Summary
Primary nociceptors relay painful touch information from the periphery to the spinal cord. While it is established that signals generated by the receptor tyrosine kinases TrkA and Ret coordinate the development of distinct nociceptive circuits, mechanisms modulating TrkA or Ret pathways in developing nociceptors are unknown. We have identified tumor necrosis factor receptor 1 (TNFR1) as a critical modifier of TrkA and Ret signaling in peptidergic and non-peptidergic nociceptors. In particular, TrkA+ peptidergic nociceptors require TNFα-TNFR1 forward signaling to suppress NGF-mediated neurite growth, survival, excitability, and differentiation. Conversely, TNFR1-TNFα reverse signaling augments the neurite growth and excitability of Ret+ non-peptidergic nociceptors. The developmental and functional nociceptive defects associated with loss of TNFR1 signaling manifest behaviorally as lower pain thresholds caused by increased sensitivity to NGF. Thus, TNFR1 exerts a dual role in nociceptor information processing by suppressing TrkA and enhancing Ret signaling in peptidergic and non-peptidergic nociceptors, respectively.
Keywords: NGF, TrkA, TNFα, TNFR1, TNFR, nociceptors, pain, DRG, sensory, signaling, Ret, development, peripheral nervous system, touch, somatosensory, reverse signaling, neurturin
Introduction
The ability to discriminate between innocuous and painful touch is critical for survival. Nociceptors are polymodal sensory neurons that relay information about noxious tactile cues from peripheral targets to the spinal cord (Julius and Basbaum 2001, Craig 2003, Basbaum et al. 2009). Importantly, nociceptors are defined by their high excitability thresholds for thermal, mechanical, or chemical stimuli, meaning that pain is perceived only when these stimuli are potent enough to trigger action potentials (Woolf and Salter 2000, Hunt and Mantyh 2001). Critical properties of nociceptors that govern the perceived intensity of a painful stimulus include density of peripheral target innervation, proper targeting of central projections, and high excitability thresholds (Pezet and McMahon 2006). It follows that if any of these components were enhanced or attenuated then the perception of pain would be altered.
Target-derived nerve growth factor (NGF) initiates signaling pathways critical for tuning nociceptor sensitivity through the regulation of survival, neurite growth, and excitability (Crowley et al. 1994, Patel et al. 2000, Chuang et al. 2001). Peripheral targets such as the skin release NGF during development, around the time that newly born nociceptors begin to innervate peripheral targets (Marmigère and Ernfors 2007, Lallemend and Ernfors 2012). Secreted NGF binds to a receptor tyrosine kinase (RTK), TrkA, on developing nociceptors to transduce intracellular signals that are required for survival and target innervation (Crowley et al. 1994, Patel et al. 2000). Deletion of genes encoding NGF or TrkA causes the death of all nociceptors during development and, as a result, pain insensitivity (Verhoeven et al. 2006, Indo et al. 1996). Conversely, hyperactivation of NGF-TrkA signaling skews pain thresholds toward hypersensitivity causing hyperalgesia or allodynia (Lewin et al. 1993, Woolf and Salter 2000, Costigan et al. 2009, Mantyh 2011). Thus, NGF signaling must be tightly controlled during development such that painful stimuli are properly interpreted.
Maturation of several functionally distinct nociceptor subclasses also requires NGF-TrkA signaling during embryonic and postnatal nociceptor development (Luo et al. 2007, Gascon et al. 2010, Marmigère and Ernfors 2007). The two major populations of mature nociceptor subclasses are peptidergic and non-peptidergic nociceptors. Peptidergic nociceptors express TrkA, calcitonin gene-related peptide (CGRP), and substance P throughout development and are responsive to target-derived NGF (Lallemend and Ernfors 2012). Non-peptidergic nociceptors do not express TrkA or neuropeptides, but rather express an RTK, Ret, which is responsive to the target-derived glial cell line-derived neurotrophic factor (GDNF) family of ligands (Airaksinen and Saarma 2002, Molliver et al. 1997). Loss of GDNF family receptor α 2 (GFRα2), a Ret co-receptor that responds to the GDNF family ligand, neurturin (NRTN), impairs peripheral target innervation and sensitivity to inflammatory pain (Lindfors et al. 2006). Interestingly, acute pain is sensed by TrkA+ peptidergic nociceptors in the absence of NRTN signaling, suggesting that TrkA+ and Ret+ nociceptors are functionally distinct from one another in adulthood despite being derived from the same early TrkA+ nociceptor lineage during embryonic development (Marmigère and Ernfors 2007, Liu and Ma 2011). Together, these studies suggest that NGF-TrkA signaling is critical for the orchestration of peptidergic and non-peptidergic nociceptive circuits, which underlie the perception of distinct forms of painful touch.
Given the broad phenotypes and functions that NGF-TrkA signaling regulates, it stands to reason that there should be factors that modulate TrkA signals and, by extension, nociceptor development and function. Clues for this hypothesis come from studies in the sympathetic nervous system, which also requires NGF-TrkA signaling for survival and target innervation (Crowley et al. 1994, Glebova and Ginty 2004). In sympathetic neurons, antagonism of NGF-TrkA signals can occur via the p75 neurotrophin receptor (p75NTR), which induces death (Deppmann et al. 2008), promotes axon pruning (Singh et al. 2008), dampens excitability (Luther and Birren 2009), and restricts post-synaptic densities (Sharma et al. 2010). As such, elimination of p75NTR is analogous to TrkA gain-of-function in sympathetic neurons. Consequently, one would expect that the loss of p75NTR in nociceptors would manifest as heightened pain sensitivity. However, numerous reports argue the opposite; loss of p75NTR results in decreased pain sensitivity and reduced cutaneous innervation by nociceptors (Lee et al. 1992, Bergmann et al. 1997). Nociceptors from p75NTR−/− mice are roughly 3-fold less sensitive to NGF (Davies et al. 1993, Lee et al. 1994), which suggests that p75NTR likely augments NGF-TrkA signaling in nociceptors.
If not p75NTR, might a different receptor gate NGF-TrkA signaling in nociceptors? It is possible that receptors structurally related to p75NTR may function as negative regulators of NGF-TrkA signals in nociceptors. Indeed, p75NTR is just one of 29 tumor necrosis factor (TNF) receptor (TNFR) superfamily members, many of which are implicated in the modulation of growth factor signaling (Deppmann and Janes 2013, Locksley et al. 2001). To test this idea, we screened for the expression of 23 TNFR family members in the dorsal root ganglia (DRG), which revealed the selective expression of three highly related TNFR family members: TNFR1, p75NTR, and death receptor 6 (DR6). Herein we pursued the role of TNFR1 signaling in nociceptor development in part because it has been shown to negatively regulate events such as neurite growth and survival in the sympathetic nervous system (Barker et al. 2001, Kisiswa et al. 2013). Similar to phenotypes observed in the sympathetic nervous system, we report that TNFR1 forward signaling can also inhibit the development of NGF-TrkA-dependent sensory circuits by negatively regulating neurite growth, cell number, and excitability of TrkA+ primary nociceptors. Moreover, loss of either TNFαor TNFR1 forward signaling drives TrkA+ nociceptors toward a premature non-peptidergic fate. Surprisingly, we found that TNFR1 plays a dichotomous role in the function of peptidergic and non-peptidergic nociceptors. While TrkA+ peptidergic nociceptors exhibit increased responsiveness to NGF as evidenced by enhanced growth and excitability in the absence of TNFR1 or TNFα, the Ret+/TrkA− population of non-peptidergic nociceptors exhibits functional impairments in NRTN-dependent neurite outgrowth and excitability due to absence of TNFR1-TNFα reverse signaling. We demonstrate that Tnf and Tnfr1 null mice are hypersensitive to several pain modalities, likely caused by enhanced nociceptor sensitivity to NGF. Finally, we provide genetic and biochemical evidence that the increased gain in nociceptive signals observed in Tnf or Tnfr1 knockouts is linked to an increase in the NGF sensitivity of TrkA+ nociceptors. These results suggest that TNFα and TNFR1 coordinate the development and function of molecularly distinct nociceptive circuits through cross-talk with TrkA and Ret to either block or promote pain sensitivity in peptidergic and non-peptidergic nociceptor populations, respectively.
Results
Characterization of TNFα and TNFR1 expression on nociceptors and their targets
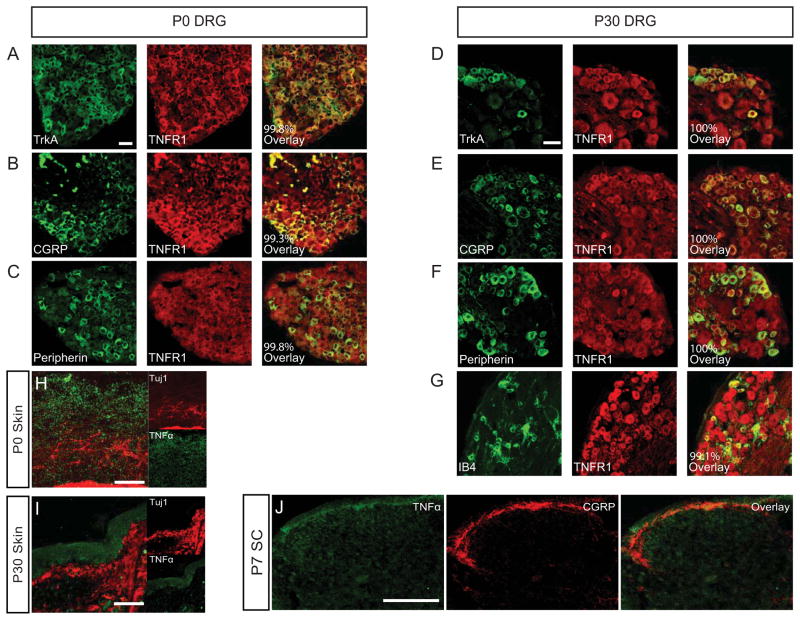
In order to identify putative factors that can antagonize NGF-TrkA dependent signaling in nociceptors, we examined the expression of 23 TNFR family members in embryonic day (E)18.5 DRG, brain, or muscle via reverse transcriptase polymerase chain reaction (RT-PCR) (Figure S1A). This analysis revealed expression of three highly related TNFR family members in the DRG: p75NTR (TNFRSF16), Dr6 (TNFRSF21), and Tnfr1 (TNFRSF1A). We focused on TNFR1, which is an understudied TNFR family member in the context of nociceptor development and function. Given that TNFR1 is robustly expressed in the DRG, which contains many different cell types, we next sought to determine whether it is expressed on nociceptors. To this end, we performed immunohistochemistry to examine percent colocalization of TNFR1 with CGRP, peripherin, or TrkA at postnatal day (P)0 (Figure 1A–C). CGRP and TrkA represent peptidergic nociceptors while peripherin labels small-diameter unmyelinated nociceptors. In each case, TNFR1 colocalizes with nociceptive markers >99% of the time (Figure S2A). In addition, we found that TNFR1 appears to be broadly expressed in the DRG as it colocalizes with parvalbumin+ proprioceptive neurons (Figure S2C).
