Abstract
A number of microRNAs (miRNAs), including miR-200 family, are aberrantly expressed in endometriosis and endometrial cancer. Here we assessed the expression and functional aspects of miR-200c in endometrial tissues (N = 52) from normal endometrial biopsies (N = 15), endometrial tissues including those exposed to hormonal therapies (N = 20), and grade I-III endometrial cancer (N = 17). miR-200c expression was elevated in normal endometrial biopsies from mid- and late-luteal phase, and in endometrial tumors as compared to endometrial tissues from peri- and postmenopausal period (P < .05) and its pattern of temporal expression displayed an inverse relationship with the expression of ZEBs. The expression of E-cadherin (CDH1) varied, but expressed at low levels, specifically in endometrial tissues and endometrial tumors. The endometrial expression of ZEBs and CDH1 in patients who were exposed to Depo-Provera and gonadotropin-releasing hormone agonist GnRHa displayed a trend toward lower expression as compared to proliferative phase; however, treatment of Ishikawa cells with 17β-estradiol, progesterone, and medroxy progesterone acetate had modest effects on the expression of miR-200c and ZEBs without affecting CDH1 expression. Gain of function of miR-200c in Ishikawa cells repressed ZEBs, as well as VEGFA, FLT1, IKKβ, and KLF9 expression at transcriptional and translational levels through direct interaction with their respective 3′untranslated regions and increased the rate of their proliferation. These results indicated that endometrial miR-200c expression undergoes dynamic changes during transition from normal into cancerous states; possibly influenced by hormonal milieu and by targeting the expression of specific genes with key regulatory functions in cellular transformation, inflammation, and angiogenesis may influence these events during normal and disease progression.
Keywords: miR-200c, expression, regulation, endometrium, menstrual cycle, cancer
Introduction
MicroRNAs (miRNAs) are a member of family of noncoding small RNAs of ∼22 nucleotide (nt) in length and through binding to miR response elements (MREs) within 3′untranslated regions (3′UTRs) of their target genes, posttranscriptionally represses their expression.1–4 Through this mechanism, miRNAs regulate many normal cellular activities ranging from cell growth and differentiation, apoptosis, metabolic activities, angiogenesis, and tissue turnover. Conversely, aberrant expression of a number of miRNAs has been closely associated with various human diseases, more specifically events related to cellular transformation, tumorigenesis, inflammation, and tissue fibrosis.1–6 In the uterus, conditional inactivation of Dicer, which is required for miRNA processing, resulted in altered expression of a large number of miRNAs, as well as histological abnormalities associated with reduction in size of oviducts and uterine horns, where direct blastocyst transfer did not result in pregnancy.7–9 In human endometrium, several studies, including next generation sequencing have identified the expression of a large number of miRNAs, some of which showed altered expression in endometriosis and endometrial cancer.10–17 Among those altered in endometriosis and endometrial cancer, include the members of miR-200 family: miR-200c/141 and miR-200b/200a/429 clusters.11 , 13 , 15 , 18–22
The expression and function of miR-200 family has been well documented in various tissues and cells, where they target the expression of many genes, including ZEBs, the transcription factors that regulate cellular transformation, more specifically epithelial-to-mesenchymal transition (EMT) during cancer development and progression through repression of adhesion molecules such as E-cadherin.2,5,23 In addition to well-established regulatory function of miR-200 on the expression of ZEB1 and ZEB2, a few studies have also validated FLT1, ETS1, SUZ12, PDGF, TGFB2, and conexin43 as direct targets of different miR-200 family members, implicating their functions in angiogenesis and tissue turnover.24–31
Endometrium during normal menstrual cycle and benign and cancerous states undergoes extensive cellular and molecular changes, including events regulated by miR-200 target genes. However, evidence suggests that miRNAs expression and regulatory function of their target genes occurs in cell- and tissue-specific manners.32,33 As such and due to limited information regarding miRNAs regulatory functions in the endometrium, we investigated the expression, regulation, and function of miR-200c and few genes targeted by miR-200 in normal endometrial biopsies and in endometrial tissues obtained from patients undergoing hysterectomy including those who received hormonal therapy, pre- and postmenopausal states, and those with endometrial cancer at different stages of the disease. Using Ishikawa cells as an in vitro model we also assessed the hormonal regulation of miR-200c and either confirmed or validated specific genes targeted by gain of function of miR-200c.
Materials and Methods
Tissue Collection
Endometrial biopsies (N = 15) were obtained from women with previously documented fertility who were requesting permanent surgical sterilization (tubal ligation) under informed consent approved by the University Florida Institutional Review Board prior. These patients were not taking any hormone therapy for the last 3 months prior to collection of the biopsies and based on histological evaluation and the last menstrual period they were from mid- (N = 2) and late-proliferative (N = 1), and early- (ES; N = 5), mid- (MS; N = 3), and late (LS; N = 4)-secretory phases of the menstrual cycle. Similarly, endometrial tissues (N = 20) were also collected from women scheduled to undergo gynecological surgery due to pelvic pain, symptomatic leiomyomas, and uterine prolapse. These patients ranged in age from 23 to 67 years (median = 42.5) and based on endometrial histology and last menstrual period they were from mid-late proliferative (N = 3) and early-mid secretory (N = 5) phases of the menstrual cycle. The tissues also included inactive endometrium from perimenopausal (N = 3) and postmenopausal women (N = 2), and patients who were exposed to gonadotropin-releasing hormone agonist (GnRHa; N = 2) and Depo-Provera (N = 5). Endometrial tissues from women diagnosed with endometrial cancer (N = 17) were obtained from the University of Florida Tissue Bank and based on histological typing conducted according to surgical staging (International Federation of Gynecology and Obstetrics [FIGO]) were from grade I to III. The patients age ranged from 40 to 90 years (median = 65). All the endometrial tissues were collected at the University of Florida affiliated Shands Hospital with prior approval from the Institutional Review Board. After collection the endometrial tissue samples were snap frozen and kept in liquid nitrogen until further analysis.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated from a small portion of the endometrial tissues using Trizol reagent, and their quantity and quality was assessed using ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, Delaware). Two μg or 10 ng of total RNA (for miRNA) was reverse transcribed using random primers for gene expression or specific stem-loop primers for miRNA expression (Applied Biosystems Foster City, California). Real-time polymerase chain reaction (PCR) was carried out using TaqMan reagents. Reactions were incubated for 10 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 minutes at 60°C, and level of messenger RNAs (mRNAs) and miRNAs expression was determined using Applied Biosystems 7300 Detection System with 18S rRNA and RNU6B used for normalization, respectively. All reactions were run in triplicate and relative expression was analyzed with the comparative cycle threshold method using 2−ΔΔCT method according to the manufacturer’s guidelines.
