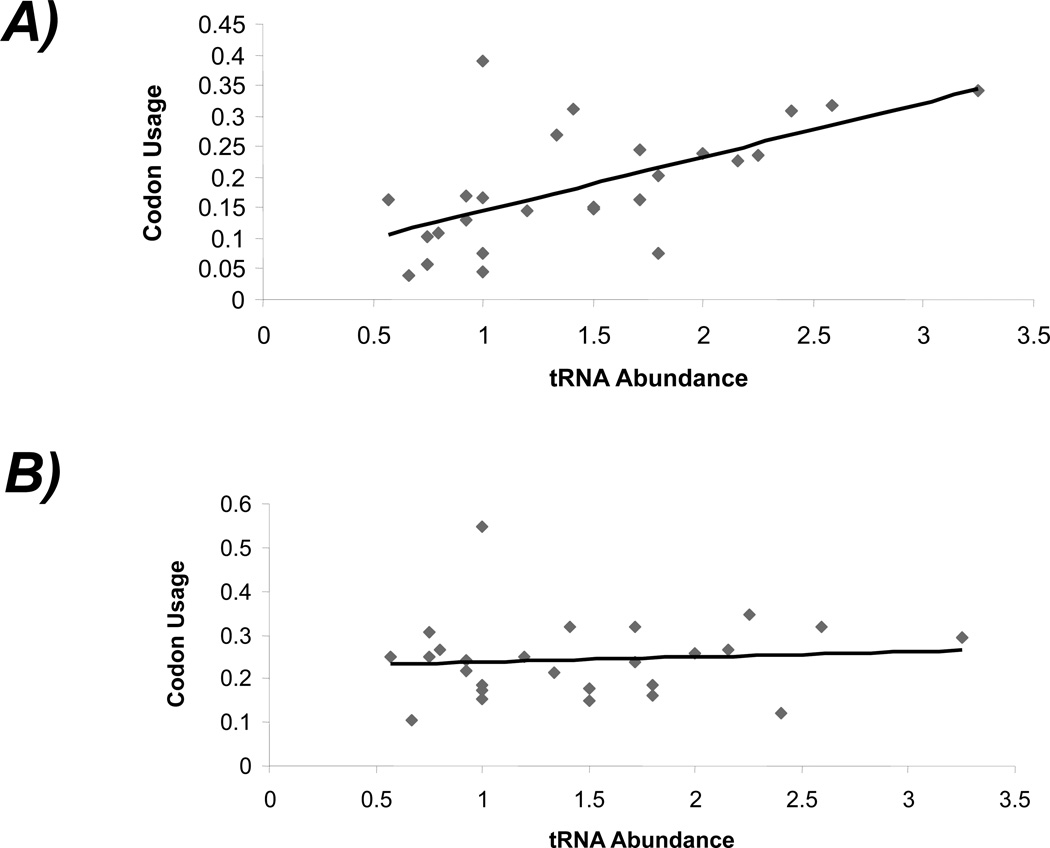
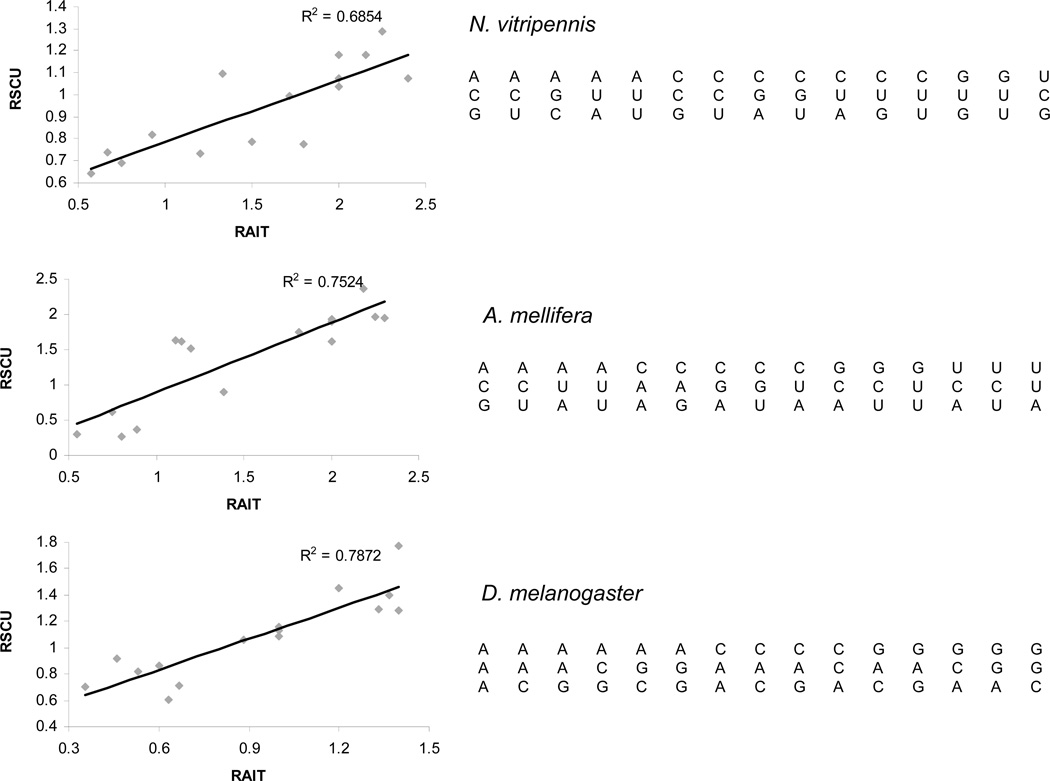
The variation of tRNA isoacceptor copies in the Nasonia genome shows differential correlation with the codon usage patterns of low expressing- and high expressing- genes. A set of 100 genes were selected from the official gene set (OGS v 1.2) of Nasonia based on representation to expressed transcripts in a non-normalized EST library (see methods). These genes show higher representation in the EST library than other genes of the official gene set suggesting that these genes may be relatively highly expressing genes compared to others. The relative synonymous codon usage (RSCU) values of these putative highly expressed genes tend to increase with the observed increase in RAIT in Nasonia. Such a tendency, however, is absent in other genes that were not qualified as ‘highly expressed’ (Figure 4). These results suggest that the highly expressed genes may be evolutionarily optimized for translational efficiency by use of codons that are more biased compared to the synonymous codons of lowly expressed genes. The codon usage bias arises due to unequal usage of synonymous codons while translating certain types of genes. Codon bias is regarded as a ‘translational selection’ strategy to control translational efficiency of genes (Dong et al. 1996; Rocha 2004; Higgs and Ran 2008). The role of genomic tRNA gene abundance on codon usage bias is known in some prokaryotes as well as eukaryotes (Kanaya et al. 1999; Rocha 2004; Higgs and Ran 2008). To determine if such a relationship between tRNA abundance and codon usage is also