Figure 1. TNFR1 is expressed by nociceptive neurons and TNFα is expressed in nociceptor targets.
(A–C) Quantification of TNFR1 colocalization the nociceptive markers TrkA, CGRP, and peripherin in the P0 DRG. Scale bar represents 30 μm.
(D–G) Quantification of TNFR1 colocalization with the same nociceptive markers above plus IB4 as a marker of non-peptidergic nociceptors in the P30 DRG. Scale bar represents 60 μm.
(H–I) TNFα is enriched mainly in the epidermis at P0 and P30. Scale bar represents 120 μm.
(J) Expression of TNFα in the marginal zone and dorsal horn neurons of the spinal cord. Scale bar represents 150 μm.
See also Figure S1–2
We performed the same analysis at P30 when most nociceptors are terminally specialized into distinct functional subclasses (Lallemend and Ernfors 2012). In addition to CGRP, TrkA, and peripherin, we examined non-peptidergic neurons by staining with the fluorescently conjugated lectin, IB4. We observed similar levels of colocalization at P30 as at P0 (i.e. at least >99% of nociceptors are positive for TNFR1) (Figure 1D–G, Figure S2B). We validated the TNFR1 antibody (Figure S1B) and found that TNFR1 is localized to both the cell bodies and axons of sensory neurons both in vivo (Figure S1C) and in vitro (Figure S1D). Together, these analyses suggest that TNFR1 is ubiquitously expressed by nociceptors across different stages of sensory circuit development, ranging from early to mature nociceptor populations.
We also sought to localize the sources of TNFα in developing nociceptive circuits. After validating the TNFα antibody (Figure S1E–G), we found that TNFα is enriched on sensory neurons in the P4 DRG (Figure S1H), in the epidermis between P0 and P30 (Figure 1H–I), and in the marginal zone and second order spinal cord neurons at P7 (Figure 1J). Importantly, we found that TNFR2, a close relative of TNFR1 that also binds TNFα, was absent from the DRG (Figure S1A) and spinal cord (Figure S1I). Taken together these data suggest that sensory neurons, peripheral targets, and central targets express TNFα, which may signal to nociceptors exclusively through TNFR1 during critical periods of nociceptor circuit development and maturation.
TNFα-TNFR1 signaling suppresses skin innervation and NGF-dependent neurite growth programs
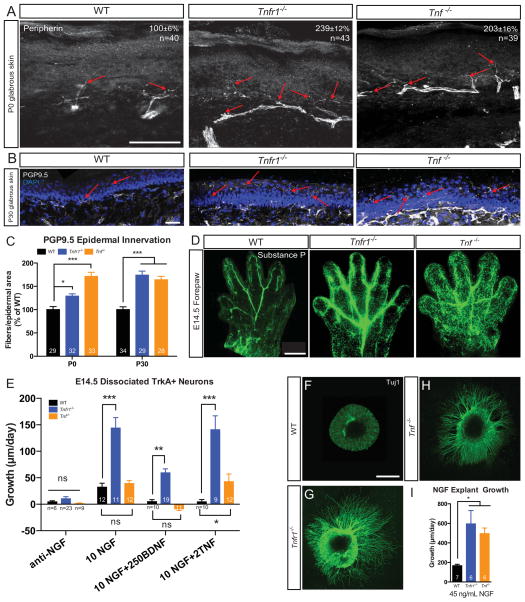
During normal developmental processes, a critical function of NGF-TrkA signaling is control of nociceptor neurite growth into peripheral targets (Patel et al. 2000). If TNFα-TNFR1 signaling antagonizes NGF-TrkA dependent growth of nociceptors into peripheral targets, we would predict that axon overgrowth into the skin would be observed in Tnfr1−/− and Tnf −/− mice. To test whether TNFα-TNFR1 signaling is indeed required, in vivo, for proper axon innervation of peripheral targets, we performed immunohistochemical analyses for nociceptive fibers in the epidermis, as described previously (Zylka et al. 2005). First, we observed that there is an increased density of small-diameter peripherin+ fibers in glabrous skin at P0 in Tnfr1−/− and Tnf −/− animals relative to WT (Figure 2A). We then stained for the pan-axonal marker protein gene product 9.5 (PGP9.5) and observed twice as many PGP9.5+ nociceptive fibers projecting into the epidermisin Tnfr1−/− or Tnf −/− mice than WT mice by P30, suggesting that TNFα and TNFR1 normally suppress neurite growth programs in the periphery (Figure 2B–C). Finally, whole-mount substance P immunostaining of the forepaw revealed that as early as E14.5, nociceptive fibers densely innervate the digits and footpad of the paw in mice lacking TNFα or TNFR1 (Figure 2D). Taken together these data suggest that the TNFα-TNFR1 pathway is critical in regulating growth and refinement of nociceptor peripheral projections in vivo.
Figure 2. Tnfr1−/− and Tnf −/− nociceptive fibers hyperinnervate the skin and are hypersensitive to NGF-dependent growth.
(A) Peripherin immunostaining of hind-paw thin glabrous skin. Arrows point to axons sprouting into the cutaneous field. Scale bar represents 50 μm. Quantification shown is peripherin+ fiber cutaneous field density normalized as percent of WT mean. 5 animals of each genotype examined.
(B–C) PGP9.5 immunostaining of thin glabrous skin and quantification of epidermal innervation. (B) P30 thin glabrous skin. Arrows point to invading fibers. (C) Quantification of the number of PGP9.5 neurites crossing into the epidermal field at P0 and P30, normalized to percent of WT. 5 animals of each genotype analyzed per time point. Scale bar represents 25 μm.
(D) Representative whole mount immunostaining of E14.5 forepaws for the peptidergic nociceptor marker substance P; n=4 paws from 4 mice stained per genotype. Scale bar represents 50 μm.
(E) In vitro neurite growth of E14.5 TrkA+ nociceptors grown in NGF and measured in compartmentalized chambers. Data shown are from 2–6 independent experiments for each condition.
(F–I) Tuj1 immunostaining (F–H) and quantification (I) of E14.5 explant outgrowth in response to 45 ng/mL NGF for 24 hours. Scale bar represents 500μm. Explants from ≥3 mice per genotype.
1-way (I) or 2-way (C,E) ANOVA, Bonferroni post-test. Data represent mean ± SEM, ns: not significant, *p<0.05, **p<0.01, ***p<0.001.
To examine the role of TNFR1 on NGF-TrkA dependent neurite growth in vitro, we established dissociated sensory neurons from WT, Tnfr1−/−, or Tnf−/− mice in microfluidic devices, which spatially isolate neuronal cell bodies and neurites (Sharma et al. 2010). We found that WT neurons bathed in NGF along with TNFα or brain-derived neurotrophic factor (BDNF) showed significantly lower rates of neurite growth than WT neurons bathed in NGF alone (Figure 2E). Strikingly, Tnfr1−/− axons grew roughly 5 times faster than WT neurons in response to 10 ng/mL NGF, suggesting that TNFR1 plays a role in suppressing NGF-dependent neurite growth (Figure 2E). While TNFα had no effect on NGF dependent neurite growth in Tnfr1−/− neurons, we observed that BDNF still slowed neurite growth in Tnfr1−/− neurons bathed in NGF, albeit to a lesser degree than BDNF treatment on WT mice neurons (Figure 2E). Interestingly, neurite growth of dissociated Tnf−/− neurons revealed separable roles for TNFα and TNFR1, suggesting that TNFR1 may suppress nociceptor neurite growth in the absence of TNFα. To assess the effect of NGF mediated neurite growth on whole ganglia from Tnfr1−/− and Tnf −/− mice, E14.5 DRG explants were cultured in NGF, which yielded results corroborating excessive NGF dependent growth in the Tnfr1−/− and Tnf −/− explants; Tnfr1−/− and Tnf −/− neurites grew ~3 times faster than WT neurites (Figure 2F–I). It is possible that paracrine, rather than autocrine, axon pruning programs are more dominant, yielding the difference in relative axon growth rates between dissociated Tnf −/− nociceptors and explant cultures. Together, these data suggest that neurons lacking Tnfr1 or Tnf are more responsive to NGF in the context of in vitro neurite growth assays due to loss of TNFα-TNFR1 forward signaling.
TNFα-TNFR1 signaling is required for proper guidance of peptidergic central projections
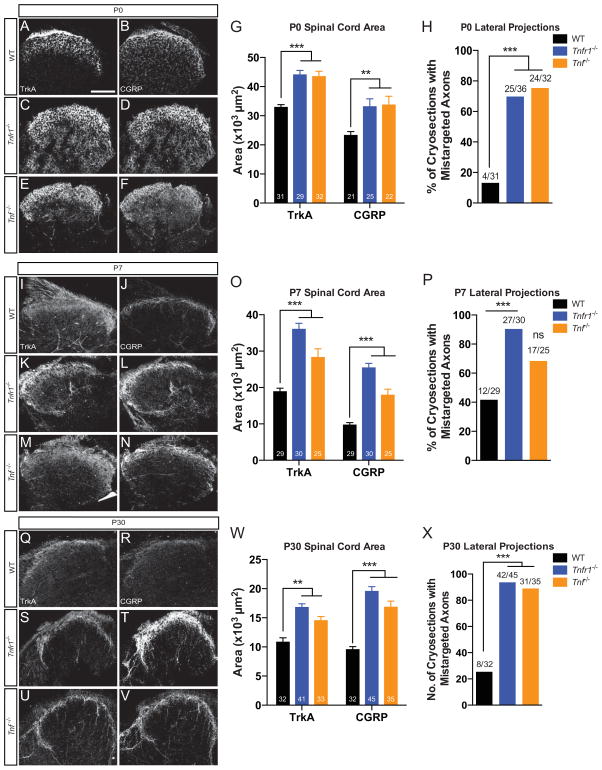
In Tnfr1−/− and Tnf −/− mice, nociceptor peripheral projections display phenotypes reminiscent of those observed in classical NGF-TrkA gain-of-function experiments demonstrating axon overgrowth into peripheral targets (Aloe et al. 1975). Therefore, we sought to test whether nociceptor central projections were disrupted similar to peripheral projections in Tnfr1−/− and Tnf −/− mice. To address this, we compared peptidergic (TrkA+, CGRP+) central projections in the spinal cord between WT, Tnfr1−/−, and Tnf −/− mice at P0, P7, and P30. We noted complete segregation of peptidergic (CGRP+) and non-peptidergic (IB4+) axons in the dorsal horn by P30 (Figure S3A–C), suggesting that non-peptidergic nociceptor fate specification is intact (Chen et al. 2006b). We found that mice lacking Tnfr1 or Tnf display a ~50% larger area occupied by TrkA+ and CGRP+ peptidergic fibers than WT lumbar dorsal horn at P0 (Figure 3A–G). Moreover, most sections contained axon bundles that misprojected within the dorsal horn and extended medially and/or laterally (Figure 3H). The same analyses performed at P7 and P30 revealed similar phenotypes suggesting that central projection defects are stable even after terminal differentiation (Figure 3I–X). The enhanced growth and misguidance of Tnfr1−/− and Tnf −/− central projections are reminiscent of other pathways that negatively modulate NGF-dependent central projections such as HoxD1 (Guo et al. 2011), suggesting that TNFα-TNFR1 signaling is also required for proper growth and refinement of nociceptor central projections.
