Abstract
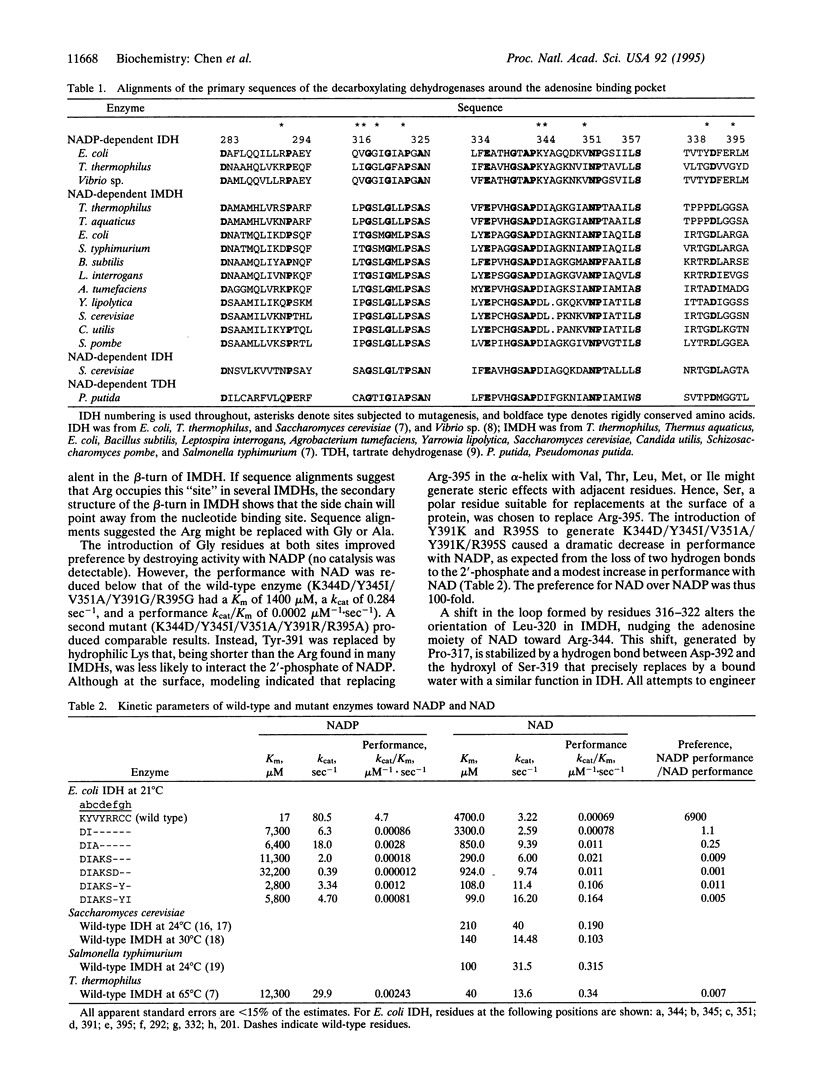
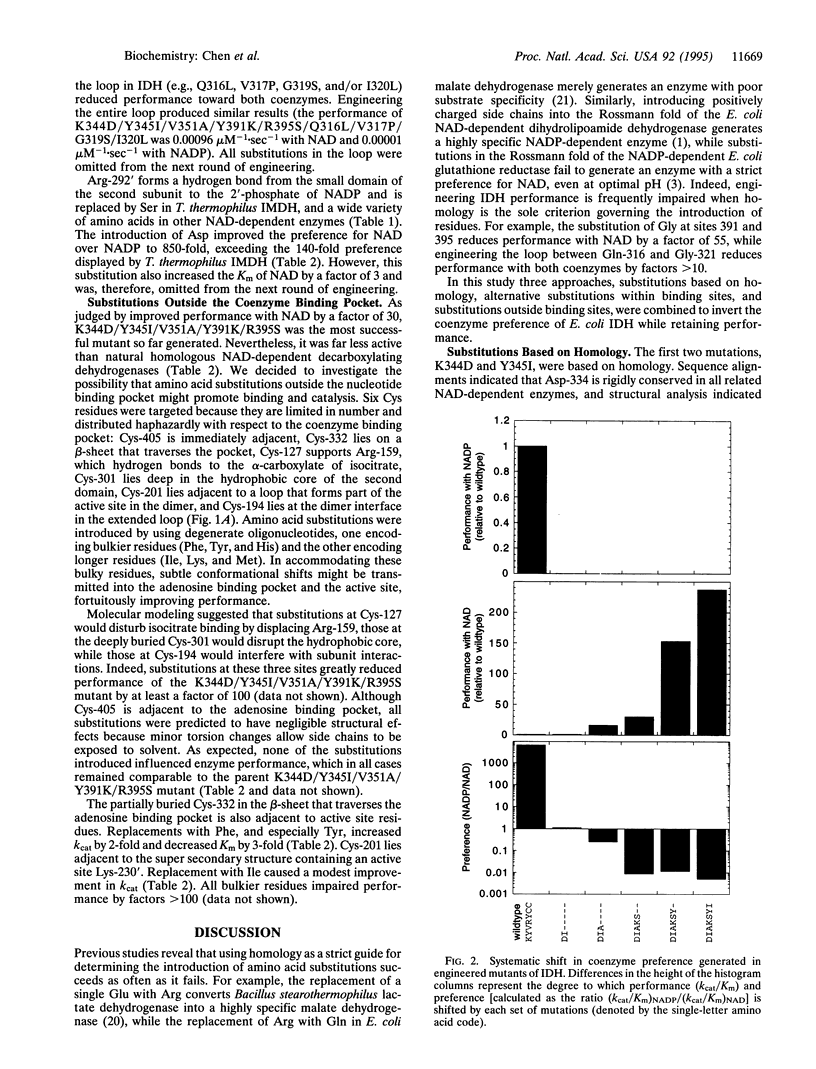
The isocitrate dehydrogenase of Escherichia coli, which lacks the Rossmann fold common to other dehydrogenases, displays a 7000-fold preference for NADP over NAD (calculated as the ratio of kcat/Km). Guided by x-ray crystal structures and molecular modeling, site-directed mutagenesis has been used to introduce six substitutions in the adenosine binding pocket that systematically shift coenzyme preference toward NAD. The engineered enzyme displays an 850-fold preference for NAD over NADP, which exceeds the 140-fold preference displayed by a homologous NAD-dependent enzyme. Of the six mutations introduced, only one is identical in all related NAD-dependent enzyme sequences--strict adherence to homology as a criterion for replacing these amino acids impairs function. Two additional mutations at remote sites improve performance further, resulting in a final mutant enzyme with kinetic characteristics and coenzyme preference comparable to naturally occurring homologous NAD-dependent enzymes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bocanegra J. A., Scrutton N. S., Perham R. N. Creation of an NADP-dependent pyruvate dehydrogenase multienzyme complex by protein engineering. Biochemistry. 1993 Mar 23;32(11):2737–2740. doi: 10.1021/bi00062a001. [DOI] [PubMed] [Google Scholar]
- Cupp J. R., McAlister-Henn L. Kinetic analysis of NAD(+)-isocitrate dehydrogenase with altered isocitrate binding sites: contribution of IDH1 and IDH2 subunits to regulation and catalysis. Biochemistry. 1993 Sep 14;32(36):9323–9328. doi: 10.1021/bi00087a010. [DOI] [PubMed] [Google Scholar]
- Dean A. M., Koshland D. E., Jr Electrostatic and steric contributions to regulation at the active site of isocitrate dehydrogenase. Science. 1990 Aug 31;249(4972):1044–1046. doi: 10.1126/science.2204110. [DOI] [PubMed] [Google Scholar]