Cell Culturing and Transfection
Ishikawa cells were cultured in Dulbecco modified Eagle medium (DMEM)/F12 supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 1% (v/v) of antibiotic and antimycotic solution. All the supplies for cell culture were purchased from Sigma-Aldrich (St Louis, Missouri), Invitrogen (Carlsbad, California), and Fisher Scientific (Pittsburgh, Pennsylvania). The cells were seeded at various densities in 6-well plates or 10 cm petri dishes and cultured in antibiotic-/antimycotic-free media until reaching 70% confluence and then transfected with 50 nmol/L of pre-miR negative control (NC) or pre-miR-200c (Ambion/Applied Biosystems) using lipofectamine 2000 (Invitrogen), according to the manufacturer’s protocol. After transfection, the cells were cultured for an additional 24 and 48 hours, respectively, and total RNA and protein were isolated and subjected to real-time PCR and Western blot analysis.
Ovarian Steroid Treatments
Ishikawa cells were cultured in 6-well plates as above for 24 hours, washed, and incubated in phenol red-free media with charcoal-stripped FBS for additional 24 hours and then treated with 17β-estradiol (E2), progesterone (P4), or medroxy progesterone acetate (MPA; Sigma) at 10−8 mol/L concentration for 6 and 24 hours. Total RNA was isolated and subjected to quantitative (Q)-RT-PCR as described above.
Luciferase Reporter Assay
Ishikawa cells were seeded in 6-well plates and transfected with pre-miR200c or NC, as described above. After 24 hours, the medium was changed and the cells were transfected with dual luciferase reporter plasmid containing 3′UTR of ZEB1, ZEB2, VEGFA, FBLN5, KLF9 (Genecopoeia, Rockville, Maryland), IKKβ (gift from Dr Gil Mor34), and NFkβ reporter plasmid containing NFkβ response element (Signosis Inc, Sunnyvale, California) using Lipofectamine 2000 (Invitrogen). For IKKβ 3′UTR reporter assay and NFkβ reporter assay, the cells were co-transfected with either IKKβ 3′UTR reporter plasmid or NFkβ reporter plasmid along with pRL-TK plasmid (Promega, Madison, Wisconsin) encoding Renilla luciferase (0.2 μg/well) as a control for assessing the transfection efficiency. Firefly and Renilla luciferase activities were measured after 48 hours using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instruction, with some modification for measuring NFkβ reporter assay and IKKβ 3′UTR luciferase activity, according to recommendation from GeneCopoeia. Firefly luciferase activity was normalized to Renilla luciferase activity and the level of induction was reported as the mean ± standard error of the mean (SEM) and compared with a ratio in cells transfected with pre-miR negative control (NC). Each experiment was done in triplicates.
Western Blot Analysis
Total protein was isolated from Ishikawa cells and following centrifugation, supernatants were collected and protein concentrations were determined by standard method (Pierce BCA Protein Assay Kit, Fisher Scientific). Aliquots of 30 µg of total protein were subjected to Western blot analysis and transferred to polyvinylidene difluoride (PVDF) membrane, and then the membranes were probed with primary antibodies (dilution:1:200) generated against ZEB1, ZEB2, VEGFA, FLT1, FBLN5, TIMP2, IKKβ, and KLF9 (Cell Signaling Technology, Danvers, Massachusetts; Abcam Inc, San Francisco, California; Santa Cruz Biotechnology, Santa Cruz, California). α-tubulin (1:2000) was used for normalization and loading control. The membranes were exposed to horseradish peroxidase–labeled secondary antiserum and immunoreactive proteins were detected with the ECL Western Blotting Substrate (Fisher Scientific).
Cell Proliferation Assay
Ishikawa cells were transfected with pre-miR-200c or pre-miR NC and the rate of their proliferation was determined after 1 to 3 days posttransfection, using CellTiter 96 Cell Proliferation Assay (Promega), according to manufacturer’s instruction. The rate of proliferation was calculated independently for days 1, 2, and 3 for pre-miR-200c transfected cells and compared to the same day control NC, respectively.
Statistical Analysis
All in vitro experiments were performed in 3 sets of independent assays in duplicates. Where appropriate, the results were reported as mean ± SEM and statistically analyzed using nonparametric Student t test for comparisons between 2 groups or analysis of variance (ANOVA) followed by post hoc pairwise multiple comparisons among several groups. Pearson correlation coefficient was used to assess correlation between miR-200c and ZEBs expression. P values of <.05 were considered significant. GraphPad Prism 4.0 software was used for all data analysis.