Figure 3. Tnfr1−/− and Tnf −/− peptidergic central projections are robust and misguided.
(A–F) TrkA and CGRP immunostaining in P0 WT (A–B), Tnfr1−/− (C–D), and Tnf −/− (E–F) lumbar spinal cord. Quantification of area (G) and lateral projections (H) within WT, Tnfr1−/−, and Tnf −/− spinal cord sections. Scale bar represents 100 μm.
(I–X) Immunostaining and quantification of P7 (I–P) and P30 (Q–X) lumbar spinal cord.
See also Figure S3.
2-way ANOVA, Bonferroni post-test (G,O,W) and Fisher’s exact test (H,P,X). 5 mice analyzed per genotype per time point.
Data represent mean±SEM, ns: not significant, *p<0.05, **p<0.01, ***p<0.001.
TNFα-TNFR1 signaling coordinates nociceptor differentiation
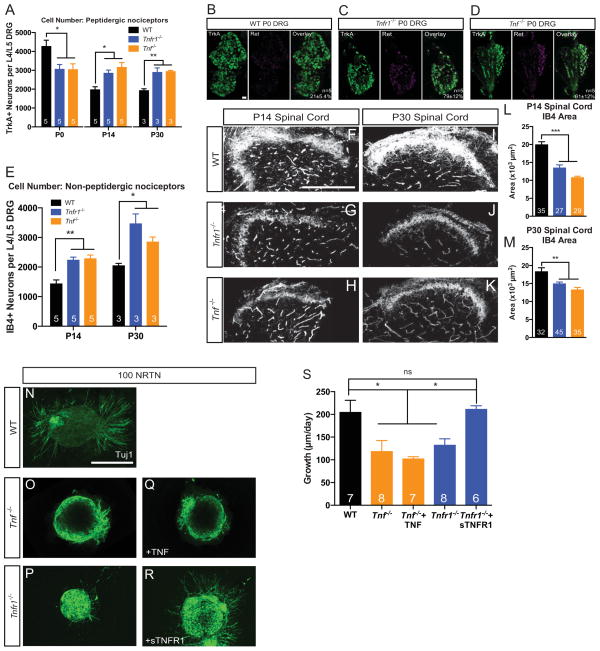
There are several possible explanations for the increase in peptidergic peripheral and central projection densities that occur in Tnfr1−/− and Tnf −/− mice: 1) an increase in the neurite density of individual nociceptors and/or 2) more nociceptors in the DRG, which would increase the overall number of fibers projecting to the skin and spinal cord. Although we have established that loss of TNFR1 signaling enhances NGF dependent axon growth, it is also possible that TNFR1 antagonizes non-axon growth aspects of NGF-TrkA signaling such as cell death and differentiation in the DRG. To test this possibility we examined cell number in the DRG by counting the number of TrkA+ nociceptors at P0, P14, and P30, time points corresponding to key regulatory events in the survival and differentiation of nociceptors (Marmigère and Ernfors 2007). Surprisingly, we observed a significant decrease in the TrkA+ nociceptor population at P0 in Tnfr1−/− and Tnf −/− mice but, relative to WT, more TrkA+ neurons at P14 and P30 (Figure 4A). It is possible that the appearance of fewer TrkA+ neurons at P0 may not be due to an altered trophic threshold for survival; rather, it may have to do with altered differentiation programs. Indeed, it is established that early TrkA+ nociceptors downregulate TrkA/CGRP and upregulate Ret to become non-peptidergic nociceptors through a process that is itself dependent on the strength of NGF-TrkA signals (Luo et al. 2007). Thus, if a Tnf or Tnfr1 deletion enhances NGF sensitivity in TrkA+ nociceptors, an increased number of early TrkA+ neurons would be driven toward a non-peptidergic fate. To examine this possibility, we assessed putative non-peptidergic cell number in the DRG by counting the number of Ret+/TrkA+ (differentiating) and IB4+ (terminally differentiated) neurons across time, as previously described (Luo et al. 2007). The number of differentiating Ret+/TrkA+ cells in Tnfr1−/− or Tnf −/− P0 DRGs was ~3 times higher than the number of differentiating neurons in WT mice (Figure 4B–D). At P14 and P30, the increase in non-peptidergic neuron number persists in Tnfr1−/− and Tnf −/− mice compared to WT (Figure 4E). These data suggest that TNFα-TNFR1 signals work during nociceptive differentiation to dampen the NGF-TrkA-dependent drive toward a non-peptidergic fate and loss of these signals leads to higher numbers of both peptidergic and non-peptidergic nociceptors.
Figure 4. Deletion of Tnfr1 or Tnf leads to the premature differentiation and impaired growth of non-peptidergic nociceptors.
(A) Quantification of the number of TrkA+ sensory neurons in the L4/L5 DRG at P0, P14, and P30.
(B–D) Representative images of Ret/TrkA colocalization in the P0 DRG from WT, Tnfr1−/−, and Tnf −/− mice. Scale bar represents 30 μm. Quantification of colocalization shown on overlay.
(E) Quantification of the number of IB4+ neurons in the L4/L5 DRG at P14 and P30.
(F–M) Immunostaining and quantification of IB4+ axons invading the spinal cord dorsal horn at P14 (F–H) and P30 (I–K) in WT, Tnfr1−/−, and Tnf −/− mice. Quantification of IB4+ axon area shown in (L–M). Scale bar represents 100 μm.
(N–S) E14.5 DRG explants cultured in 100 ng/mL of NRTN (N–P), 100 ng/mL NRTN + 2 ng/mL TNF (Q), or 100 ng/mL NRTN + 5 μg/mL sTNFR1 (R) for 24 hours. Explants from ≥3 mice per condition. Scale bar represents 100 μm.
1-way ANOVA, Bonferroni post-test (A,L,M) or Tukey post-test (S). 2-way ANOVA, Bonferroni post-test (E). Number of mice analyzed is indicated in A,E. 5 mice analyzed in L–M.
Data represent mean±SEM, ns: not significant, *p<0.05, **p<0.01, ***p<0.001.
TNFR1-TNFα signaling is required for non-peptidergic nociceptor axon extension into the spinal cord and NRTN-dependent neurite outgrowth
We next sought to determine whether TNFα and TNFR1 specifically influence developmental processes in TrkA+ neurons or can more broadly affect other nociceptor pools such as Ret-expressing non-peptidergic neurons. To address this we used WT, Tnfr1−/− and Tnf −/− mice to evaluate whether the extent of non-peptidergic central projections would reflect the robust expansion into the dorsal horn observed for peptidergic nociceptor central projections. Surprisingly, there was approximately a 30–50% reduction in the area devoted to non-peptidergic, IB4+ axons in the dorsal horn at P14 and P30 in Tnfr1−/− or Tnf −/− mice relative to WT mice (Figure 4F–M). Having observed an increase in the number of these non-peptidergic, IB4+ cells in Tnfr1−/− and Tnf −/− mice, this finding suggests that TNFα-TNFR1 signaling is required for the maintenance of non-peptidergic nociceptor central projections.
A well-established signaling pathway implicated in promoting the growth of non-peptidergic nociceptive axons is through the ligand, co-receptor, and RTK: NRTN, GFRα2, and Ret, respectively (Stucky et al. 1999, Airaksinen and Saarma 2002, Lindfors et al. 2006, Luo et al. 2007). Therefore, we next asked whether loss of TNFα or its receptor could influence Ret-dependent neurite growth. To this end, we cultured E14.5 DRG explants from WT, Tnfr1−/−, and Tnf −/− mice in 100 ng/mL NRTN. Surprisingly, in response to NRTN, neurite outgrowth was significantly lower in Tnfr1−/− and Tnf −/− explants compared to WT (Figure 4N–P), consistent with in vivo observations of decreased density of non-peptidergic axons in lamina IIi of the spinal cord. We initially attempted to rescue this impaired neurite outgrowth by activating TNFα-TNFR1 forward signaling through incubation of Tnf −/− explants in TNFα+NRTN (Figure 4Q), which yielded no significant differences when compared to the growth of Tnf −/− explants grown solely in NRTN (Figure 4S). Given that a recent study implicated TNFR1-TNFα reverse signaling (Sun & Fink 2007) in the growth of sympathetic axons (Kisiswa et al. 2013), we hypothesized that perhaps TNFα-TNFR1 reverse signaling, rather than forward signaling, is responsible for non-peptidergic neurite outgrowth. We tested this idea by incubating E14.5 explant cultures in NRTN as well as selective re-addition of soluble TNFR1 (sTNFR1) to Tnfr1−/− explants to activate TNFR1-TNFα reverse signaling pathways. Consistent with a reverse signaling paradigm, sTNFR1 addition to Tnfr1−/− explants restored neurite growth to WT levels(Figure 4R–S). These data suggest that in contrast to the repressive effects of TNFα-TNFR1 forward signaling on NGF-TrkA dependent growth, a TNFR1-TNFα reverse signaling program enhances NRTN-GFRα2-Ret dependent neurite growth in non-peptidergic nociceptors.
