Abstract
The host interferon (IFN) antiviral response involves a myriad of diverse biochemical pathways that disrupt virus replication cycles at many different levels. As a result, viruses have acquired and evolved genes that antagonize the host antiviral proteins. IFNs inhibit viral infections in part through the 2′,5′-oligoadenylate (2-5A) synthetase (OAS)/RNase L pathway. OAS proteins are pathogen recognition receptors that exist at different basal levels in different cell types and that are IFN inducible. Upon activation by the pathogen-associated molecular pattern viral double-stranded RNA, certain OAS proteins synthesize 2-5A from ATP. 2-5A binds to the antiviral enzyme RNase L causing its dimerization and activation. Recently, disparate RNA viruses, group 2a betacoronaviruses, and group A rotaviruses, have been shown to produce proteins with 2′,5′-phosphodiesterase (PDE) activities that eliminate 2-5A thereby evading the antiviral activity of the OAS/RNase L pathway. These viral proteins are members of the eukaryotic-viral LigT-like group of 2H phosphoesterases, so named for the presence of 2 conserved catalytic histidine residues. Here, we will review the biochemistry, biology, and implications of viral and cellular 2′,5′-PDEs that degrade 2-5A. In addition, we discuss alternative viral and cellular strategies for limiting the activity of OAS/RNase L.
Double-Stranded RNA Signaling Through 2′,5′-Oligoadenylate Synthetase to RNase L in the Interferon Antiviral Response
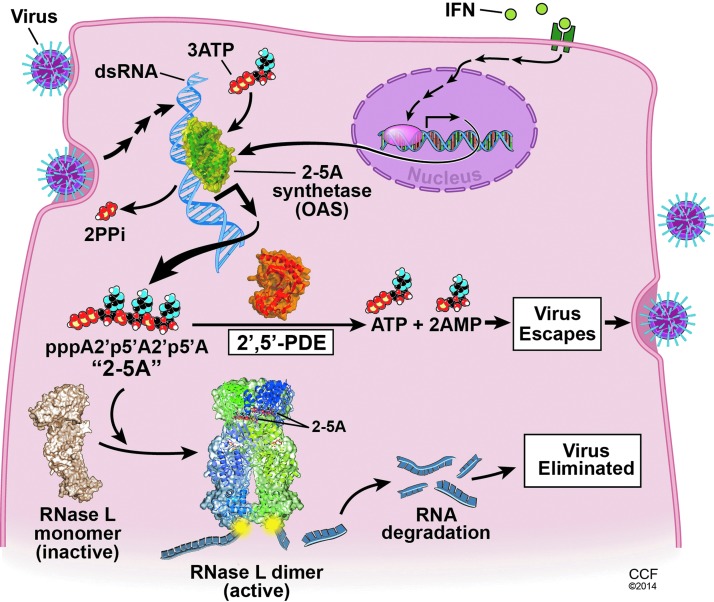
Double-stranded RNA (dsRNA) is a common pathogen-associated molecular pattern of both RNA and DNA viruses that triggers innate immune responses in the infected host cell. The replication of many families of viruses with single-stranded RNA genomes, including the Picornaviridae, Coronaviridae, Orthomyxoviridae, Paramyxoviridae, and Rhabdoviridae, requires synthesis of RNA of opposite polarity to the genomic RNA strand (the anti-genome) that generates dsRNA by annealing of the positive- and negative-sense RNA strands (Knipe and Howley 2007). Other viruses, such as the Reoviridae family that includes rotaviruses, have segmented dsRNA genomes (Knipe and Howley 2007). Double-stranded structures also occur in some viral RNAs that are otherwise single stranded (Maitra and others 1994; Han and Barton 2002). Also, certain DNA viruses, such as vaccinia virus in the family Poxviridae, produce dsRNA from annealing of complementary single-stranded RNAs produced by symmetrical transcription of the viral genome (Colby and Duesberg 1969). DsRNA initiates signaling pathways resulting in transcription of type I interferon (IFN) genes through either TLR3 present in endosomal membranes or cytoplasmic RIG-I-like receptors (RLR) [reviewed in Wilkins and Gale (2010)]. In addition, viral dsRNA directly triggers a cellular antiviral response through the 2′,5′-oligoadenylate (2-5A) synthetase (OAS)/RNase L pathway (Fig. 1) [reviewed in Silverman (2007)].