present in Nasonia and honey bee, we compared codon usage of 1:1 orthologous copies of ribosomal protein genes (RPGs) with the tRNA abundance (RAIT values). The RPGs are major house keeping genes and regarded as translationally efficient and equipped with optimized codons (Ben-Dor et al., 2007; Sharp and Li, 1987;) as they are required for protein translation at all times and spaces. Our data shows that the RSCU values of only a subset of codons (n = 15) of the RPGs show correlation with the relative abundance (RAIT values) of the cognate tRNA genes in these insects. But, these codons are different among Nasonia, honey bee and Drosophila suggesting that different residues are selected for codon optimization of RPGs in different insects. However, these codons included both the frequently used (preferred) and the rarely used (un-preferred) codons that corresponded to the abundant tRNA genes and the rare tRNA genes in the genome (Figure 5). Taken together, our results support a conclusion that the genomic abundance of tRNA genes may be linked with translation selection of Nasonia protein coding genes. And these evolutionary links between tRNA copy number and codon usage pattern may also exist in similar genes in other insect species.
Figure 4.
Contrasting patterns of the relationship of tRNA gene abundance in Nasonia genome with the codon usage between: A) a group of 100 genes predicted to be highly expressed and B) all other genes of Nasonia. Codon usage was measured by RSCU (relative synonymous codon usage) and tRNA abundance was measured by RAIT (relative abundance of isoacceptor tRNAs).
Figure 5.