TNFα and TNFR1 are required to differentially regulate the excitability of peptidergic and non-peptidergic nociceptors
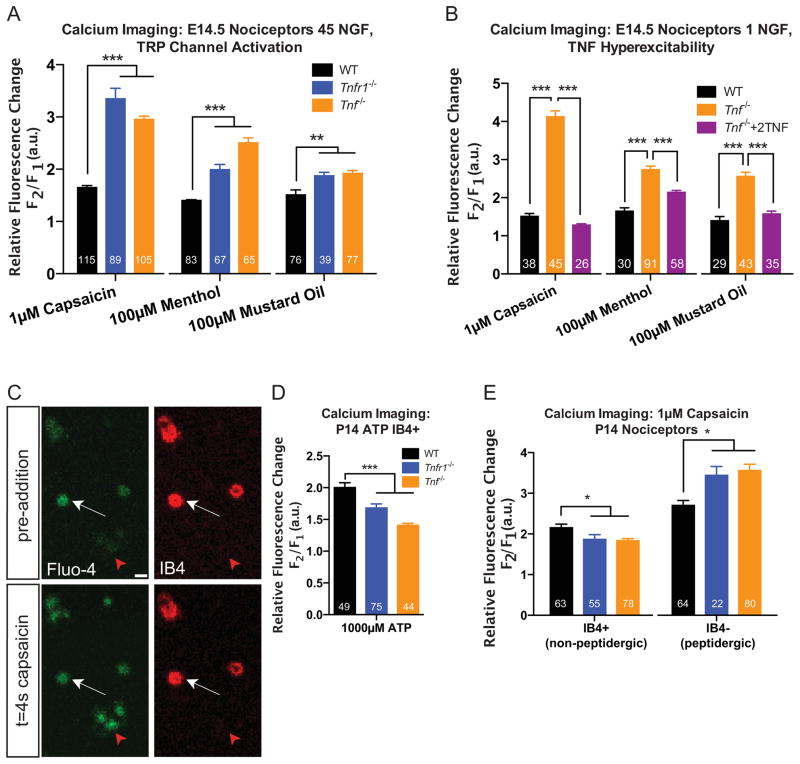
Nociceptors sense thermal, mechanical, and chemical stimuli through the expression of different ion channels such as the transient receptor potential (TRP) family of ion channels (Clapham 2003, Moran et al. 2011). Importantly, RTK signaling modulates the excitability of ion channels in nociceptors. For instance, NGF-TrkA signaling can enhance the expression and sensitivity of TRP ion channels in peptidergic nociceptors (Chuang et al. 2001, Zhang et al. 2005, Ji et al. 2002, Luo et al. 2007) whileNRTN-GFRα2-Ret signaling regulates expression of the ATP-gated ion channel, P2X purinoceptor 3 in non-peptidergic nociceptors (Wang et al. 2013). To test whether Tnfr1−/− and Tnf −/− peptidergic nociceptors exhibit altered excitability, we performed calcium imaging in cultured E14.5 TrkA+ sensory neurons, similar to previous studies (Liu et al. 2009). Neurons were stimulated with mustard oil, menthol, and capsaicin, which are specific agonists of the temperature sensitive TRP channels, TRPA1, TRPM8, and TRPV1, respectively (Caterina et al. 1997, Peier et al. 2002, Bandell et al. 2004). Upon stimulation with any of the three agonists, Tnfr1−/− and Tnf −/− TrkA+ neurons were hypersensitive to each of the chemical analogues relative to WT neurons (Figure 5A). We also conducted a rescue experiment to determine whether TNFα is sufficient to quench the hyperexcitability of Tnf −/− nociceptors. We incubated E14.5 nociceptors from WT and Tnf −/− mice in 1 ng/mL NGF and observed hyperexcitability of Tnf −/− nociceptors relative to WT, analogous to the effects observed in 45 ng/mL NGF. However, when we selectively added 2 ng/mL TNFα to Tnf −/− nociceptors cultured in 1 ng/mL NGF, we were able to restore the TRP channel excitability to WT levels (Figure 5B). This suggests that TNFα-TNFR1 forward signaling negatively regulates the excitability of TRP channels in TrkA+ nociceptors and most prominently, TRPV1. In addition, the reported fluorescence fold changes are somewhat less robust than those reported in the literature due to a longer working distance in the z-plane during live imaging sessions (Figure S4A–C). This did not influence our ability to accurately assess relative excitability between genotypes (Figure S4D).
Figure 5. TNFα/TNFR1 signaling differentially regulates the excitability of peptidergic and non-peptidergic nociceptors.
(A) Fluo-4 in vitro calcium imaging of E14.5 nociceptors. The changes in fluorescence of E14.5 TrkA+ nociceptors were measured (cultured in 45 ng/mL NGF for 3 days in vitro) after acute addition of the indicated TRP channel agonist. 2 experiments per condition, at least 3 mice per experiment are reported.
(B) Rescue experiment demonstrating that TNFα is sufficient to quench the hyperexcitability of Tnf −/− nociceptors. E14.5 nociceptors from WT or Tnf −/− mice were cultured in 1 ng/mL NGF (with or without TNF for Tnf −/− neurons) for 2 days in vitro and prepared for calcium imaging as in (A). 2 experiments per condition are reported; at least 3 mice per experiment were used.
(C) Representative image of P14 calcium imaging strategy examining non-peptidergic nociceptor excitability. IB4+ and IB4− neurons are shown before and after agonist addition. Red arrow: IB4− neuron; white arrow: IB4+ neuron. Scale bar represents 20 μm.
(D) Quantification of fluorescence changes of P14 nociceptors (cultured in 45 ng/mL NGF for 1 day in vitro)in response to 1000 μM ATP.
(E) Excitability of P14 IB4+ and IB4− nociceptors responding to capsaicin. 2–3 experiments per condition, 3 mice per experiment.
See also Figure S4.
Statistics determined by 1-way (A–B, D–E) ANOVA, Bonferroni post-test. Data represent mean±SEM, *p<0.05, **p<0.01, ***p<0.001.
We also tested whether the excitability of non-peptidergic nociceptors is altered in the absence of TNFα or TNFR1 since NRTN is known to regulate sensitivity of P2X ion channels (Wang et al. 2013). We employed a strategy that permits recording fluorescence changes exclusively in non-peptidergic nociceptors by incubating the cultures with Alexa 568-conjugated IB4 before application of a chemical agonist (Figure 5C) (Gerevich et al. 2004). In response to ATP stimulation, Tnfr1−/− and Tnf −/− P14 IB4+ nociceptors were less excitable than WT IB4+ neurons (Figure 5D). Because non-peptidergic nociceptors also express TRPV1 (Chen et al. 2006b, Luo et al. 2007), we tested whether a similar effect was observed after capsaicin stimulation. Indeed, IB4+ neurons from Tnfr1−/− or Tnf −/− nociceptors were hypoexcitable to capsaicin stimulation compared to WT while IB4− nociceptors were hyperexcitable (Figure 5E). As observed for axon growth, these data suggest opposing regulatory roles for TNFα and TNFR1 in attenuating excitability of TrkA+ peptidergic nociceptors or enhancing excitability of Ret+ non-peptidergic nociceptors.
Tnf −/− and Tnfr1−/− mice are hypersensitive to thermal and mechanical stimuli
NGF-TrkA signaling controls nociceptor target innervation, cell survival, fate specification, excitability, and as a result, pain sensitivity thresholds (Pezet and McMahon 2006). Overactive NGF signaling therefore causes hyperalgesia largely through pathological hyperactivation of pathways regulating target innervation and excitability (Smelter and Hochberg 2013, Hefti et al. 2006, Pezet and McMahon 2006). We found that excessive NGF signaling and loss of TNFR1 or TNFα signaling phenocopy one another with respect to hyperinnervation of the skin and spinal cord (Figures 2–3), hypersensitivity to TRP channel agonists (Figure 5), as well as a greater overall number of nociceptors (Figure 4A, E). Thus, we hypothesized that developmental loss of TNFα-TNFR1 signaling would translate to increased sensitivity to pain analogous to injection with NGF (Lewin et al. 1993), which can be assessed by behavioral assays (for review, see Sandkühler 2009). To test this hypothesis, we first sought to precisely corroborate our calcium imaging data by subjecting the mice to a variety of different temperatures at prescribed intervals. Toward this end, we performed the tail flick assay where tails of WT or mutant mice were submerged in water ranging from 5°C to 50°C for no more than 20 seconds and the latency to tail withdrawal was measured. Tnfr1−/− and Tnf −/− mice were significantly more sensitive to every temperature examined including neutral temperatures between 25°C and 37°C (Figure 6A). Hyperexcitability to all temperatures is consistent with the sensitization of TRPV1, TRPM8, and TRPA1 observed in our calcium imaging experiments (Figure 5A) since Tnfr1−/− and Tnf −/− mice were more responsive to hot, cool, and freezing temperatures, respectively, in the behavioral assays. Tnfr1−/− and Tnf −/− mice also responded more quickly than WT in the Hargreaves radiant heat test, which applies heat with no tactile cue (Figure 6B), further corroborating hypersensitivity to heat in the absence of TNFα or TNFR1. These data are consistent with the notion that TNFα-TNFR1 forward signaling contributes to proper perception of temperature by dampening nociceptor sensitivity thresholds.
Figure 6. Tnfr1−/− and Tnf −/− mice are hypersensitive to thermal and mechanical stimuli.
(A–E) Behavioral sensitivity of WT, Tnfr1−/−, and Tnf −/− mice in response to the tail flick (A), Hargreaves radiant heat (B), acetone drop (C), hot plate (D), and von Frey (E) tests measuring reflexive (A–B) or centrally-mediated (C–D) thermal pain thresholds or mechanical acuity (E).
n=6 mice per data point (aged P26-P56), if n>6 mice, it is indicated. 2-way (A,D,E) or 1-way (B,C) ANOVA, Bonferroni post-test. All mice shown are on a B6;129s mixed background. WT mice are background matched, non-littermate controls for Tnfr1−/− and Tnf −/− mice.
See also Figure S5.
Data represent mean±SEM, ns: not significant, *p<0.05, **p<0.01, ***p<0.001.
In addition to measuring reflexive pain, we utilized two tests that interrogate centrally-mediated hot- and cold-sensing abilities: the hot plate test and the acetone drop test. These tests quantify grooming responses indicative of which temperatures mice perceive as noxious. For the hot-plate test we examined several temperatures ranging from mild to noxious heat. Both ligand and receptor null mice also displayed similar increases in grooming behaviors in the acetone drop test, which simulates cold sensation (Figure 6C). Mice lacking Tnfr1 and Tnf had lower pain thresholds for heat and consequently, an altered perception of thermal stimuli when compared to WT mice (Figure 6D). In addition to enhanced reflexive acuity to pain, Tnfr1−/− and Tnf −/− mice display skewed thresholds of which temperatures are noxious and which are not, as they confuse the distinction between mild and injurious temperatures in both tests.