FIG. 1.
Viral activation and antagonism of the OAS/RNase L antiviral pathway in the host cell determines the outcome of infection. dsRNA:OAS1 (PDB ID code 4IG8) (Donovan and others 2013); porcine RNase L in a complex with natural 2-5A and AMP-PNP ligands (PDB ID code 4O1P) (Huang and others 2014); and rat AKAP7 central domain (PDB ID code 2VFY) as an example of a 2′,5′-PDE in the 2H phosphoesterase superfamily (Gold and others 2008). OAS, 2′,5′-oligoadenylate (2-5A) synthetase; dsRNA, double-stranded RNA; PDE, phosphodiesterase.
There are multiple OAS genes encoding many different OAS proteins, including isoforms resulting from alternative splicing events [reviewed in Justesen and others (2000) and Kristiansen and others (2011)]. However, some OAS proteins do not synthesize 2-5A and these species are presumably unrelated to activation of RNase L. In humans, there are 4 OAS genes, OAS1, OAS2, OAS3, and OASL, all of which are IFN inducible and map to chromosome 12q24.1 (OAS1–3) and 12q24.2 (OASL) [reviewed in Kristiansen and others (2011)]. OASL is also directly induced by viral infection through transcription factor IRF3 (Melchjorsen and others 2009). The human OAS proteins, OAS1 (p40/p46), OAS2 (p69/p71), and OAS3 (p100) have 1, 2, and 3 catalytic units, respectively, and synthesize 2-5A [p3(A2′p)nA, n≥2] from ATP upon activation by dsRNA binding (Kerr and Brown 1978; Benech and others 1985; Marie and Hovanessian 1992; Marie and others 1997; Rebouillat and others 1999; Sarkar and others 1999). In contrast, human OASL (p59) contains 2 C-terminal ubiquitin-like repeats and lacks the ability to synthesize 2-5A (Hartmann and others 1998; Rebouillat and others 1998), but nevertheless has antiviral activity against the picornavirus, encephalomyocarditis virus (Marques and others 2008). In mice, there are 8 Oas1 genes, mouse (m)Oas1a to mOas1h on mouse chromosome 5 (Mashimo and others 2003; Perelygin and others 2006; Kristiansen and others 2011). Of the mOAS1 isoforms, however, only mOAS1a and mOAS1g are believed to be enzymatically active based on results of functional assays and/or amino acid sequence analysis (Kakuta and others 2002; Mashimo and others 2003). mOAS1b is the product of the flavivirus resistance gene (Flv), and although it is enzymatically inactive (in fact it suppresses 2-5A synthesis) it is nevertheless a potent inhibitor of flavivirus replication (Mashimo and others 2002; Perelygin and others 2002; Elbahesh and others 2010; Courtney and others 2012). mOAS1d is expressed in oocytes, lacks enzymatic activity, and inhibits mOAS1a (Yan and others 2005). In addition, mOAS1d−/− mice have reduced fertility suggesting that OAS1d, and other enzymatically inactive isoforms of mOAS1, may partially protect oocytes from detrimental effects of RNase L during oogenesis and early embryonic development. Mice have 4 additional Oas genes, which produce 3 enzymatically active (mOAS2, mOAS3 and mOASL2) and one inactive (mOASL1) protein (Kakuta and others 2002). Instead, mOASL1 inhibits the translation of IRF7 mRNA and is a negative regulator of type I IFN synthesis (Lee and others 2013). The mouse OAS proteins are constitutively expressed at different levels in a wide range of different primary cell types and are also highly induced by IFN (Zhao and others 2013). In mouse brain, mOas1a, mOas2, and mOas3 mRNAs are constitutively expressed at about 10-fold higher levels in microglia than in astrocytes and about 100-fold higher than in neurons or oligodendrocytes. Also, mOas1a, mOas2, and mOas3 mRNAs are expressed at 10- to 1,000-fold higher levels in bone marrow macrophages than in hepatocytes, indicating high levels in some inflammatory cell types. In a separate study, high levels of Oasl2 mRNA expression were also observed in peritoneal macrophages (Sorgeloos and others 2013).