Results
The Expression of miR-200c is Altered During Endometrial Transition into Disease States
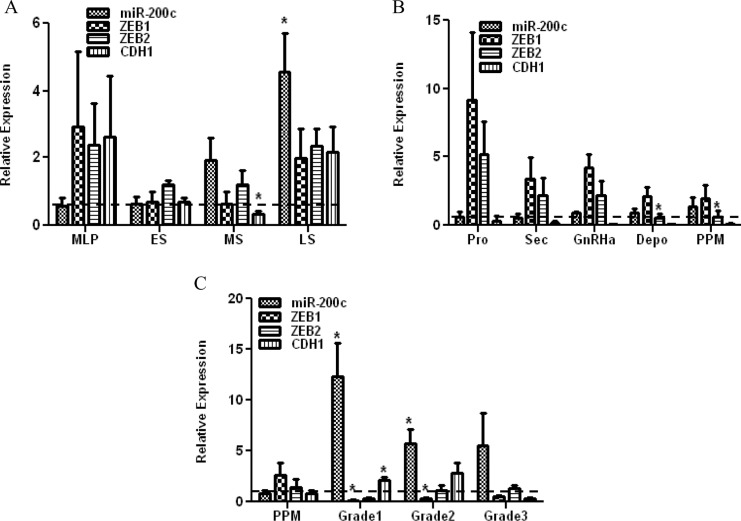
We first determined the expression of miR-200c in endometrial tissues (N = 52) consisting of biopsies obtained from normal menstrual cycle, endometrial tissue from untreated and patients exposed to different hormonal therapies, and grade I-III endometrial cancer. The relative expression of miR-200c varied among these cohorts with endometrial biopsies and was higher in mid-secretory (MS; P = .052) and late-secretory (LS) phase (P = .050), as compared to mid-late proliferative (MLP; Figure 1A). The relative expression of miR-200c did not significantly change (ANOVA) in endometrial tissues from secretory (Sec) phases of the menstrual cycle, peri- and postmenopausal period (PPM) with inactive endometrium, or women who were exposed to GnRHa and Depo-Provera (Depo; Figure 1B), when compared to the proliferative (Pro) phase. However, the level of miR-200c expression was significantly elevated in grade I and II endometrial tumors as compared to PPM period (P = .0079, P = .0376; Figure 1C). We next examined the expression of ZEB1 and ZEB2 and their downstream target genes, E-cadherin (CDH1), in these tissues and found variable level of expression in endometrial biopsies, which displayed a trend toward lower levels in ES and MS (Figure 1A). In endometrial tissues, the highest level of ZEB1 and ZEB2 was detected during proliferative phase, with a gradual reduction in tissues from secretory phase, GnRHa, Depo, and PPM groups (Figure 1B). In endometrial tumors from grade I and II, the level of ZEB1, but not ZEB2, was significantly lower (P = .007, P = .005) with increased CDH1 levels (P = .0317, P = .0734), as compared to tissues from PPM (Figure 1C). Although there was no statistically significant correlation between miR-200c and ZEBs expression among these tissues, the pattern of miR-200c expression, specifically in endometrial tissues, and endometrial tumors, was inversely related to ZEBs, specifically ZEB1 expression (Figure 1B and C).
Figure 1.
Relative expression of miR-200c, ZEB1, and ZEB2, in endometrial biopsies (A) from mid-late proliferative phase (MLP), early- (ES), mid- (MS), and late (LS)-secretory phase of the menstrual cycle and endometrial tissues (B) from proliferative (Pro) and secretory (Sec) phases of the menstrual cycle, peri- and postmenopausal period (PPM), women exposed to GnRH agonist (GnRHa) and Depo-Provera (Depo), and grade I, II, and III endometrial cancer (C). The results are presented as mean ± standard error of the mean (SEM) and analyzed using analysis of variance (ANOVA) or nonparametric student t test (*P < .05 as compared to levels in mid-late proliferative phase [MLP] in Figure A, proliferative phase [Pro] in Figure B and peri- and postmenopausal period [PPM] in Figure C).
Ovarian Steroids had Variable Effects on the Expression of miR-200c and Target Genes
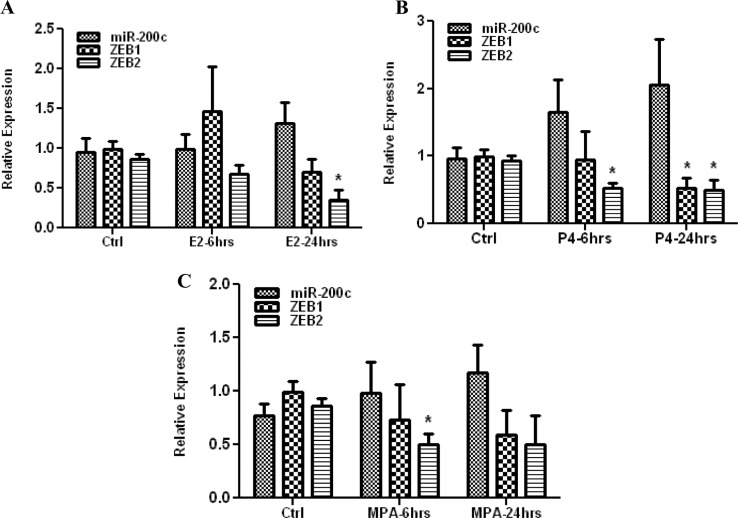
We next assessed the influence of ovarian steroids on miR-200c, ZEBs, and CDH1 expression using Ishikawa cell as an in vitro model. As shown in Figure 2, treatments with E2, P4, and MPA had variable effects on the expression of miR-200c, which tend to increase with time in all the cases; however, comparisons did not reach statistical significance. The expression of ZEBs was repressed by E2, P4, and MPA in a time-dependent manner, with E2 inhibition of ZEB2 after 6 hours, P4 inhibition of ZEB1 and ZEB2 after 24 hours, and MPA inhibition of ZEB2 after 6 hours (P < .05; Figure 2). There was no significant difference between P4 and MPA effects on miR-200c and ZEBs expression. In all the cases, there was no significant change in CDH1 expression (data not shown) which was expressed at very high levels.
Figure 2.
Relative expression of miR-200c, ZEB1, and ZEB2 in Ishikawa cells treated with 17-β Estradiol (E2) (A), progesterone (P4) (B), or medroxy progesterone acetate (MPA) (C) for 6 and 24 hours as compared to untreated control (Ctrl). The results are presented as mean ± standard error of the mean (SEM) and analyzed by analysis of variance (ANOVA) and nonparametric student t test (*P < .05 from their respective Ctrl).
