Abstract
PCR-restriction fragment length polymorphism analysis of heat shock protein 70 genes discriminates most neotropical Leishmania species, as well as Trypanosoma cruzi. The assay, combined with capillary electrophoresis in a microchip device, may be applied directly on clinical samples with a high sensitivity, hence supporting clinical and epidemiological monitoring of leishmaniasis.
Leishmaniasis is endemic in 88 countries, causing a burden estimated at 2,357,000 disability adjusted life years and 59,000 deaths (12). The disease is characterized by a considerable clinical and epidemiological pleomorphism, which is linked—besides host factors—with the important diversity of Leishmania species and their vectors. Clinical and epidemiological monitoring requires rapid and high throughput tools for species typing. This can be achieved with PCR assays combining a high detection level with the adequate discriminatory power. Currently, a few genetic targets are available: ribosomal DNA internal transcribed spacers (ITS) (5), gp63 genes (10), miniexon genes (7), and kinetoplast DNA (3). However, there are still very few studies on their direct application to human tissues as well as their clinical and epidemiological validation in the New World, where several species can be sympatric (6, 11). We report here a complementary assay based on PCR amplification of the repeated heat shock protein 70 genes (1, 8), followed by restriction fragment length polymorphism analysis (hsp70 PCR-RFLP).
Development of hsp70 PCR assay.
Reported sequences of Leishmania (Viannia) braziliensis and Leishmania (Leishmania) mexicana hsp70 genes (accession no. AF291716 and M87878) were aligned with the CLUSTAL W 1.8 program. The primers were designed from the conserved region between Leishmania species with PRIMER PREMIER software: Hsp70sen (5′ GACGGTGCCTGCCTACTTCAA 3′) and Hsp70ant (5′ CCGCCCATGCTCTGGTACATC 3′). hsp70 PCR amplification was carried out in a 50-μl solution containing 1× Taq polymerase buffer (Eurogentec, Seraing, Belgium), 1.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, 5% dimethyl sulfoxide, 20 pmol of primers, and 2.5 U of Taq DNA polymerase (Eurogentec). The reaction mixtures were amplified in a MJ Research PTC-100 cycler with a heated lid at 94°C for 5 min followed by 33 cycles, each consisting of 30 s at 94°C, 1 min at 61°C, and 3 min at 72°C, and a final extension step of 10 min at 72°C.
hsp70 PCR on laboratory samples.
We analyzed 59 reference strains (Table 1) of species reported in Latin America: L. (V.) braziliensis, Leishmania (Viannia) peruviana, Leishmania (Viannia) lainsoni, Leishmania (Viannia) guyanensis, Leishmania (Viannia) panamensis, Leishmania (Leishmania) amazonensis, and Leishmania (Leishmania) infantum [synonym Leishmania (Leishmania) chagasi]. Promastigotes were cultivated and harvested (10) and were DNA purified with DNAzol (Gibco, Merelbeke, Belgium). After hsp70 PCR, a single 1,300-bp product was observed in all Leishmania strains, corresponding to the expected size. The same product was encountered in Trypanosoma cruzi, but no amplification was detected with DNA from humans Mycobacterium tuberculosis, or Sporothrix schenckii.
TABLE 1.