To assay another form of tactile sensitivity, we tested mechanical acuity in Tnfr1−/− and Tnf −/− mice. Reflexive mechanical sensitivity can be probed with von Frey filaments of varying diameter to determine the threshold sensitivity of a mechanical force. In response to mild mechanical forces both Tnfr1−/− and Tnf −/− mice were more acutely sensitive to von Frey filament stimulation than WT (Figure 6F), suggesting that TNFα-TNFR1 signaling can also shape and suppress perception of mechanical force in addition to reflexively- and centrally-mediated thermal pain. These data were independently confirmed using littermate controls aged 6–7 weeks of age (Figure S5A–E). Thus, we propose that the TNFα-TNFR1 forward signaling pathway functions to tune primary nociceptors to properly interpret polymodal tactile stimuli.
Deletion of Tnfr1 rescues pain phenotypes associated with Ngf heterozygosity
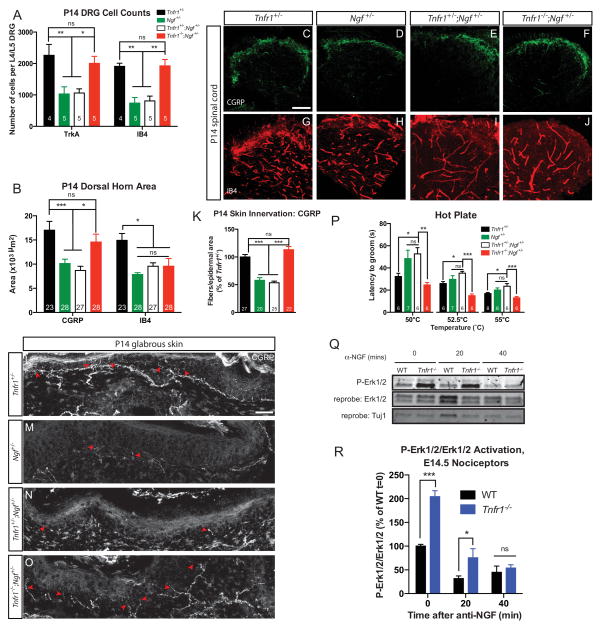
Our in vitro and in vivo data demonstrate a consistent role for TNFα-TNFR1 antagonism of NGF-TrkA function. Based on these data we speculated that by modulating one pathway we might be able to correct phenotypes observed in the other. While Ngf −/− mice die perinatally (Crowley et al. 1994), it is established that nociceptors in mice heterozygous for Ngf have roughly 50% less cell survival and target innervation (Crowley et al. 1994, Brennan et al. 1999). If the role of TNFα-TNFR1 signaling is to antagonize NGF-TrkA signaling in nociceptors, then we would predict that ablation of Tnfr1 would rescue pain insensitivity phenotypes that might be associated with loss of one Ngf allele. First, we performed an analysis of the number of TrkA+ and IB4+ nociceptors in the P14 L4 or L5 DRG of Tnfr1+/−, Ngf +/−, Tnfr1+/−;Ngf +/−, and Tnfr1−/−;Ngf +/− mice. Consistent with the loss of 50% of target-derived NGF, Ngf +/− and Tnfr1+/−;Ngf +/− mice exhibited roughly 50% fewer TrkA+ and IB4+ neurons at P14 compared to Tnfr1+/− mice (Figure 7A). Tnfr1−/−;Ngf +/− mice, by contrast, exhibited TrkA+ and IB4+ neuron numbers similar to Tnfr1+/−, which suggests that loss of Tnfr1 increases the NGF-sensitivity of TrkA+ nociceptors, thereby rescuing the impaired cell survival caused by Ngf heterozygosity.
Figure 7. Loss of Tnfr1 can compensate for heterozygous Ngf deletion.
(A) Quantification of the number of TrkA+ and IB4+ neurons per L4/L5 DRG at P14. One DRG used per animal, number of DRGs is indicated.
(B–J) Analysis of the P14 L4/L5 spinal cord dorsal horn innervation density of CGRP (C–F) and IB4 (G–J) fibers and quantification (B). 5 animals analyzed per genotype, number of sections analyzed is indicated. Scale bar represents 50 μm.
(K–O) CGRP+ fiber density in P14 footpad (thick) glabrous skin. (L–O) Representative images of CGRP+ peptidergic nociceptor fibers invading the hindpaw footpad of P14 mice. Arrowheads point to invading fibers. Scale bar represents 25 μm. (K) Quantification of peptidergic nociceptor skin innervation. 5 mice analyzed per genotype.
(P) Hot plate behavioral analysis at 50, 52.5, and 55°C. Behavioral analysis performed on mice from a mixed, B6;129 background. Mice analyzed are littermate controls except Ngf+/−, which are background matched.
(Q–R) Western blot (Q) and quantification (R) showing the decay in P-Erk1/2 signal after E14.5 nociceptors from WT and Tnfr1−/− mice were deprived of NGF via incubation with anti-NGF for the indicated times. Nociceptors were cultured in 45 ng/mL NGF for 1 DIV. Data are from 3 independent experiments using cultured nociceptors from 4–6 mice per experiment per genotype. 2-way ANOVA, Holm-Sidak post-test.
See also Figure S6.
Statistics determined by 1-way (K,P) or 2-way (A,B) ANOVA, Tukey post-test. Data represent mean ± SEM, ns: not significant, *p<0.05, **p<0.01, ***p<0.001.
Next, we analyzed the central projections into the spinal cord dorsal horn in Tnfr1+/−, Ngf +/−, Tnfr1+/−;Ngf +/−, and Tnfr1−/−;Ngf +/− mice. As expected (Crowley et al. 1994), there was roughly a 50% decrease in the area occupied by CGRP+ fibers in the dorsal horn at P14 in Ngf +/− and Tnfr1+/−;Ngf +/− mice relative to Tnfr1+/− mice (Figure 7B–F). Homozygous Tnfr1 deletion can rescue this fiber deficiency in Tnfr1−/−;Ngf +/− mice, providing additional evidence for functional antagonism between TNFR1 and TrkA in vivo. Although the number of non-peptidergic nociceptors in the DRG is rescued by homozygous elimination of Tnfr1, the area occupied by non-peptidergic axons in the dorsal horn was identical between Ngf +/−, Tnfr1+/−;Ngf +/−, and Tnfr1−/−;Ngf +/− mice (Figure 7B, G–J). This is consistent with the idea that TNFR1 positively regulates the maintenance of IB4+ fibers and is consistent with the notion that TNFR1-TNFα reverse signaling might be required for the full elaboration of non-peptidergic axons through cooperation with NRTN signaling.
We next examined innervation of glabrous skin from the hindpaw footpad of P14 Tnfr1+/−, Ngf +/−, Tnfr1+/−;Ngf +/−, and Tnfr1−/−;Ngf +/− mice by quantifying the number of CGRP+ fibers projecting into the epidermis (Figure 7K–O). This analysis revealed a 50% reduction in epidermal innervation by CGRP+ fibers in Ngf +/− and Tnfr1+/−;Ngf+/− mice compared to Tnfr1+/− mice. As expected, the skin of Tnfr1−/−;Ngf +/− mice exhibited peptidergic fiber innervation comparable to that of Tnfr1+/− mice, suggesting that TNFR1 opposes NGF-mediated growth of peptidergic fibers in the periphery.
We performed behavioral analyses on Tnfr1+/−, Ngf +/−, Tnfr1+/−;Ngf +/−, and Tnfr1−/−;Ngf +/− mice to determine if modulating nociceptor circuitry translated to changes in pain perception. Ngf +/− and Tnfr1+/−;Ngf +/− mice were less sensitive than Tnfr1+/− mice when performing the hot plate task at 50, 52.5, and 55°C (Figure 7P). As expected, the latency to onset of rapid, sustained grooming was rescued in Tnfr1−/−;Ngf +/− mice when compared to the less sensitive Ngf +/− or Tnfr1+/−;Ngf +/− mice. These loss-of-function data indicate that ablation of TNFR1 can sensitize TrkA signaling in nociceptors and thereby compensate for reduced NGF availability.
In addition to our genetic approach, we sought to use a biochemical approach to definitively test whether TNFR1 indeed functions as a bona fide antagonist of NGF-TrkA signaling in developing nociceptors. To test this, we cultured E14.5 sensory neurons in NGF for 24 hours and assayed for the activation of signaling molecules downstream of NGF-TrkA. We found that Tnfr1−/− sensory neurons exhibit increased activation of the NGF-TrkA downstream effectors Erk1/2 after 24 hours in vitro relative to WT sensory neurons (Figure 7Q). Moreover, we conducted an NGF deprivation time course to measure whether Tnfr1−/− sensory neurons exhibit sustained activation of the NGF-TrkA pathway after the removal of NGF. Indeed, Tnfr1−/− neurons maintained hyperactivation of P-Erk1/2, at intervals after NGF withdrawal (Figure 7R) despite decaying at the same rate (Figure S6). These data more directly demonstrate that TNFR1 attenuates the strength of NGF-TrkA trophic signals in developing nociceptors.
Discussion
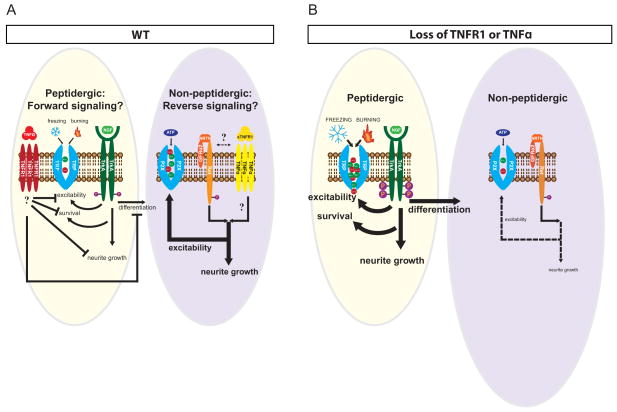
This study describes dual roles for TNFα and TNFR1 signaling in the coordination of peptidergic and non-peptidergic nociceptive circuits during PNS development through forward and reverse signaling paradigms. We demonstrate that, as early as E14.5 and as late as P30, forward TNFα-TNFR1 signals are required to negatively regulate the neurite growth, differentiation, and excitability of peptidergic nociceptors. In contrast, TNFR1-TNFα reverse signaling is required for full target innervation by and excitability of non-peptidergic nociceptors between P14 and P30. We propose that these differential functions are principally dependent on the complement of RTK signaling and trophic factor responsiveness resident in different nociceptor populations, specifically NGF-TrkA and NRTN-GFRα2-Ret in peptidergic and non-peptidergic nociceptors, respectively (Figure 8A–B).