miR-200c Targets the Expression of ZEBs, VEGFA, FLT1, IKKβ, KLF9, and FBLN5 and Alters Cellular Proliferation
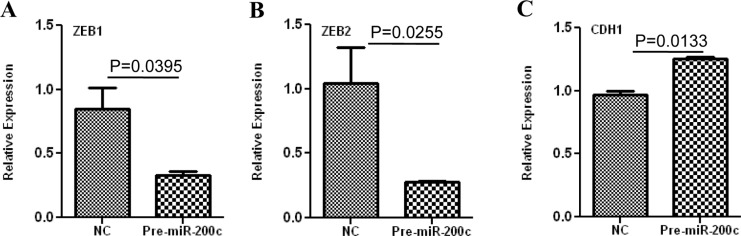
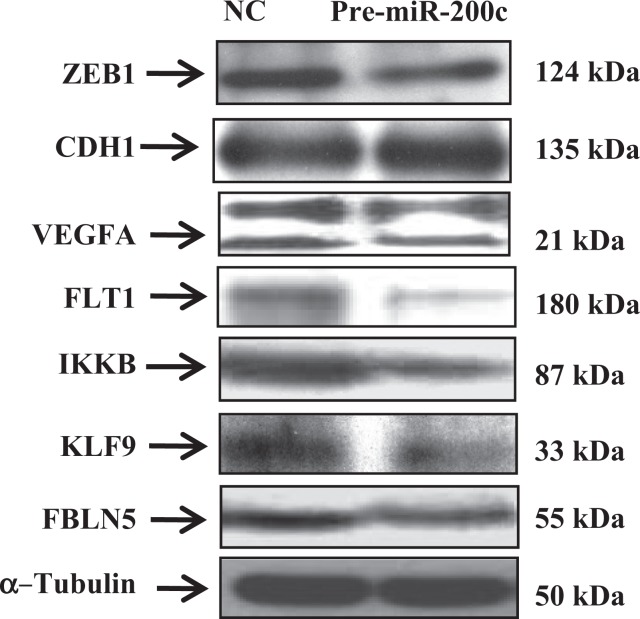
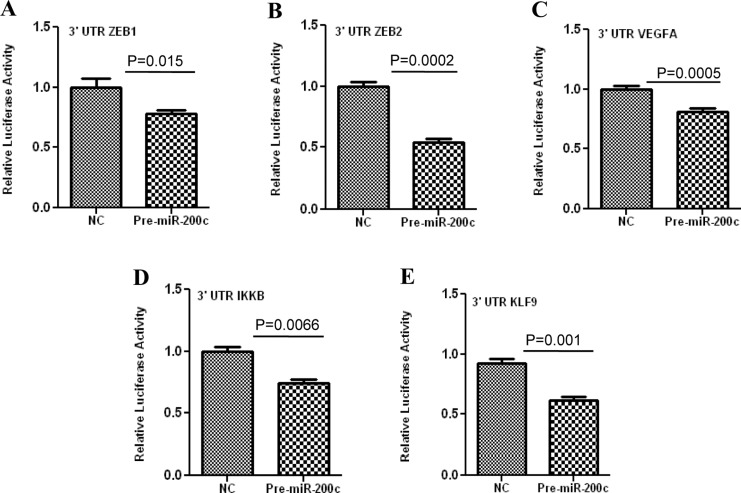
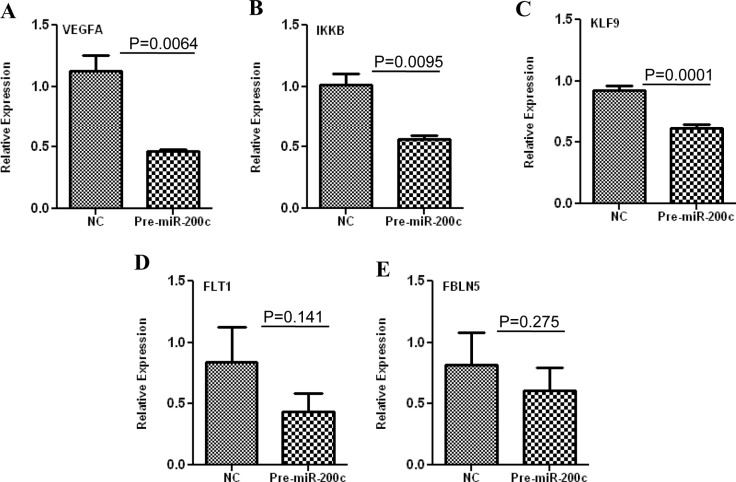
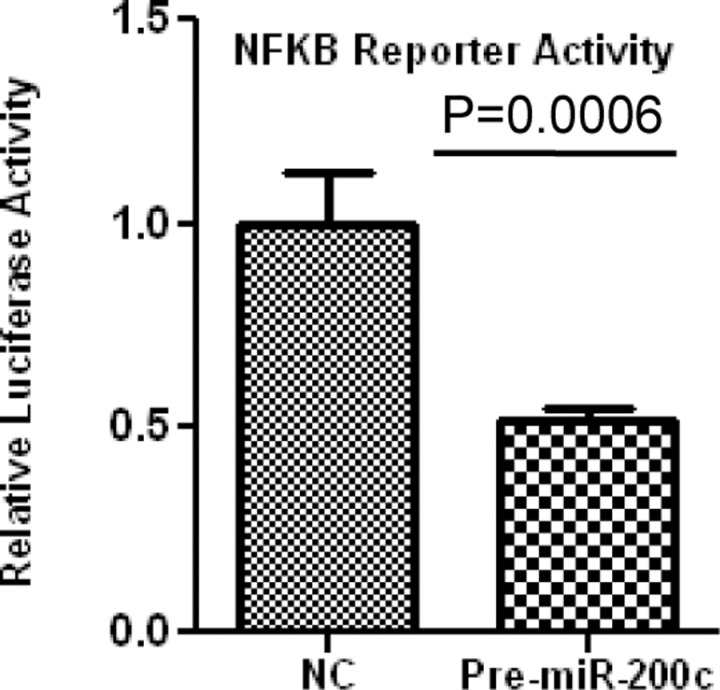
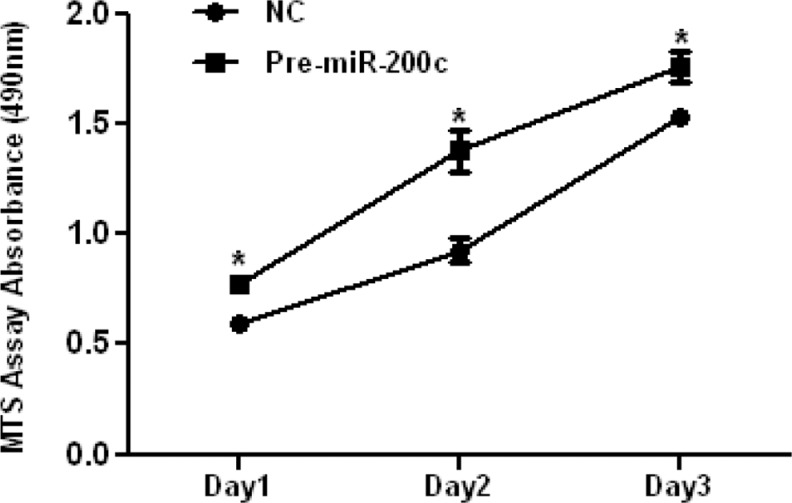
ZEBs are well established as direct targets of miR-200 family, including miR-200c; however, miRNAs expression and regulatory function of their target genes has been shown to occur in cell- and tissue-specific manners.32,33 As such using Ishikawa cells, we also found that gain of function of miR-200c repressed the expression of ZEB1 and ZEB2 at mRNA levels (Figure 3A and B), and ZEB1 at protein level (Figure 5); however, ZEB2 expression was too low and could not be detected by Western blot analysis. Overexpression of miR-200c in Ishikawa cells was also accompanied by elevated expression of CDH1 at both mRNA (Figure 3C) and protein levels (Figure 5). Gain of function of miR-200c repressed ZEBs expression through direct interaction with their respective 3′UTR as determined by luciferase reporter assay (Figure 6A and B). In addition to ZEBs, miR-200c is predicted to target the expression of VEGFA, FLT1, IKKβ, KLF9, FBLN5, and TIMP2 (Table 1). As such we extended our investigation and further demonstrated that gain of function of miR-200c also repressed the expression of VEGFA, IKKβ, and KLF9 at mRNA (Figure 4A-C) and protein (Figure 5) levels and reduced the expression of FLT1 and FBLN5 at protein (Figure 5), but not at mRNA levels (Figure 4D and E) without affecting TIMP2 expression either at mRNA or protein level (Supplementary Figure 1). Gain of function of miR-200c repressed VEGFA, IKKβ, and KLF9 expression through direct interaction with their respective 3′UTRs (Figure 6C-E), but not with FBLN5 3′UTR (Supplementary Figure 1) as determined by luciferase reporter assay. Since miR-200c directly regulated the expression of IKKβ, a key cofactor in NFkβ activation, we examined and found that gain of function of miR-200c significantly decreased NFkβ activity in Ishikawa cells (Figure 7). In addition, gain of function of miR-200c increased the rate of Ishikawa cell proliferation during the 3 days of incubation as compared to pre-miR negative control (NC) transfected cells (Figure 8).
Figure 3.
Relative expression of ZEB1 (A), ZEB2 (B), and CDH1 (C), in Ishikawa cells transfected with pre-miR-200c or pre-miR negative control (NC) determined by real-time polymerase chain reaction (PCR). The results are presented as mean ± standard error of the mean (SEM) and analyzed using nonparametric student t test with P values indicated by corresponding lines.
Figure 5.