Correlation patterns between RAIT (x-axis) and RSCU (y-axis) of orthologous ribosomal protein genes in N.vitripennis, A. mellifera and D. melanogaster. The specific codons that show correlation to the cognate tRNA copy number are shown at the right side of the graph.
Discussion
In this study, we identified and analyzed the nuclear tRNA genes of Nasonia and honey bee in a comparative manner. The tRNA genes show three distinct patterns of sequence evolution: the highly conserved, the moderately conserved and the least conserved tRNAs. The anticodon, along with the acceptor arm, contributes to the sequence variation among the least conserved tRNAs in these species. The first nucleotide of the anticodon triplet is more variable as expected. This is because the third nucleotide of codons generally determines degeneracy of codons. However, we observed variations in the second position of anticodons in some tRNAs of these species (data not shown). This may account for possible second nucleotides of codons as the wobble position. The second wobble positions are identified in some species and it is believed that second wobble may have a role in stabilizing the codon-anticodon binding between mRNA and tRNA (Lehmann and Libchaber 2008). We also observed the presence of intron sequences in three tRNA genes of these insects. It seems from our limited analysis that only Ile-, Leu- and Tyr- tRNAs may have introns in other insects such as fruit flies, mosquitoes, silk worm and beetle. The presence of an intron in a tRNA gene plays a critical role such as base modification (pseudouridine formation) of the anticodon triplet (Grosjean et al 1997; Johnson and Abelsonn 1983; Choffat et al. 1988; van Tol and Beier 1988) that affects codon degeneracy in the species. The tRNA genes that contain intron sequences vary from species to species (Genomic tRNA database: http://gtrnadb.ucsc.edu/). It will be interesting to understand why insects tend to have the same three tRNAs (Ile, leu and Tyr) that contain introns in their copies.
The sequence changes observed in the internal promoter regions (A- box and B-box) may affect the transcription of tRNA genes. However, in some cases, especially in insects, tRNA expression has been shown to be influenced by flanking regulatory sequences (Trivedi et al. 1999). Thus, the immediate flanking sequences of tRNA genes may be critical for maturation of active tRNAs. The maturation of the 5’-end of the tRNA precursor is performed by endonuclease RNase P whereas the 3’-terminus is processed by different endonucleolytic cleavage processes (Kirsebom 2007; Altman 2007). The genomic position of a tRNA gene may affect the biogenesis of its precursors. This is because the precursor sequences contain some flanking sequences of the gene. We observed differential genomic distributions of tRNA genes clusters in Nasonia and honey bee. It is however not known how such cluster patterns affect the tRNA expression and maturation.
Our analysis also provides some evidence that the tRNA gene abundance in Nasonia is correlated with highly expressed genes. We analyzed all the protein coding genes predicted from the Nasonia genome and found contrasting patterns of correlation of a small subset (n =100) of genes that seem to be highly expressed based on over-representation among ESTs. Unfortunately, we have not been able to identify the functional categories or gene ontologies associated with such genes. We speculate that these genes may be involved in some house keeping activities as these seem to be highly expressed. Thus, correlation of codon usage with rare and abundant tRNA copies in the genome suggests that specific codons may be preferred over other synonymous codons during translation of these genes. We also observed correlation between tRNA genes and codon usage patterns of ribosomal protein genes in Nasonia, honey bee and fruit fly. While such correlations have been observed in many other species (Ikemura 1985; Percudani et al. 1997; Hani and Feldmann 1998; Dittmar et al. 2006; Charles et al. 2006), in some organisms, codon usage can’t be explained based on tRNA gene copy number (Kanaya et al. 2001).
Methods
The tRNA genes of Drosophila melanogaster were used as query sequences to search the genome sequences of Nasonia (v1.0) and honey bee (v 4.0) for similar sequences (e-value threshold 0.01). The sequences of all the blast hits were then extracted using the NasoniaBase and BeeBase genome browsers and were run separately in the ARAGORN (Laslett and Canback 2004) and tRNAscanSE (Lowe and Eddy 1997) programs to identify tRNA genes. We also employed similar procedures to extract specific tRNA gene sequences of other selected arthropods where genome sequences are available. They include silk worm {Contigs(NCBI);10/1/2003}, body louse (PhumU1, Jan. 2007), beetle (Tcas_2.0; 14/09/2005), and water flea (Dpul JAZZ 1.0; 09/01/2006). The total number of tRNA genes annotated in some of these insects and their genomic positions were obtained from their respective genome sequence websites.