Strains of Leishmania analyzed in this studya
| Species | International code | Origin | Pathology |
|---|---|---|---|
| L. (V.) braziliensis | MHOM/BO/00/CUM27 | Bolivia | M |
| MHOM/BO/00/CUM29 | Bolivia | M | |
| MHOM/BO/94/CUM43 | Bolivia | M | |
| MHOM/BO/00/CUM45 | Bolivia | M | |
| MHOM/BO/94/CUM49 | Bolivia | M | |
| MHOM/BO/94/CUM52 | Bolivia | M | |
| MHOM/BO/00/CUM68 | Bolivia | M | |
| MHOM/BO/00/CUM152 | Bolivia | M | |
| MHOM/PE/93/LC2143 | Peru | C | |
| MHOM/PE/93/LC2176 | Peru | C | |
| MHOM/PE/00/LC2177 | Peru | C | |
| MHOM/PE/00/LC2320 | Peru | M | |
| MHOM/PE/94/LC2368 | Peru | M | |
| MHOM/BO/94/CUM41 | Bolivia | C | |
| MHOM/BO/94/CUM153 | Bolivia | C | |
| MHOM/BO/94/CUM42 | Bolivia | C | |
| MHOM/BO/00/LC2123 | Peru | C | |
| MHOM/BO/00/CUM97 | Bolivia | C | |
| MHOM/PE/00/LC2355 | Peru | C | |
| MHOM/PE/00/LC2284 | Peru | C | |
| MHOM/PE/00/LC2367 | Peru | C | |
| L. (V.) peruviana | MHOM/PE/90/HB22 | Peru | C |
| MHOM/PE/90/HB44 | Peru | C | |
| MHOM/PE/90/HB67 | Peru | C | |
| MHOM/PE/90/HB83 | Peru | C | |
| MHOM/PE/90/LCA09 | Peru | C | |
| MHOM/PE/90/LH249 | Peru | C | |
| MHOM/PE/90/LH827 | Peru | C | |
| MHOM/PE/90/LC1015 | Peru | C | |
| MHOM/PE/90/LCA04 | Peru | C | |
| L. (V.) guyanensis | MHOM/PE/91/LC1446 | Peru | C |
| MHOM/PE/91/LC1447 | Peru | C | |
| MHOM/PE/91/LC1448 | Peru | C | |
| MHOM/PE/94/LC2309 | Peru | C | |
| MHOM/PE/00/LC2797 | Peru | C | |
| MHOM/BR/75/M5378 | Brazil | C | |
| MHOM/GF/85/LEM699 | French Guyana | C | |
| IPRN/PE/00/Lp52 | Peru | Lutzomyia peruensis | |
| MHOM/PE/00/LH941 | Peru | C | |
| MHOM/PE/00/LH705 | Peru | C | |
| L. (V.) panamensis | MHOM/PA/71/LS94 | Panama | C |
| MCHO/PA/00/M4039 | Panama | Choloepus | |
| L. (V.) lainsoni | MHOM/BO/95/CUM71 | Bolivia | C |
| MHOM/BO/94/CUM78 | Bolivia | C | |
| MHOM/BO/94/CUM88 | Bolivia | C | |
| MHOM/BO/95/CUM129 | Bolivia | C | |
| MHOM/PE/92/LC1581 | Peru | M | |
| MHOM/PE/00/LH619 | Peru | C | |
| MHOM/PE/93/LC2029 | Peru | C | |
| MHOM/PE/00/LC2190 | Peru | C | |
| MHOM/PE/00/LH1154 | Peru | C | |
| MHOM/PE/00/LH762 | Peru | C | |
| L. (L.) amazonensis | MHOM/BO/00/CEN001 | Bolivia | C |
| MHOM/BO/00/CEN018 | Bolivia | C | |
| MPRO/BR/77/LV78 | Brazil | Proechimys | |
| IFLA/BR/67/PH8 | Brazil | Lutzomyia flaviscutellata | |
| MHOM/BR/73/M2269 | Brazil | C | |
| L. (L.) infantum | MHOM/FR/1978/LEM75 | France | V |
| T. cruzi | CANIII | Brazil | Chagas' disease |
C, M, and V, cutaneous, mucocutaneous, and visceral leishmaniasis, respectively. Species identification was determined by isoenzyme electrophoresis (2).
hsp70 PCR-RLFP on laboratory samples.
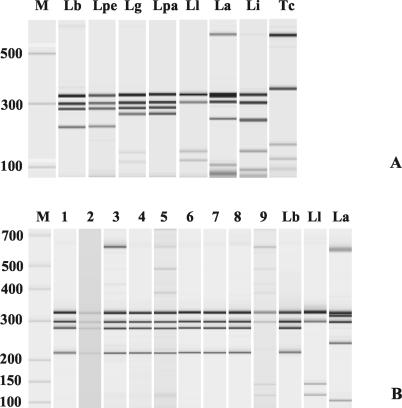
hsp70 PCR products were ethanol precipitated and resuspended in 20 μl of water. Digestion with restriction enzymes was performed according to the suppliers' recommendations in a final volume of 10 μl. Electrophoretic resolution was first performed in 3% agarose, by using 9 μl of digestion products, and then in microchips (2100 Bioanalyzer capillary electrophoresis system; Agilent Technologies, Karlsruhe, Germany) (LabChip 1500 or 7500; Caliper Technologies, Mountain View, Calif.) with only 1 μl of digests because of their high sensitivity and discriminatory power. From the five restriction enzymes tested, AsuI, TaqI, AluI, and AvaI distinguished L. (L.) amazonensis from all Leishmania samples of subgenus Viannia, while HaeIII (BsuRI) distinguished all species but L. (V.) peruviana (Fig. 1A), a species found only in the Peruvian highlands (11). However, hsp70 sequencing data revealed a BsiI restriction site differentiating that species from L. (V.) braziliensis. There was no intraspecies polymorphism as with assays targeting rDNA ITS or gp63 (5, 10). T. cruzi, in which a 1,300-bp hsp70 amplicon was also encountered, showed a different cleavage pattern (Fig. 1A). This might be particularly useful for identifying mixed infections of Leishmania spp. and T. cruzi, which can be quite frequent in Latin America (4). Analytical sensitivity of the PCR-RFLP assay was higher with capillary (3 parasites/μl before PCR) than with agarose electrophoresis (30 parasites/μl).