Western blot analysis of ZEB1, CDH1, VEGFA, IKKβ, KLF9, FLT1 and FBLN5 in Ishikawa cells transfected with pre-miR-200c and pre-miR negative control (NC) with α-Tubulin serving as loading control.
Figure 6.
Firefly luciferase assay with pZEX-MT01 or pGL3-Luc constructs carrying a 3′ untranslated region (3′ UTR) fragment of ZEB1 (A), ZEB2 (B), VEGFA (C), IKKβ (D), and KLF9 (E), respectively. Ishikawa cells were co-transfected with firefly luciferase reporters, Renilla luciferase transfection control plasmid (in case of IKKβ ), pre-miR-200c, or pre-miR negative control (NC). The ratio of firefly: Renilla was determined and reported as relative luciferase activity as compared to empty vector and analyzed using non-parametric student t-test with p values indicated by corresponding lines.
Table 1.
The miR-200c seed sequence and its complementary binding site on respective 3′ untranslated region (UTR) of its predicated target genes (represented in bold letters)
| Position 369-375 of ZEB1 3′UTR | 5′AUUGUUUUAUCUUAUCAGUAUUA… |
| hsa-miR-200c | 3′AGGUAGUAAUGGGCCGUCAUAAU… |
| Position 454-461 of ZEB2 3′UTR | 5′CGCACUACAAUGCAUCAGUAUUA… |
| hsa-miR-200c | 3′AGGUAGUAAUGGGCCGUCAUAAU… |
| Position 901-907 of IKBKB 3′UTR | 5′UCUCACAGCAUCUAGCAGUAUUA… |
| hsa-miR-200c | 3′AGGUAGUAAUGGGCCGUCAUAAU… |
| Position 1298-1304 of VEGFA 3′UTR | 5′AGUAGGGUUUUUUUUCAGUAUUC… |
| hsa-miR-200c | 3′AGGUAGUAAUGGGCCGUCAUAAU… |
| Position 68-74 of FLT1 3′UTR | 5′GAAACUAGCUUUUGCCAGUAUUA… |
| hsa-miR-200c | 3′AGGUAGUAAUGGGCCGUCAUAAU… |
| Position 3041-3047 of KLF9 3′UTR | 5′AGUUUUAUUUUUGUACAGUAUUA… |
| hsa-miR-200c | 3′AGGUAGUAAUGGGCCGUCAUAU… |
| Position 2447-2452 of TIMP2 3′UTR | 5′AAAUCUUUGCUUGAUAAGUAUUA… |
| hsa-miR-200c | 3′AGGUAGUAAUGGGCCGUCAUAAU… |
| Position 222-227 of FBLN5 3′UTR | 5′GACACAGUUAUCAAAAAGUAUUA… |
| hsa-miR-200c | 3′AGGUAGUAAUGGGCCGUCAUAAU… |
Figure 4.
Relative expression of VEGFA (A), IKKβ (B), KLF9 (C), FLT1 (D) and FBLN5 (E) in Ishikawa cells transfected with pre-miR-200c and pre-miR negative control (NC) determined by real-time polymerase chain reaction (PCR). The results are presented as mean ±SEM and analyzed using nonparametric unpaired student t-test with p values indicated by corresponding lines.
Figure 7.
Firefly luciferase assay with pTF-Luc reporter constructs carrying NFkβ response element to determine NFkβ activation following pre-miR-200c transfection. Ishikawa cells were co-transfected with firefly luciferase reporter, Renilla luciferase transfection control plasmid, pre-miR-200c, or pre-miR negative control (NC). The ratio of firefly–Renilla was determined and reported as relative luciferase activity and analyzed using nonparametric unpaired student t test and P values is indicated by corresponding line.
Figure 8.