The genomic distribution of tRNA genes was determined based on their start and end coordinates. Clusters of tRNA copies in each genome were identified by determining the intergenic distances. To perform an unbiased comparison, we set a criterion to identify the clusters in different species, wherein we choose the clusters that contained at least three tRNA genes and where the distance between neighboring genes is limited within 10 kb. From the list of clusters obtained by above criteria, we separated the relatively longer clusters by choosing the specific clusters that contained six or more tRNA genes.
Nucleotide diversity of tRNA gene sequences was estimated using the DnaSP program (Rozas et al. 2003). We also independently measured nucleotide diversity of the loop, arm and promoter regions of tRNA sequences between Nasonia and honey bee. They include the acceptor arm, D-arm, T-arm, anticodon loop and A-box and B-box sequences. While the arm and loop sequences play critical roles in the aminoacylation of cognate amino acids during translation, the A- and B-box sequences serve as internal promoters for transcription of the tRNAs (Goodenbour and Pan, 2006).
The phylogenetic analysis was performed using the MEGA 3.0 program (Kumar et al. 2004) by the minimal evolution method, and a bootstrap test of the inferred phylogeny was performed with 1000 replications. ClustalW incorporated in MEGA 3.0 was used, with default set of parameters, to perform all multiple alignments. The ancestral DNA sequences were constructed using a maximum likelihood method with the BASEML program (Yang 2007).
We wanted to determine if the tRNA gene abundance has any relationship with the codon usage pattern in Nasonia. We compared the relative abundance of isoacceptor tRNAs (RAIT) in the genome with the relative synonymous codon usage (RSCU) of 1:1 orthologous ribosomal protein genes (RPG) of Nasonia, honey bee and fruit fly. RAIT values were calculated by dividing the observed number of tRNA isoacceptor genes with the average number of tRNAs within an isoacceptor family in that gene group. RSCU was determined using the CodonW program (http://codonw.sourceforge.net/). RSCU values refer to the number of times a particular codon is observed, relative to the number of times that the codon would be observed in the absence of any codon usage bias. The orthologous RPGs were determined by reciprocal blasts using the annotated RPG sequences from FlyBase.
We also compared the correlation of tRNA gene abundance with lowly expressed and highly expressed genes in Nasonia. The highly expressed genes in Nasonia were predicted from relative transcript abundance in two non-normalized EST (expressed sequence tag) libraries of Nasonia (Accession numbers: ES632969-ES651267). The ESTs were matched, by BLAT (Kent, 2002), against the rRNA sequences of all Nasonia genes (OGS v1.2). The BLAT results were filtered with criteria that included: 1) only the best alignment for each EST and the genes scoring at most 1% worse than the best were kept and 2) each alignment had at least 95% identity. Using this procedure, a set of 100 genes were predicted as highly expressed in Nasonia. Each of these 100 genes showed sequence alignments with more than 28 ESTs, unlike other genes predicted in the complete listing of OGS v 1.2. We compared the RAIT values with the cumulative RSCU values of these 100 genes and with cumulative RSCU values of the rest of the genes in OGS v1.2.
The two non-normalized EST libraries of N. vitripennis (one for late larval stages and one for pupal and adult stages) were generated as follows. Total RNA extractions were performed using RNeasy Mini Kit (Qiagen Inc., Valencia, CA) and cDNA libraries were constructed and sequenced as previously described (Hunter et al 2003). This resulted in 27,553 total reads. However, we noticed a high level of chimeric sequences present in the library, where fragments of multiple genes were joined together. In order to remove these, we ran BLAST searches (Altschul et al 1990) of the 100 bp on both ends of each EST against the N. vitripennis genome. ESTs with ends that matched different scaffolds of over 40 kb in length were removed. Additionally, we removed all ESTs which matched mitochondrial sequences, resulting in 18,688 high quality ESTs. These were submitted to GenBank (Accession numbers: ES632969-ES651267).