FIG. 1.
hsp70 PCR-RFLP patterns (HaeIII) after capillary electrophoresis (Bioanalyzer). M, size markers. (A) Reference strains of L. (V.) braziliensis (Lb), L. (V.) peruviana (Lpe), L. (V.) guyanensis (Lg), L. (V.) panamensis (Lpa), L. (V.) lainsoni (Ll), L. (L.) amazonensis (La), L. (L.) infantum (Li), and T. cruzi (Tc). (B) Biopsy specimens from Bolivian patients with leishmaniasis. Lanes 1 to 8, biopsies with L. (V.) braziliensis; lane 9, biopsy specimen with L. (V.) lainsoni. Reference strains of L. (V.) braziliensis (Lb), L. (V.) lainsoni (Ll), and L. (L.) amazonensis (La) are shown. M, size markers.
Clinical samples.
Thirty-four biopsy samples (4 mm) from Bolivian patients with clinical suspicion of tegumentary leishmaniasis (cutaneous and mucosal) were obtained with informed consent from the Isiboro secure area, between 1994 and 2000. Frozen biopsy specimens were lysed at 65°C for 3 h in 50 μl of TNE buffer (25 mM Tris, 100 mM NaCL, 5 mM EDTA [pH 8]) containing 5% sodium dodecyl sulfate and 200 μg of proteinase K/μl. After ethanol precipitation, DNA pellets were resuspended in 15 μl of buffer TE (10 mM Tris, 1 mM EDTA [pH 7.4]), and 2 μl was used for hsp70 PCR.
First, the capacity to detect parasites was considered. Sensitivity was compared with other methods, by using a laboratory case definition (positivity with microscopy, culture, or hsp70 PCR itself) and scored as follows (Table 2): 100% (hsp70 PCR), 92.9% (axenic culture), 80.9% (intradermal reaction of Montenegro [IDRM]), and 28.6% (microscopy). hsp70 PCR is thus slightly more sensitive than similar assays designed for species identification (85 to 89.7%; 6, 11). The relatively high sensitivity observed here for axenic culture can be explained by the fact that in CUMETROP, several aspirates are taken from the same patient to increase the isolation rate. The specificity of hsp70 PCR was 100%, but routinely, any positive sample should be digested to confirm a Leishmania pattern (versus T. cruzi, for instance). Concordance was highest between PCR and culture (kappa = 0.82).
TABLE 2.
Diagnostic performance of 34 hsp70 PCRs on samples from Bolivian patients with clinical suspicion of tegumentary leishmaniasis compared to microscopy, culture, and IDRMa
| Clinical status | No. of results
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Micro- scopy
|
Culture
|
hsp70 PCR
|
Totalb | IDRM
|
Total | |||||
| + | − | + | − | + | − | + | − | |||
| Cases | 8 | 20 | 26 | 2 | 28 | 0 | 28 | 17 | 4 | 21 |
| Noncases | 0 | 6 | 0 | 6 | 0 | 6 | 6 | 0 | 5 | 5 |
| Total samples | 8 | 26 | 26 | 8 | 28 | 6 | 34 | 17 | 9 | 26 |
IDRM available for only 26 patients. Case definition based on positivity by microscopy, culture, or hsp70 PCR.
Total results from microscopy, culture, and hsp70 PCR.
Second, species identification was performed by cutting the hsp70 amplicons of the 28 PCR-positive samples with HaeIII; 26 and 28 patterns were detected in agarose gels and microchips, respectively. Species identification was L. (V.) braziliensis (27 samples, including the two that were only PCR positive) and L. (V.) lainsoni (1 sample) (Fig. 1B).
Conclusion.