Figure 8. Model of TNFα-TNFR1 signaling properties in NGF-responsive and NRTN-responsive peptidergic and non-peptidergic nociceptors.
(A) Model of TNFR1 and TrkA signaling events in WT animals. TNFα-TNFR1 forward signals suppress NGF-TrkA dependent excitability, neurite growth, survival, and differentiation. These signals lead to proper specialization of some TrkA+ peptidergic nociceptors as Ret+ non-peptidergic neurons, which are approximately equal in number to peptidergic cells in young adulthood (P30). Non-peptidergic nociceptors are dependent on TNFR1-TNFα reverse signaling for maximal neurite outgrowth and excitability.
(B) Loss of TNFα or TNFR1 signaling causes a gain-of-function of NGF-TrkA signals, hyperactivating TrkA excitability, neurite growth, cell survival, and differentiation pathways (thick arrows). Consequently, more TrkA+ neurons are driven to toward a non-peptidergic fate earlier during development, which results in roughly 50% more IB4+ neurons by young adulthood (P30). In contrast, non-peptidergic nociceptors are deficient in excitability and NRTN-dependent neurite growth in the absence of TNFα or TNFR1 (dashed arrows) possibly through loss of a reverse signaling mechanism.
TNFα-TNFR1 forward signaling attenuates NGF-TrkA dependent constructive processes in peptidergic nociceptors
Our in vivo and in vitro results provide evidence that TNFα and TNFR1 are required for proper development of nociceptive circuits through antagonism of NGF-TrkA signaling. Several lines of evidence support this assertion including: (1) In vivo, Tnfr1−/− and Tnf −/− mice exhibit an increased density of nociceptive fibers both peripherally and centrally (Figures 2A–D, Figure 3). (2) In vitro, Tnfr1−/− and Tnf −/− neurons are more sensitive to NGF-mediated neurite growth (Figures 2E–I). (3) In vitro calcium imaging suggests that TNFα-TNFR1 signaling normally suppresses the sensitivity of several TRP channels in TrkA+ peptidergic nociceptors since TrkA+ nociceptors are hyperexcitable in the absence of TNFα-TNFR1 signaling (Figure 5A) but can be rescued via reactivation of this pathway (Figure 5B). (4) We show that homozygous deletion of Tnfr1 is sufficient to compensate for neurite growth and behavioral pain deficits caused by the loss of a single Ngf allele (Figure 7). (5) Finally, we provide biochemical evidence of TNFR-RTK antagonism in neurons, by showing that TNFR1 suppresses NGF-TrkA activation (Figure 7Q–R).
Our findings that TNFα-TNFR1 signaling and NGF-TrkA signaling mutually oppose one another in nociceptors are analogous to the broad antagonistic function of p75NTR in sympathetic neurons (Kaplan & Miller 2000, Deppmann et al. 2008). For instance, p75NTR prevents axon overgrowth (Yeo et al. 1997, Singh et al. 2008), modulates electrical properties (Luther and Birren 2009), and expedites developmental cell death (Deppmann et al. 2008) in sympathetic neurons. While p75NTR does not appear to play this role in sensory neurons (Lee et al. 1992), we suggest that TNFα and TNFR1 may have assumed the antagonistic role toward NGF-TrkA processes in developing nociceptors. Indeed, loss of Tnfr1 leads to hyperactivation of trophic pathways such as P-Erk1/2, which is consistent with an increase in non-peptidergic nociceptor number and peptidergic nociceptor target innervation, as suggested previously (Zhong et al. 2007, Newbern et al. 2011). This apparently direct TNFα-TNFR1 suppression of downstream neurotrophic signals is reminiscent of a similar relationship with the insulin receptor (Steinberg et al. 2006). Thus, it is interesting to speculate that other TNFR family members might also serve as pro-refinement mechanisms in sensory neurons. DR6 is one likely candidate given its robust enrichment in the DRG (Figure S1A) and its pronounced role in sensory neuron degeneration (Nikolaev et al. 2009). One avenue of future investigation will be to ask whether TNFR family members can cooperate to facilitate pro-refinement functions in the somatosensory system.
TNFα and TNFR1 enhance non-peptidergic nociceptor growth and excitability
Despite increases in the number of IB4+ neurons in mice lacking TNFα or TNFR1, it appears that rather than Ret antagonism, TNFR1 signaling cooperates with Ret in non-peptidergic nociceptors. Several lines of evidence support this synergistic relationship including: (1) In vivo, a reduction in IB4+ fibers occupying lamina IIi at P14 and P30, despite the presence of more non-peptidergic neurons in the DRG at P14 and P30 (Figure 4F–M). (2) In vitro, DRG explants from Tnfr1−/− or Tnf −/− micedisplay impaired axon outgrowth in response to NRTN (Figure 4N–P, S). (3) Addition of sTNFR1, but not sTNF, rescues the impaired neurite outgrowth of explants cultured in NRTN (Figure 4Q–S), (4) In vitro calcium imaging assays reveal that Tnf −/− and Tnfr1−/− non-peptidergic nociceptors are hypoexcitable to acute ATP and capsaicin stimulation (Figure 5D–E). (5) The inability of a Tnfr1 deletion to rescue deficient non-peptidergic central projections to lamina IIi caused by Ngf heterozygosity (Figure 7B, G–J). These results point to a dichotomous function for TNFα and TNFR1 signals in two populations of functionally disparate neurons and suggest that TNFα and TNFR1 may enhance growth and excitability through reverse signaling.
What is the molecular basis for predominantly TNFα-TNFR1 forward signaling in peptidergic nociceptors but TNFR1-TNFα reverse signaling in non-peptidergic nociceptors? One possibility is that contextual molecular differences bias production of sTNFα or sTNFR1. For instance, just as NGF-TrkA signals upregulate the expression of a disintegrin and metalloprotease 17 (ADAM17) in sympathetic neurons (Kommaddi et al. 2011), it is possible that a similar paradigm holds in TrkA+ sensory neurons. One could envision that strongly trophic TrkA+ nociceptors upregulate ADAM17, which would cleave membrane-bound TNFα to produce sTNFα and induce TNFα-TNFR1 forward signals in peptidergic nociceptors. Upon committing to a non-peptidergic fate however, ADAM17 or other metalloproteases might preferentially shed TNFR1 from the cell surface to generate sTNFR1. Although further studies on these possibilities are necessary, it is a compelling idea to consider the number of permutations of TNFR family members antagonizing or synergizing with different RTKs to promote vastly different functional outcomes such as what we observe in peptidergic (TrkA+) and non-peptidergic (Ret+) nociceptors. As such, our findings likely extend beyond neuronal populations specifically, and to RTK and TNFR family member function, generally.
Cell fate specification of TrkA+ sensory neurons is coordinated by TNFα-TNFR1 signaling
All newly born nociceptors are TrkA+ but mature to become several diverse subpopulations expressing various combinations of the RTKs TrkA, Ret, and Met during adulthood (Lallemend and Ernfors 2012, Liu and Ma 2011, Gascon et al. 2010, Luo et al. 2007). These fate specification events are reported to be dependent on hierarchal NGF-TrkA modulation of the RTK, Ret, and the transcription factor, Runx1 (Lallemend and Ernfors 2012, Gascon et al. 2010, Luo et al. 2007, Chen et al. 2006b). Thus, it stands to reason that perturbing TrkA signals in developing sensory neurons would perturb the fate specification of several different nociceptor lineages. How then is such remarkable diversity coordinated from the standpoint of an early TrkA-expressing nociceptor? We propose that TNFR1 plays a pivotal role in the specification of different nociceptor subtypes based on the following observations: (1) Elimination of TNFα-TNFR1 signaling leads to higher numbers of TrkA+ nociceptors in adulthood, consistent with previous reports that TNFα serves a death-promoting developmental role in sensory neurons (Figure 4A) (Barker et al. 2001). (2) Loss of TNFR1 or TNFα also results in a higher proportion of nociceptors moving toward a non-peptidergic fate commitment at P0 (Figure 4B–D). (3) Consistent with this idea, at P14 and P30 there are more IB4+ neurons in Tnfr1 and Tnf knockout animals than WT mice in the L4/L5 DRG (Figure 4E).
Previous work has demonstrated that loss of key transcription factors impairs differentiation and function of sensory neurons (Chen et al. 2006a, Chen et al. 2006b, Maricich et al. 2009, Wende et al. 2012). While the importance of TrkA and Met in regulating nociceptor differentiation has already been established (Luo et al. 2007, Gascon et al. 2010), to our knowledge TNFR1 is the only non-RTK reported to influence sensory neuron fate specification. We find that loss of TNFα-TNFR1 signaling is more akin to enhancement of TrkA-dependent differentiation as more non-peptidergic nociceptors are generated in the absence of TNFα or TNFR1. These observations are consistent with our biochemical data that reveal loss of TNFR1 leads to hyperactive Erk1/2 signaling (Figure 7Q–R) and previous reports implicating Erk/MAPK signals in fate specification (Zhong et al. 2007). Importantly, while we specifically focus on the development of non-peptidergic nociceptors derived from TrkA+ precursors, there are several other distinct populations of TrkA-derived cells that are likely affected by loss of TNFα or TNFR1 such as those expressing mrg family members regulating pain, itch, or even low threshold mechanosensation (Dong et al. 2001, Liu et al. 2009, Li et al. 2011, Vrontou et al. 2013, Abraira and Ginty 2013). In future studies it will be interesting to examine how TNFα and TNFR1 influence the development and specification of several important sensory neuron subtypes critical for varied modes of tactile perception.