Supplementary Material
Acknowledgement
This work was supported in part by grant RO1-AI059342 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA
References
- Altman S. A view of RNase P. Mol Biosyst. 2007;3:604–607. doi: 10.1039/b707850c. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ben-Dor A, Bruhn L, Friedman N, Nachman I, Schummer M, Yakhini Z. Tissue classification with gene expression profiles. J Comput Biol. 2007;7:559–583. doi: 10.1089/106652700750050943. [DOI] [PubMed] [Google Scholar]
- Björk GR, Jacobsson K, Nilsson K, Johansson MJ, Byström AS, Persson OP. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedergren RJ, LaRue B, Sankoff D, Lapalme G, Grosjean H. Convergence and minimal mutation criteria for evaluating early events in tRNA evolution. Proc Natl Acad Sci U S A. 1980;77:2791–2795. doi: 10.1073/pnas.77.5.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles H, Calevro F, Vinuelas J, Fayard JM, Rahbe Y. Codon usage bias and tRNA over-expression in Buchnera aphidicola after aromatic amino acid nutritional stress on its host Acyrthosiphon pisum. Nucl. Acids Res. 2006;34:4583–4592. doi: 10.1093/nar/gkl597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choffat Y, Suter B, Behra R, Kubli E. Pseudouridine modification in the tRNA(Tyr) anticodon is dependent on the presence, but independent of the size and sequence, of the intron in eucaryotic tRNA(Tyr) genes. Mol Cell Biol. 1988;8:3332–3337. doi: 10.1128/mcb.8.8.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenbour JM, Pan T. Diversity of tRNA genes in eukaryotes. Nucl. Acids Res. 2006;34:6137–6146. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Szweykowska-Kulinska Z, Motorin Y, Fasiolo F, Simos G. Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: a review. Biochimie. 1997;79:293–302. doi: 10.1016/s0300-9084(97)83517-1. [DOI] [PubMed] [Google Scholar]
- Hani J, Feldmann tRNA genes and retroelements in the yeast genome. Nucl. Acids Res. 1998;26:689–696. doi: 10.1093/nar/26.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs PG, Ran W. Coevolution of codon usage and tRNA genes leads to alternative stable states of biased codon usage. Mol Biol Evol. 2008;25:2279–2291. doi: 10.1093/molbev/msn173. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Hunter WB, Dang PM, Bausher MG, Chaparro JX, McKendree W, Shatters RG, Jr, McKenzie CL, Sinisterra XH. Aphid biology: expressed genes from alate Toxoptera citricida, the brown citrus aphid. J Insect Sci. 2003;3:23. doi: 10.1093/jis/3.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PF, Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983;302:681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol. 2001;53:290–298. doi: 10.1007/s002390010219. [DOI] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kudo Y, Ikemura T. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene. 1999;238:143–155. doi: 10.1016/s0378-1119(99)00225-5. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom LA. RNase P RNA mediated cleavage: substrate recognition and catalysis. Biochimie. 2007;89:1183–1194. doi: 10.1016/j.biochi.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucl. Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann J, Libchaber A. Degeneracy of the genetic code and stability of the base pair at the second position of the anticodon. RNA. 2008;14:1264–1269. doi: 10.1261/rna.1029808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl. Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R, Pavesi A, Ottonello S. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J Mol Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- Rocha EP. Codon usage bias from tRNA's point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004;14:2279–2286. doi: 10.1101/gr.2896904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Saks ME, Sampson JR. Evolution of tRNA recognition systems and tRNA gene sequences. J Mol Evol. 1995;40:509–518. doi: 10.1007/BF00166619. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1895. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun FJ, Caetano-Anollés G. Transfer RNA and the origins of diversified life. Sci Prog. 2008;91:265–284. doi: 10.3184/003685008X360650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi A, Young LS, Ouyang C, Johnson DL, Sprague KU. A TATA element is required for tRNA promoter activity and confers TATA-binding protein responsiveness in Drosophila Schneider-2 cells. J Biol Chem. 1999;274:11369–11375. doi: 10.1074/jbc.274.16.11369. [DOI] [PubMed] [Google Scholar]
- van Tol H, Beier H. All human tRNATyr genes contain introns as a prerequisite for pseudouridine biosynthesis in the anticodon. Nucl. Acids Res. 1988;16:1951–1966. doi: 10.1093/nar/16.5.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.