Our study brings new and original aspects to the field of Leishmania genetic characterization. Microchip capillary electrophoresis increases the performance of PCR-RFLP assays. hsp70 genes represent an adequate target for sensitive typing of neotropical Leishmania species in host tissues. They bring complementary information to other markers: (i) encoding for a major antigen (9), they allow probing of the genetic variability of molecules possibly involved in immunopathology, and (ii) presenting a lower rate of genetic variation than gp63 genes or rDNA ITS, for instance, they may be applied at other taxonomical levels (combining species and genus typing). This new marker also paves the way to future multigenic PCR-based approaches, essential for direct population studies in the host. Further work should be undertaken to compare on the same clinical samples the sensitivity, specificity, and discrimination power of the different PCR-RFLP assays currently available and to confirm the performance of hsp70 PCR-RFLP in other trypanosomatids.
Acknowledgments
This study received financial support from TDR (grant 00476), EC (grant IC18-CT96-0123), Belgian technical cooperation (02-0001-BOL/00/003), the Belgian Agency for Cooperation and Development (DGCD), and FWO (Flemish fund for Scientific research 1.5.047.02).
We thank C. Barnabé, Institut de recherche pour le Développement, Montpellier, France, for the gift of T. cruzi CANIII DNA.
REFERENCES
- 1.Arora, S. K., G. S. Kapoor, and S. Sehgal. 1998. Heterogeneity in heat shock protein genes in Leishmania isolates. Immunol. Cell Biol. 76:186-189. [DOI] [PubMed] [Google Scholar]
- 2.Bañuls, A. L. 1998. Apport de la génétique évolutive à la taxonomie et à l'épidémiologie du genre Leishmania. Ph.D. thesis. University of Montpellier, Montpellier, France.
- 3.Bastrenta, B., R. Buitrago, F. Vargas, F. Le Pont, M. Torrez, M. Flores, N. Mita, and S. F. Breniere. 2002. First evidence of transmission of Leishmania (Viannia) lainsoni in a Sub Andean region of Bolivia. Acta Trop. 83:249-253. [DOI] [PubMed] [Google Scholar]
- 4.Bastrenta, B., N. Mita, R. Buitrago, F. Vargas, M. Flores, M. Machane, N. Yacsik, M. Torrez, F. Le Pont, and S. F. Brenière. 2003. Human mixed infections of Leishmania spp. and Trypanosoma cruzi in a Sub Andean Bolivian area: identification by polymerase chain reaction/hybridization and isoenzyme. Mem. Inst. Oswaldo Cruz 98:255-264. [DOI] [PubMed] [Google Scholar]
- 5.Cupolillo, E., G. Grimaldi, Jr., H. Momen, and S. M. Beverley. 1995. Intergenic region typing (IRT), a rapid molecular approach to the characterization and evolution of Leishmania. Mol. Biochem. Parasitol. 73:145-155. [DOI] [PubMed] [Google Scholar]
- 6.Marfurt, J., A. Nasereddin, I. Niederwieser, C. L. Jaffe, H. P. Beck, and I. Felger. 2003. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J. Clin. Microbiol. 41:3147-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marfurt, J., I. Niederwieser, N. D. Makia, H. P. Beck, and I. Felger. 2003. Diagnostic genotyping of Old and New World Leishmania species by PCR-RFLP. Diagn. Microbiol. Infect. Dis. 46:115-124. [DOI] [PubMed] [Google Scholar]
- 8.Quijada, L., M. Soto, C. Alonso, and J. M. Requena. 1997. Analysis of post-transcriptional regulation operating on transcription products of the tandemly linked Leishmania infantum hsp70 genes. J. Biol. Chem. 272:4493-4499. [DOI] [PubMed] [Google Scholar]
- 9.Rico, A. I., S. O. Angel, C. Alonso, and J. M. Requena. 1999. Immunostimulatory properties of the Leishmania infantum heat shock proteins HSP70 and HSP83. Mol. Immunol. 36:1131-1139. [DOI] [PubMed] [Google Scholar]
- 10.Victoir, K., A. L. Banuls, J. Arevalo, A. Llanos-Cuentas, R. Hamers, S. Noël, S. De Doncker, D. Le Ray, M. Tibayrenc, and J. C. Dujardin. 1998. The gp63 gene locus, a target for genetic characterization of Leishmania belonging to subgenus Viannia. Parasitology 117:1-13. [PubMed] [Google Scholar]
- 11.Victoir, K., S. De Doncker, L. Cabrera, E. Alvarez, J. Arevalo, A. Llanos-Cuentas, D. Le Ray, and J. C. Dujardin. 2003. Detection and direct identification of Leishmania species in biopsies from patients with American tegumentary leishmaniasis. Trans. R. Soc. Trop. Med. Hyg. 97:80-87. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization. 2002. The world health report 2002. Reducing risks, promoting healthy life. World Health Organization, Geneva, Switzerland. [Online.] http://www.who.int/whr/2002/en.