TNFα-TNFR1 forward signaling gates NGF-TrkA signaling to suppress pain
Our data suggest that TNFR1 acts as a gatekeeper on nociceptive neurons where it exerts control over several processes underlying stimulus perception. Importantly, our data also imply that TNFα-TNFR1 signaling within nociceptors is, in turn, controlled by the levels of TNFα produced by tissues that nociceptors innervate such as the skin and spinal cord. TNFα-TNFR1 signals modify nociceptor information processing by attenuating NGF-TrkA signals and refining components of the primary nociceptive circuit such that nociceptors properly interpret painful touch. Our study and findings from other groups have demonstrated that elimination of NGF or TrkA signaling results in nociceptive impairment (Crowley et al. 1994, Silos-Santiago et al. 1995, Indo et al. 1996). Conversely, excessive NGF-TrkA activation causes nociceptor sensitization (Pezet and McMahon 2006, Lane et al. 2010). Thus, deletion of Tnf or Tnfr1 is analogous to increasing the sensitivity of nociceptors to NGF stimulation, effectively increasing the “gain in pain” signals that are transmitted to the CNS (Figure 6), as suggested previously (Woolf and Salter 2000). In support of this notion, we find that loss of TNFR1 can rescue the defective nociceptive perception observed in mice heterozygous for Ngf. This rescue can be observed for cell number, central projection, and peripheral innervation density as well as behavior (Figure 7). Importantly, we present biochemical evidence that suggests these phenotypes are due to hyperactive NGF-TrkA signals in nociceptors.
This study supports the emergent concept that development of the peripheral nervous system requires a balance of interacting pro-refinement and pro-constructive cues. When this equation becomes unbalanced we observe disorders such as congenital insensitivity to pain or TNFR-associated periodic syndrome (TRAPS), an autoinflammatory disease marked by recurrent pain and fever (Ozen & Bilginer 2013). While the idea that TNFR family members negatively regulate the Trk family of RTKs isn’t a new one, the argument that the infamously pro-inflammatory TNFα-TNFR1 pathway opposes pain perception is novel. In fact, we report that nearly every major hallmark of the NGF-TrkA pathway is augmented when Tnfr1 or Tnf is ablated, suggesting a broad, general, and robust antagonism of TrkA signaling in nociceptors. Surprisingly, while TNFα and TNFR1 are analgesic signals in TrkA+ nociceptors, they serve an algesic role in Ret+ nociceptors, which highlights the importance of molecular context in the formation of developing neural circuits.
Methods
Animals
All experiments were carried out in compliance with the Association for Assessment of Laboratory Animal Care policies and approved by the University of Virginia Animal Care and Use Committee. WT mice are on a B6;129 mixed background. Tnfr1a−/− and Tnf −/− mice were purchased from Jackson Labs and backcrossed to a B6;129 mixed background for ≥4 generations. Ngf+/− mice were a gift from David Ginty and were maintained on a B6;129s mixed background.
In vitro calcium imaging
Calcium imaging was performed with Fluo-4 AM (Life Technologies), as previously described (Suo et al. 2014), with some modifications. Briefly, mass cultures of nociceptors were established in NGF, loaded with Fluo-4, then imaged before acute treatment with 1μM capsaicin (MP Biomedicals), 100μM L-menthol (MP Biomedicals), or 100μM mustard oil (Acros Organics). The change in fluorescence after treatment with a compound was measured relative to baseline fluorescence and a relative fold change was computed. One chemical treatment was used per experiment. P14 cultures were also loaded with IB4-568 (Life Technologies).
Behavioral Assays
Mice were tested between 4–8 weeks (Figure 6), between 4–6 weeks (Figure 7P), or between 6–7 weeks (Figure S5) during the light cycle. Before testing, mice were habituated to handling. Behavioral experiments in Figure 6 were performed on mixed background matched (B6;129s), non-littermate controls. Figure 7 behavioral experiments Ngf+/− rescue experiments were performed with littermate controls except Ngf+/− mice, which were background matched on the B6;129 mixed background.
Behavioral tests were performed with at least 20 minutes between tests and no more than 3 tests per day. One trial per mouse is reported for each data point.
Tail flick
Mice were manually restrained while the posterior 1/3 of the mouse’s tail was submerged in a water bath (Fisher, Isotemp) maintained at the designated temperature ±0.3°C until mouse flicked or reacted to the temperature.
Hot plate
Mice were placed on plates maintained at the given temperature (Columbus Instruments, Hotplate analgesia meter) and restricted to move within the area of an inverted 1000 mL beaker. The latency to vigorously lick/groom was recorded.
Hargreaves
Mouse was manually restrained and its hind-paw was placed over an infrared light source (Ugo Basile) until reflexive removal of paw.
von Frey
Mice were placed on a thin mesh screen and restrained within slightly opaque red containers during the test duration. Mechanical filaments of varying diameter (Bioseb, Touch Test) were administered to hind-paws beneath the apparatus and the percent response of paw lifting per 5 trials per filament was recorded.
Acetone drop
50μL of acetone was applied to the hind-paw and permitted to evaporate for 10 seconds. The duration of grooming per 120 seconds after the evaporation was recorded. Grooming was judged as forepaw/hindpaw/stomach licking, facial grooming, and hindpaw dragging.
Quantitation of Images
Quantification of skin innervation was performed as described previously (Zylka et al. 2005). DRG analyses were performed largely as described (Luo et al. 2007). Spinal cord quantification analyses were performed as previously described (Guo et al. 2011).
RT-PCR, immunostaining, cell culture, explants, dissociated neuron neurite growth, and western blotting
Detailed protocols found in the extended experimental procedures.
Supplementary Material
Highlights.
TNFR1 forms a rheostat with RTKs to tune tactile perception in nociceptors
TNFR1 forward signaling antagonizes TrkA to regulate growth, excitability, and fate
Ret+ nociceptors require TNFα reverse signaling for axon growth and excitability
Imbalanced TrkA/TNFR1 signaling results in severe behavioral pain phenotypes
Acknowledgments
We thank Pamela Neff and J. Stuart Cauley for technical assistance. We thank members of the Deppmann lab for helpful discussions and comments on the manuscript. We thank members of the Keck Center for helpful advice with microscopy. We thank Barry Condron, David Ginty, Ali Güler, David Hill, Wenqin Luo, Brian Pierchala, Iggy Provencio, and Nikki Watson for helpful comments on the manuscript. MW was supported by 5T32GM008328 and 5T32GM008136. DH and SK received support from the Center for Undergraduate Excellence. This work was supported by the Sloan Foundation, UVa Fund for Excellence in Science and Technology, and NIH-NINDS (R01NS072388) awarded to CD.
Footnotes
Respective Contributions
MW conducted and analyzed all experiments except the TNFRSF RT-PCR analysis (DH) and quantification of neurite growth in microfluidic chambers (SK). DH and SK also conducted some immunostaining experiments. SE contributed to immunostaining and behavioral assays. AS provided support for biochemistry experiments. CC provided advice and equipment for behavioral testing. MW and CD conceived the experiments and wrote the manuscript with input from co-authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abraira VE, Ginty DD. The sensory neurons of touch. Neuron. 2013;79:618–39. doi: 10.1016/j.neuron.2013.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Airaksinen MS, Saarma M. The GDNF family: Signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–94. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- 3.Aloe L, Mugnaini E, Levi-Montalcini R. Light and electron microscopic studies on the excessive growth of sympathetic ganglia in rats injected daily from birth with 6-OHDA and NGF. Arch Ital Biol. 1975;113:326–53. [PubMed] [Google Scholar]
- 4.Bandell M, Story GM, et al. Noxious cold ion channel TRPA1 is activated by pungent compounds and bradykinin. Neuron. 2004;41:849–57. doi: 10.1016/s0896-6273(04)00150-3. [DOI] [PubMed] [Google Scholar]
- 5.Barker V, Middleton G, et al. TNFα contributes to the death of NGF-dependent neurons during development. Nat Neurosci. 2001;4:1194–8. doi: 10.1038/nn755. [DOI] [PubMed] [Google Scholar]
- 6.Basbaum AI, Bautista DM, et al. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–84. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergmann I, Priestley JV, et al. Analysis of cutaneous sensory neurons in transgenic mice lacking the low affinity neurotrophin receptor p75. Eur J Neurosci. 1997;9:18–28. doi: 10.1111/j.1460-9568.1997.tb01349.x. [DOI] [PubMed] [Google Scholar]
- 8.Brennan C, Rivas-Plata K, et al. The p75 neurotrophin receptor influences NT-3 responsiveness of sympathetic neurons in vivo. Nat Neurosci. 1999;2:699–705. doi: 10.1038/11158. [DOI] [PubMed] [Google Scholar]
- 9.Caterina MJ, Schumacher MA, et al. The capsaicin receptor: A heat-activated ion channel in the pain pathway. Nature. 1997;389:816–24. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 10.Chen AI, De Nooij JC, et al. Graded activity of transcription factor Runx3 specifies the laminar termination pattern of sensory axons in the developing spinal cord. Neuron. 2006;49:395–408. doi: 10.1016/j.neuron.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 11.Chen CL, Broom DC, et al. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron. 2006;49:365–77. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 12.Chuang HH, Prescott ED, et al. Bradykinin and nerve growth factor release the capsaicin receptor from PtdIns(4,5)P2-mediated inhibition. Nature. 2001;411:957–62. doi: 10.1038/35082088. [DOI] [PubMed] [Google Scholar]
- 13.Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–24. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 14.Costigan M, Scholz J, et al. Neuropathic pain: A maladaptive response of the nervous system to damage. Annu Rev Neurosci. 2009;32:1–32. doi: 10.1146/annurev.neuro.051508.135531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig ADB. Pain mechanisms: Labeled lines versus convergence in central processing. Annu Rev Neurosci. 2003:1–30. doi: 10.1146/annurev.neuro.26.041002.131022. [DOI] [PubMed] [Google Scholar]
- 16.Crowley C, Spencer SD, et al. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell. 1994;76:1001–1011. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- 17.Davies AM, Lee KF, et al. p75-deficient trigeminal sensory neurons have an altered response to NGF but not to other neurotrophins. Neuron. 1993;11:565–74. doi: 10.1016/0896-6273(93)90069-4. [DOI] [PubMed] [Google Scholar]
- 18.Deppmann CD, Janes K. Cytokine-cytokine crosstalk and cell-death decisions. In: Lavrik IN, editor. Systems Biology of Apoptosis. New York, NY: Springer Publishers; 2013. pp. 163–80. [Google Scholar]