- Dean A. M., Koshland D. E., Jr Kinetic mechanism of Escherichia coli isocitrate dehydrogenase. Biochemistry. 1993 Sep 14;32(36):9302–9309. doi: 10.1021/bi00087a007. [DOI] [PubMed] [Google Scholar]
- Garnak M., Reeves H. C. Purification and properties of phosphorylated isocitrate dehydrogenase of Escherichia coli. J Biol Chem. 1979 Aug 25;254(16):7915–7920. [PubMed] [Google Scholar]
- Haeffner-Gormley L., Chen Z., Zalkin H., Colman R. F. Importance of lysine-286 at the NADP site of glutamate dehydrogenase from Salmonella typhimurium. Biochemistry. 1992 Sep 1;31(34):7807–7814. doi: 10.1021/bi00149a010. [DOI] [PubMed] [Google Scholar]
- Hsu Y. P., Kohlhaw G. B. Leucine biosynthesis in Saccharomyces cerevisiae. Purification and characterization of beta-isopropylmalate dehydrogenase. J Biol Chem. 1980 Aug 10;255(15):7255–7260. [PubMed] [Google Scholar]
- Hurley J. H., Dean A. M., Koshland D. E., Jr, Stroud R. M. Catalytic mechanism of NADP(+)-dependent isocitrate dehydrogenase: implications from the structures of magnesium-isocitrate and NADP+ complexes. Biochemistry. 1991 Sep 3;30(35):8671–8678. doi: 10.1021/bi00099a026. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Dean A. M., Sohl J. L., Koshland D. E., Jr, Stroud R. M. Regulation of an enzyme by phosphorylation at the active site. Science. 1990 Aug 31;249(4972):1012–1016. doi: 10.1126/science.2204109. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Dean A. M. Structure of 3-isopropylmalate dehydrogenase in complex with NAD+: ligand-induced loop closing and mechanism for cofactor specificity. Structure. 1994 Nov 15;2(11):1007–1016. doi: 10.1016/s0969-2126(94)00104-9. [DOI] [PubMed] [Google Scholar]
- Ishii A., Suzuki M., Sahara T., Takada Y., Sasaki S., Fukunaga N. Genes encoding two isocitrate dehydrogenase isozymes of a psychrophilic bacterium, Vibrio sp. strain ABE-1. J Bacteriol. 1993 Nov;175(21):6873–6880. doi: 10.1128/jb.175.21.6873-6880.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn G. D., Barnes L. D., Atkinson D. E. Yeast diphosphopyridine nucleotide specific isocitrate dehydrogenase. Binding of ligands. Biochemistry. 