The effect of gain of function of miR-200c on Ishikawa cell proliferation. The cells were transfected with pre-miR-200c or pre-miR negative control (NC) and cell proliferation was determined using MTS assay on indicated days. The results are shown as mean ± standard error of the mean (SEM) and analyzed using nonparametric student t test (*P < .05 as independently compared to corresponding negative control for each day).
Discussion
Using a large cohort of endometrial tissues from normal, benign, and cancerous conditions our results indicated that the endometrial miR-200c expression undergoes dynamic changes that reflected possible influence of hormonal milieu and its correlation with disease progression. The endometrial expression of miR-200c to some extent inversely correlated with the expression of ZEBs, a well-established target of miR-200 family, however the pattern of their expression did not reflect such regulatory interactions, specifically in endometrial cancer, since loss of miR-200 family and overexpression of ZEBs accompanied by loss of CDH1 has been closely associated with cellular transformation and tumorigenesis in various tissues. In this context, a previous study reported the presence of immunoreactive ZEB1 only in endometrial stromal cells which increased during the secretory phase35 and in low-grade endometrial cancer with aberrant expression in epithelial-derived tumor.36 Real-time PCR analysis in our study indicated a trend toward lower expression of ZEBs in normal and benign endometrial tissues during the secretory phase and all grades of endometrial cancer, whereas our results with miR-200c expression in endometrial cancer is supported by two recent reports showing elevated levels of all miR-200 family, including miR-200c in all stages of endometrial cancer.22,37 Since miRNA regulatory functions have been reported to be tissue and cell specific,32,33 differences in miR-200 and ZEBs expression in normal, benign and cancer states may reflect such specificity, not only in regulating the specific gene/genes at given stage, but also in differences in expression within other tissues. Although transcriptional decay of miRNA target genes may not correspond to the level of miRNA expression, correlating translational repression of a given target genes may provide some possibility of establishing a functional relationship between miRNA and their target genes.1
Using Ishikawa cells with epithelial characteristics as an in vitro model, we found low levels of ZEBs, specifically ZEB2 and high level of CDH1 expression, corresponding to ZEB1 levels reported in endometrial epithelial cell layer.38 However, overexpressing or blocking miR-200c which targets ZEBs expression did not cause any noticeable changes in Ishikawa cell morphology, possibly due to high basal expression of CDH1. With regard to the hormonal influence, ovarian steroids modestly altered the expression of miR-200c and ZEBs, with P4 and MPA increasing miR-200c expression while inhibiting ZEBs, which reflected the patterns seen at tissue level corresponding to natural hormonal milieu. Treatment of isolated human myometrial smooth muscle cells or ovariectomized mice with P4 or estrogen has been shown to increase the expression of ZEB1 35 whereas P4 inhibited miR-200b and miR-429 expression in T47D, breast cancer cell line.31 Several studies have demonstrated differential influence of ovarian steroids on miRNAs expression in several cell lines, including endometrial cancer cells, through a mechanism possibly involving transcriptional regulation of their processing enzymes.39
There is considerable evidence in support of miR-200 family regulation of ZEBs in several cell types from different origins.31,35,40–44 We also confirmed that ZEBs are direct targets of miR-200c in Ishikawa cells. The importance of endometrial expression and regulation of ZEBs may be related to their regulatory influence on the expression of number of genes functionally involved in cell cycle progression, differentiation, and cellular transformation, specifically EMT during tumorigenesis.45–47 Endometrium during normal menstrual cycle, progression into pre- and postmenopausal states, endometriosis, and endometrial cancer, undergoes significant morphological changes involving many cellular events regulated by miR-200 target genes. We demonstrated that miR-200c either directly or through indirect mechanism regulates the expression of VEGFA, FLT1, IKKβ, KLF9, and FBLN5, without affecting TIMP2 expression. This finding may be biologically significant due to a wide range of cellular activities regulated by these genes; KLF9 serves as transcription factor, VEGF, FLT1, and FBLN5 are angiogenic factors; IKKβ, is an inflammatory mediator; FBLN5 supply the extracellular matrix protein essential for elastic fiber assembly and vasculogenesis; and TIMP2 is physiological inhibitor of MMPs known to regulate inflammation, matrix degradation, and cellular invasion. Although our finding with miR-200c regulation of KLF9 is the first of such validation, others reported VEGFA, FLT1, ETS1, SUZ12, PDGF, TGF-β2, and conexin43, as well as Notch pathway components, such as Jagged1 (JAG1) and mastermind-like coactivators, MAMLl2 and MAML3, as direct targets of different member of miR-200 family.24–30,41
Because of the lack of adequate endometrial biopsies and endometrial cancer tissues, we could not determine the coexpression of VEGFA, FLT1, IKKβ, and KLF9 in the same tissues; however, several studies reported their expression in human endometrium with altered or aberrant expression in endometriosis and endometrial cancer.48–58 The biological significance of VEGFA and their receptors (FLT1 and VEGFR1), in various activities, including angiogenesis, is well established, including in normal endometrium throughout the menstrual cycle, dysfunctional uterine bleeding, and endometrial repair and endometrial cancer.53,54,57 Additionally, IKKβ by serving as a cofactor for activation of NFkβ pathway plays a key role in regulating the expression of many proinflammatory mediators, including interleukin 8.48,59 Several miRNAs have been predicted to target IKKβ and in addition to our finding with miR-200c a recent report also demonstrated IKKβ as a direct target of miR-199a.34,59 Our results also suggest that miR-200c via downregulation of IKKβ may regulate NFkβ activation, which has been shown to upregulate Snail expression and induce EMT.60 KLF9 serves as a key regulator of genes functionally involved in cellular proliferation, differentiation, apoptosis, and tumorigenesis whose expression, and to some extent function, has been well documented in the uterus with altered expression in endometriosis and endometrial cancer.56,61,62 We found that KLF9 is a direct target of miR-200c, and since gain of function of miR-200c increased the rate of Ishikawa cell proliferation, we speculate that this increase in the cell proliferation is mediated through KLF9 repression, similar to the results reported earlier using KLF9 siRNA approach.62 Since several other miRNAs are predicted to target the expression of these genes, further investigation is needed for establishing the complex regulatory interaction among these genes and miRNAs that target their expression.