- 19.Deppmann CD, Mihalas S, et al. A model for neuronal competition during development. Science. 2008;320:369–73. doi: 10.1126/science.1152677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong X, Han S, et al. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106:619–32. doi: 10.1016/s0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- 21.Gascon E, Gaillard S, et al. Hepatocyte growth factor-Met signaling is required for Runx1 extinction and peptidergic differentiation in primary nociceptive neurons. J Neurosci. 2010;30:12414–23. doi: 10.1523/JNEUROSCI.3135-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerevich Z, Borvendeg SJ, et al. Inhibition of N-type voltage-activated calcium channels in rat dorsal root ganglion neurons by P2Y receptors is a possible mechanism of ADP-induced analgesia. J Neurosci. 2004;24:797–807. doi: 10.1523/JNEUROSCI.4019-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glebova NO, Ginty DD. Heterogeneous requirement of NGF for sympathetic target innervation in vivo. J Neurosci. 2004;24:743–51. doi: 10.1523/JNEUROSCI.4523-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo T, Mandai K, et al. An evolving NGF-Hoxd1 signaling pathway mediates development of divergent neural circuits in vertebrates. Nat Neurosci. 2011;14 :31–6. doi: 10.1038/nn.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hefti FF, Rosenthal A, et al. Novel class of pain drugs based on antagonism of NGF. Trends Pharmacol Sci. 2006;27:85–91. doi: 10.1016/j.tips.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Hunt S, Mantyh P. The molecular dynamics of pain control. Nat Rev Neurosci. 2001;2:83–91. doi: 10.1038/35053509. [DOI] [PubMed] [Google Scholar]
- 27.Indo Y, Tsuruta M, et al. Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat Genet. 1996;13:485–8. doi: 10.1038/ng0896-485. (1996) [DOI] [PubMed] [Google Scholar]
- 28.Ji RR, Samad TA, et al. p38 MAPK activation by NGF in primary sensory neurons after inflammation increases TRPV1 levels and maintains heat hyperalgesia. Neuron. 2002;36:57–68. doi: 10.1016/s0896-6273(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 29.Julius D, Basbaum AI. Molecular mechanisms of nociception. Nature. 2001:413–203. 10. doi: 10.1038/35093019. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan DR, Miller FD. Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol. 2000;10:381–91. doi: 10.1016/s0959-4388(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 31.Kommaddi RP, Thomas R, et al. Trk-dependent ADAM17 activation facilitates neurotrophin survival signaling. FASEB J. 2011;25:2061–70. doi: 10.1096/fj.10-173740. [DOI] [PubMed] [Google Scholar]
- 32.Kisiswa L, Osório C, et al. TNFα reverse signaling promotes sympathetic axon growth and target innervation. Nat Neurosci. 2013;16:865–73. doi: 10.1038/nn.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lallemend F, Ernfors P. Molecular interactions underlying the specification of sensory neurons. Trends Neurosci. 2012;35:373–81. doi: 10.1016/j.tins.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Lane NE, Schnitzer TJ, et al. Tanezumab for the treatment of pain from osteoarthritis of the knee. N Engl J Med. 2010;363:1521–31. doi: 10.1056/NEJMoa0901510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee KF, Li E, et al. Targeted mutation of the gene encoding the low affinity NGF receptor p75 leads to deficits in the peripheral sensory nervous system. Cell. 1992;69:737–49. doi: 10.1016/0092-8674(92)90286-l. [DOI] [PubMed] [Google Scholar]
- 36.Lewin GR, Ritter AM, et al. Nerve Growth Factor-induced Adult Rat Hyperalgesia in the Neonatal and Adult Rat. J Neurosci. 1993;13:2136–48. doi: 10.1523/JNEUROSCI.13-05-02136.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Rutlin M, et al. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell. 2011;147:1615–1627. doi: 10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindfors PH, Võikar V, et al. Deficient nonpeptidergic epidermis innervation and reduced inflammatory pain in glial cell line-derived neurotrophic factor family receptor α2 knock-out mice. J Neurosci. 2006;26:1953–60. doi: 10.1523/JNEUROSCI.4065-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q, Tang Z, et al. Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell. 2009;139:1353–65. doi: 10.1016/j.cell.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Ma Q. Generation of somatic sensory neuron diversity and implications on sensory coding. Curr Opin Neurobiol. 2011;21:52–60. doi: 10.1016/j.conb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Locksley RM, Killeen N, et al. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 42.Luo W, Wickramasinghe SR, et al. A hierarchical NGF signaling cascade controls Ret-dependent and Ret-independent events during development of nonpeptidergic DRG neurons. Neuron. 2007;54:739–54. doi: 10.1016/j.neuron.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 43.Luther JA, Birren SJ. p75 and TrkA signaling regulates sympathetic neuronal firing patterns via differential modulation of voltage-gated currents. J Neurosci. 2009;29:5411–24. doi: 10.1523/JNEUROSCI.3503-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mantyh PW, Koltzenburg M, et al. Antagonism of nerve growth factor-TrkA signaling and the relief of pain. Anesthesiology. 2011;115:189–204. doi: 10.1097/ALN.0b013e31821b1ac5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maricich SM, Wellnitz SA, et al. Merkel cells are essential for light-touch responses. Science. 2009;324:1580–2. doi: 10.1126/science.1172890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marmigère F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007;8:114–27. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- 47.Molliver DC, Wright DE, et al. IB4-binding DRG neurons switch from NGF to GDNF dependence in early postnatal life. Neuron. 1997;19:849–61. doi: 10.1016/s0896-6273(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 48.Moran MM, McAlexander MA, et al. Transient receptor potential channels as therapeutic targets. Nat Rev Drug Discov. 2011;10:601–20. doi: 10.1038/nrd3456. [DOI] [PubMed] [Google Scholar]
- 49.Newbern JM, Li X, et al. Specific functions for ERK/MAPK signaling during PNS development. Neuron. 2011;69:91–105. doi: 10.1016/j.neuron.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nikolaev A, McLaughlin T, et al. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–9. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Ozen S, Bilginer Y. A clinical guide to autoinflammatory diseases: familial Mediterranean fever and next-of-kin. Nat Rev Rheumatol. 2013;10:1–13. doi: 10.1038/nrrheum.2013.174. [DOI] [PubMed] [Google Scholar]
- 52.Patel TD, Jackman A, et al. Development of sensory neurons in the absence of NGF/TrkA signaling in vivo. Neuron. 2000;25:345–57. doi: 10.1016/s0896-6273(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 53.Peier AM, Moqrich A, et al. A TRP channel that senses cold stimuli and menthol. Cell. 2002;108:705–15. doi: 10.1016/s0092-8674(02)00652-9. [DOI] [PubMed] [Google Scholar]
- 54.Pezet S, McMahon SB. Neurotrophins: Mediators and modulators of pain. Annu Rev Neurosci. 2006;29:507–38. doi: 10.1146/annurev.neuro.29.051605.112929. [DOI] [PubMed] [Google Scholar]
- 55.Sandkühler J. Models and mechanisms of hyperalgesia and allodynia. Physiol Rev. 2009;89:707–58. doi: 10.1152/physrev.00025.2008. [DOI] [PubMed] [Google Scholar]
- 56.Sharma N, Deppmann CD, et al. Long-distance control of synapse assembly by target-derived NGF. Neuron. 2010;67:422–34. doi: 10.1016/j.neuron.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silos-Santiago I, Molliver DC, et al. Non-TrkA-expressing small DRG neurons are lost in TrkA deficient mice. J Neurosci. 1995;15:5929–42. doi: 10.1523/JNEUROSCI.15-09-05929.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh KK, Park KJ, et al. Developmental axon pruning mediated by BDNF-p75NTR-dependent axon degeneration. Nat Neurosci. 2008;11:649–58. doi: 10.1038/nn.2114. [DOI] [PubMed] [Google Scholar]
- 59.Smelter E, Hochberg ME. New treatments for osteoarthritis. Curr Opin Rheumatol. 2013;3:310–6. doi: 10.1097/BOR.0b013e32835f69b4. [DOI] [PubMed] [Google Scholar]
- 60.Snider WD, McMahon SB. Tackling pain at the source: New ideas about nociceptors. Neuron. 1998;20:629–32. doi: 10.1016/s0896-6273(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 61.Steinberg GR, Michell BJ, et al. Tumor necrosis factor alpha-induced skeletal muscle insulin resistance involves suppression of AMP-kinase signaling. Cell Metab. 2006;4:465–74. doi: 10.1016/j.cmet.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Stucky CL, Lewin GR. Isolectin B4-positive and -negative nociceptors are functionally distinct. J Neurosci. 1999;19:6497–505. doi: 10.1523/JNEUROSCI.19-15-06497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun M, Fink PJ. A new class of reverse signaling costimulators belongs to the TNF family. J Immunol. 2007;179:4307–12. doi: 10.4049/jimmunol.179.7.4307. [DOI] [PubMed] [Google Scholar]
- 64.Suo D, Park J, et al. Coronin-1 is a neurotrophin endosomal effector that is required for developmental competition for survival. Nat Neurosci. 2014;17:36–45. doi: 10.1038/nn.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verhoeven K, Timmerman V, et al. Recent advances in hereditary sensory and autonomic neuropathies. Curr Opin Neurol. 2006;19:474–80. doi: 10.1097/01.wco.0000245370.82317.f6. [DOI] [PubMed] [Google Scholar]
- 66.Vrontou S, Wong AM, et al. Genetic identification of C fibres that detect massage-like stroking of hairy skin in vivo. Nature. 2013;493:669–73. doi: 10.1038/nature11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang T, Jing X, et al. Neurturin overexpression in skin enhances expression of TRPM8 in cutaneous sensory neurons and leads to behavioral sensitivity to cool and menthol. J Neurosci. 2013;33:2060–70. doi: 10.1523/JNEUROSCI.4012-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wende H, Lechner SG, et al. The transcription factor c-Maf controls touch receptor development and function. Science. 2012;335:1373–6. doi: 10.1126/science.1214314. [DOI] [PubMed] [Google Scholar]
- 69.Woolf CJ, Salter MW. Neuronal Plasticity: Increasing the Gain in Pain. Science. 2000;288:1765–1768. doi: 10.1126/science.288.5472.1765. [DOI] [PubMed] [Google Scholar]
- 70.Yeo TT, Chua-Couzens J, et al. Absence of p75NTR causes increased basal forebrain cholinergic neuron size, choline acetyltransferase activity, and target innervation. J Neurosci. 1997;17:7594–605. doi: 10.1523/JNEUROSCI.17-20-07594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang X, Huang J, et al. NGF rapidly increases membrane expression of TRPV1 heat-gated ion channels. EMBO J. 2005;24:4211–23. doi: 10.1038/sj.emboj.7600893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong J, Li X, et al. Raf kinase signaling functions in sensory neuron differentiation and axon growth in vivo. Nat Neurosci. 2007;10:598–607. doi: 10.1038/nn1898. [DOI] [PubMed] [Google Scholar]
- 73.Zylka MJ, Rice FL, et al. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron. 2005;45:17–25. doi: 10.1016/j.neuron.2004.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.