1971 Oct 12;10(21):3945–3951. doi: 10.1021/bi00797a023. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Miyazaki K., Oshima T. Co-enzyme specificity of 3-isopropylmalate dehydrogenase from Thermus thermophilus HB8. Protein Eng. 1994 Mar;7(3):401–403. doi: 10.1093/protein/7.3.401. [DOI] [PubMed] [Google Scholar]
- Nicholls D. J., Miller J., Scawen M. D., Clarke A. R., Holbrook J. J., Atkinson T., Goward C. R. The importance of arginine 102 for the substrate specificity of Escherichia coli malate dehydrogenase. Biochem Biophys Res Commun. 1992 Dec 15;189(2):1057–1062. doi: 10.1016/0006-291x(92)92311-k. [DOI] [PubMed] [Google Scholar]
- Nishiyama M., Birktoft J. J., Beppu T. Alteration of coenzyme specificity of malate dehydrogenase from Thermus flavus by site-directed mutagenesis. J Biol Chem. 1993 Mar 5;268(7):4656–4660. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Stoddard B. L., Dean A., Koshland D. E., Jr Structure of isocitrate dehydrogenase with isocitrate, nicotinamide adenine dinucleotide phosphate, and calcium at 2.5-A resolution: a pseudo-Michaelis ternary complex. Biochemistry. 1993 Sep 14;32(36):9310–9316. doi: 10.1021/bi00087a008. [DOI] [PubMed] [Google Scholar]
- Thorsness P. E., Koshland D. E., Jr Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem. 1987 Aug 5;262(22):10422–10425. [PubMed] [Google Scholar]
- Tipton P. A., Beecher B. S. Tartrate dehydrogenase, a new member of the family of metal-dependent decarboxylating R-hydroxyacid dehydrogenases. Arch Biochem Biophys. 1994 Aug 15;313(1):15–21. doi: 10.1006/abbi.1994.1352. [DOI] [PubMed] [Google Scholar]
- Wilks H. M., Hart K. W., Feeney R., Dunn C. R., Muirhead H., Chia W. N., Barstow D. A., Atkinson T., Clarke A. R., Holbrook J. J. A specific, highly active malate dehydrogenase by redesign of a lactate dehydrogenase framework. Science. 1988 Dec 16;242(4885):1541–1544. doi: 10.1126/science.3201242. [DOI] [PubMed] [Google Scholar]