In summary, using a cohort of normal, benign, and cancerous endometrial tissues we found a dynamic temporal pattern of miR-200c expression and confirmed and/or validated a number of genes, including ZEBs, VEGFA, FLT1, IKKβ, and KLF9 and FBLN5, as direct and/or indirect targets of miR-200c regulatory function. Considering the importance of these gene products in various cellular activities ranging from cell growth, differentiation, inflammation, angiogenesis, cellular transformation, and tissue turnover that led either to cellular transformation and tumorigenesis or tissue fibrosis, miR-200 family including miR-200c could play a key role in regulating these processes in normal endometrium and in disorders such as endometriosis, abnormal uterine bleeding, and endometrial cancer (Figure 9). While the goal of this study was to better understand the regulatory function of miR-200c in endometrial tissue, future investigation is required to address the role of miR-200c and other members of this family in endometrium and its diseased states such as endometriosis and endometrial cancer.
Figure 9.
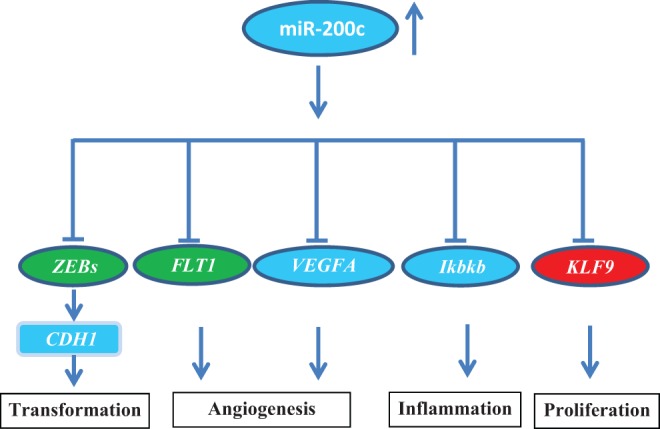
Schematic presentation of miR-200c and specific genes either directly and/or indirectly regulated by miR-200c in endometrium. Through such regulatory functions altered expression miR-200c could influence cell proliferation, migration, inflammation, and tissue turnover, events with central roles in normal cellular activities as well as in the outcome of disorders such as endometriosis and endometrial cancer and associated symptoms.
Acknowledgments
The authors thank Dr Barry Ripps for providing some of the endometrial biopsies used in this study. This project was supported by grants HD37432 and HD58664 from Eunice Kennedy Shriver National Institute of Health. Presented in part at the 59th annual meeting of the Society for Gynecological Investigation, San Diego CA, March 21-24, 2012.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This work was funded by NIH Grants: RO1HD37432 and RO1HD58664.
References
- 1. Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331(6017):550–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dykxhoorn DM. MicroRNAs and metastasis: little RNAs go a long way. Cancer Res. 2010;70(16):6401–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379 [DOI] [PubMed] [Google Scholar]
- 4. Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27(45):5959–5974 [DOI] [PubMed] [Google Scholar]
- 5. Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop—a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11(9):670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469(7330):336–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gonzalez G, Behringer RR. Dicer is required for female reproductive tract development and fertility in the mouse. Mol Reprod Dev. 2009;76(7):678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hong X, Luense LJ, McGinnis LK, Nothnick WB, Christenson LK. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology. 2008;149(12):6207–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nagaraja AK, Andreu-Vieyra C, Franco HL, et al. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22(10):2336–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boren T, Xiong Y, Hakam A, et al. MicroRNAs and their target messenger RNAs associated with endometrial carcinogenesis. Gynecol Oncol. 2008;110(2):206–215 [DOI] [PubMed] [Google Scholar]
- 11. Cohn DE, Fabbri M, Valeri N, et al. Comprehensive miRNA profiling of surgically staged endometrial cancer. Am J Obstet Gynecol. 2010;202(6):656–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hawkins SM, Buchold GM, Matzuk MM. Minireview: the roles of small RNA pathways in reproductive medicine. Mol Endocrinol. 2011;25(8):1257–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hawkins SM, Creighton CJ, Han DY, et al. Functional MicroRNA Involved in Endometriosis. Mol Endocrinol. 2011;25(5):821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Myatt SS, Wang J, Monteiro LJ, et al. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70(1):367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, et al. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23(2):265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13(11):797–806 [DOI] [PubMed] [Google Scholar]
- 17. Ramon LA, Braza-Boils A, Gilabert-Estelles J, et al. MicroRNAs expression in endometriosis and their relation to angiogenic factors. Hum Reprod. 2011;26(5):1082–1090 [DOI] [PubMed] [Google Scholar]
- 18. Castilla MA, Moreno-Bueno G, Romero-Perez L, et al. Micro-RNA signature of the epithelial-mesenchymal transition in endometrial carcinosarcoma. J Pathol. 2011;223(1):72–80 [DOI] [PubMed] [Google Scholar]
- 19. Cochrane DR, Howe EN, Spoelstra NS, Richer JK. Loss of miR-200c: a marker of aggressiveness and chemoresistance in female reproductive cancers. J Oncol. 2010;2010:821717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filigheddu N, Gregnanin I, Porporato PE, et al. Differential expression of microRNAs between eutopic and ectopic endometrium in ovarian endometriosis. J Biomed Biotechnol. 2010;2010:369549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuokkanen S, Chen B, Ojalvo L, Benard L, Santoro N, Pollard JW. Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human endometrium. Biol Reprod. 2010;82(4):791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee JW, Park YA, Choi JJ, et al. The expression of the miRNA-200 family in endometrial endometrioid carcinoma. Gynecol Oncol. 2011;120(1):56–62 [DOI] [PubMed] [Google Scholar]
- 23. Davalos V, Esteller M. Opening the treasure chest of miR-200 s family members. Cell Cycle. 2009;8(14):2141–2142 [PubMed] [Google Scholar]
- 24. Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem. 2011;286(3):2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39(5):761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kong D, Li Y, Wang Z, et al. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27(8):1712–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Korpal M, Ell BJ, Buffa FM, et al. Direct targeting of Sec23a by miR-200 s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17(9):1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roybal JD, Zang Y, Ahn YH, et al. miR-200 Inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol Cancer Res. 2011;9(1):25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uhlmann S, Zhang JD, Schwager A, et al. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene. 2010;29(30):4297–4306 [DOI] [PubMed] [Google Scholar]
- 30. Wang B, Koh P, Winbanks C, et al. miR-200a prevents renal fibrogenesis through repression of TGF-beta2 expression. Diabetes. 2011;60(1):280–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc Natl Acad Sci U S A. 2010;107(48):20828–20833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dahiya N, Sherman-Baust CA, Wang TL, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS One. 2008;3(6):e2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Inomata M, Tagawa H, Guo YM, Kameoka Y, Takahashi N, Sawada K. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113(2):396–402 [DOI] [PubMed] [Google Scholar]
- 34. Chen R, Alvero AB, Silasi DA, et al. Regulation of IKK[beta] by miR-199a affects NF-[kappa]B activity in ovarian cancer cells. Oncogene. 2008;27(34):4712–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spoelstra NS, Manning NG, Higashi Y, et al. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res. 2006;66(7):3893–3902 [DOI] [PubMed] [Google Scholar]
- 36. Singh M, Spoelstra NS, Jean A, et al. ZEB1 expression in type I vs type II endometrial cancers: a marker of aggressive disease. Mod Pathol. 2008;21(7):912–923 [DOI] [PubMed] [Google Scholar]
- 37. Snowdon J, Zhang X, Childs T, Tron VA, Feilotter H. The microRNA-200 family is upregulated in endometrial carcinoma. PLoS One. 2011;6(8):e22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spoelstra NS, Manning NG, Higashi Y. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res. 2006;66(7):3893–3902 [DOI] [PubMed] [Google Scholar]
- 39. Yoshimoto N, Toyama T, Takahashi S, et al. Distinct expressions of microRNAs that directly target estrogen receptor alpha in human breast cancer. Breast Cancer Res Treat. 2011;130(1):331–339 [DOI] [PubMed] [Google Scholar]
- 40. Bendoraite A, Knouf EC, Garg KS, et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol Oncol. 2010;116(1):117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brabletz S, Bajdak K, Meidhof S, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30(4):770–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dykxhoorn DM, Wu Y, Xie H, et al. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4(9):e7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601 [DOI] [PubMed] [Google Scholar]
- 44. Xia H, Ng SS, Jiang S, et al. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun. 2010;391(1):535–541 [DOI] [PubMed] [Google Scholar]
- 45. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119(6):1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890 [DOI] [PubMed] [Google Scholar]
- 47. Vandewalle C, Comijn J, De CB, et al. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33(20):6566–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen SU, Lee H, Chang DY, et al. Lysophosphatidic acid mediates interleukin-8 expression in human endometrial stromal cells through its receptor and nuclear factor-kappaB-dependent pathway: a possible role in angiogenesis of endometrium and placenta. Endocrinology. 2008;149(11):5888–5896 [DOI] [PubMed] [Google Scholar]
- 49. Graesslin O, Cortez A, Fauvet R, Lorenzato M, Birembaut P, Darai E. Metalloproteinase-2, -7 and -9 and tissue inhibitor of metalloproteinase-1 and -2 expression in normal, hyperplastic and neoplastic endometrium: a clinical-pathological correlation study. Ann Oncol. 2006;17(4):637–645 [DOI] [PubMed] [Google Scholar]
- 50. Hannan NJ, Paiva P, Meehan KL, Rombauts LJ, Gardner DK, Salamonsen LA. Analysis of fertility-related soluble mediators in human uterine fluid identifies VEGF as a key regulator of embryo implantation. Endocrinology. 2011;152(12):4948–4956 [DOI] [PubMed] [Google Scholar]
- 51. Huang F, Liu Q, Wang H, Zou Y. Effect of GnRH II and GnRH I on secretion of VEGF by eutopic and ectopic endometrial stromal cells of endometriosis patients. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2010;35(5):409–418 [DOI] [PubMed] [Google Scholar]
- 52. King AE, Critchley HO, Kelly RW. The NF-kappaB pathway in human endometrium and first trimester decidua. Mol Hum Reprod. 2001;7(2):175–183 [DOI] [PubMed] [Google Scholar]
- 53. Maybin JA, Hirani N, Jabbour HN, Critchley HO. Novel roles for hypoxia and prostaglandin E2 in the regulation of IL-8 during endometrial repair. Am J Pathol. 2011;178(3):1245–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maybin JA, Hirani N, Brown P, Jabbour HN, Critchley HO. The regulation of vascular endothelial growth factor by hypoxia and prostaglandin Falpha during human endometrial repair. J Clin Endocrinol Metab. 2011;96(8):2475–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Okada H, Okamoto R, Tsuzuki T, Tsuji S, Yasuda K, Kanzaki H. Progestins inhibit estradiol-induced vascular endothelial growth factor and stromal cell-derived factor 1 in human endometrial stromal cells. Fertil Steril. 2011;96(3):786–791 [DOI] [PubMed] [Google Scholar]
- 56. Simmen RC, Pabona JM, Velarde MC, Simmons C, Rahal O, Simmen FA. The emerging role of Kruppel-like factors in endocrine-responsive cancers of female reproductive tissues. J Endocrinol. 2010;204(3):223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Taylor RN, Yu J, Torres PB, et al. Mechanistic and therapeutic implications of angiogenesis in endometriosis. Reprod Sci. 2009;16(2):140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ulukus M, Ulukus EC, Tavmergen Goker EN, Tavmergen E, Zheng W, Arici A. Expression of interleukin-8 and monocyte chemotactic protein 1 in women with endometriosis. Fertil Steril. 2009;91(3):687–693 [DOI] [PubMed] [Google Scholar]
- 59. Dai L, Gu L, Di W. MiR-199a attenuates endometrial stromal cell invasiveness through suppression of the IKK{beta}/NF-kB pathway and reduced interleukin-8 expression. Mol Hum Reprod. 2011;18(3):136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Julien S, Puig I, Caretti E, et al. Activation of NF-kappaB by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene. 2007;26(53):7445–7456 [DOI] [PubMed] [Google Scholar]
- 61. Du H, Sarno J, Taylor HS. HOXA10 inhibits Kruppel-like factor 9 expression in the human endometrial epithelium. Biol Reprod. 2010;83(2):205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Simmons CD, Pabona JM, Heard ME, et al. Kruppel-like factor 9 loss-of-expression in human endometrial carcinoma links altered expression of growth-regulatory genes with aberrant proliferative response to estrogen. Biol Reprod. 2011;85(2):378–385 [DOI] [PMC free article] [PubMed] [Google Scholar]