As mention above, the only known enzymatic function of OAS proteins is to synthesize 2-5A and related oligonucleotides. In addition, the only well established function of 2-5A is to activate RNase L, a protein that is widely constitutive expressed in many cell types and tissues and which is also upregulated by IFN in some mouse cell types (Jacobsen and others 1983; Zhou and others 1993, 1997). 2-5A (trimeric and longer species) specifically binds with high affinity to the inactive, monomeric form of RNase L causing it to dimerize into its enzymatically active state (Dong and Silverman 1995). RNase L has 3 major domains: an N-terminal 9 ankyrin repeat domain, a protein kinase-like or pseudokinase domain, and a C-terminal ribonuclease domain (Hassel and others 1993). Recently, the laboratory of Frank Sicheri solved the x-ray crystal structure of a dimeric form of RNase L bound to 2-5A and the ATP mimetic, AMP-PNP (in collaboration with the Silverman laboratory and others) (Huang and others 2014). The structure showed that 2-5A interacts with both the ankyrin repeat and protein kinase domains. ATP binding to the kinase domain causes a closed conformation of the kinase domain that is essential for RNA cleavage. RNase L cleaves single-stranded RNA, both cellular and viral, predominantly after UpU and UpA dinucleotides leaving a 5′-OH and a 2′,3′-cyclic phosphate on the cleavage products (Wreschner and others 1981b). Activation of RNase L can lead to autophagy (Chakrabarti and others 2012; Siddiqui and Malathi 2012) and apoptosis (Castelli and others 1997, 1998; Zhou and others 1997), both of which impact viral yields. Small RNAs generated from cleavage of viral and cellular RNAs by RNase L signal to RLRs, RIG-I, and MDA5, to amplify type I IFN productions in some cell types, such as mouse fibroblasts and human hepatoma Huh7 cells (Malathi and others 2007, 2010). In contrast, macrophages have high levels of both OAS (as mentioned above) and RNase L leading to extensive RNA degradation, apoptosis, and decreased (rather than increased) type I IFN production (Banerjee and others 2014).
Cell Type-Specific Evasion of the IFN Antiviral Response by Mouse Hepatitis Virus ns2
ns2 is a 30 kDa cytoplasmic nonstructural accessory protein encoded in the genomes of group 2a betacoronaviruses (Bredenbeek and others 1990; Zoltick and others 1990; Snijder and others 2003). This group includes the prototype coronavirus, mouse hepatitis virus (MHV), and human coronaviruses OC43 (an agent of the common cold) (Mazumder and others 2002; Snijder and others 2003) in addition to enteric HEC4408 (Han and others 2006) and several other mammalian coronaviruses. Interestingly, the coronavirus replicase proteins, which are encoded in 2 very long open reading frames (pp1a, pp1ab) in the 5′ two-thirds of the genome, and the structural proteins, encoded in the 3′ one-third of the genome, are highly conserved, whereas the accessory proteins differ among the various genera and subgroups of coronaviruses (Snijder and others 2003). Because accessory proteins often act as antagonists of the host response, especially the type I IFN response, this observation suggests that each genera or subgroup of coronavirus may use a different mechanism of host antagonism.
ns2 was predicated to be a member of a LigT-like family (family II) of the 2H phosphoesterase superfamily by analysis of sequence homology (Mazumder and others 2002). The 2H phosphoesterases are characterized by a pair of conserved His-x-Thr/Ser motifs (described in more detail below). Based on homology, ns2 was initially predicted to have cyclophosphodiesterase activity, as were other members of the LigT-like family (Mazumder and others 2002; Snijder and others 2003; Roth-Cross and others 2009). However, attempts to demonstrate this activity using 2′,3′ cAMP, 3′,5′ cAMP, and ADP-ribose 1,″2″ cyclic phosphate as substrates were unsuccessful (Zhao and others 2012).
MHV primarily infects the central nervous system (brain and spinal cord) and the liver inducing acute encephalitis and hepatitis followed by chronic demyelination, providing a model for human demyelinating diseases such as multiple sclerosis and also for viral hepatitis. Initial attempts at finding a function for ns2 were unsuccessful as a naturally occurring deletion mutant in the MHV, strain JHM.WU (JHM.WU-Δns2), exhibited no detectable defects in cell culture (Schwarz and others 1990) and was not attenuated in terms of replication or pathogenesis in the central nervous system [S. Perlman, J Leibowitz, personal communication; Zhao and others (2011)]. Site directed mutants of MHV strain A59 (A59), with amino acid substitutions in each of the 2 predicted catalytic His residues, H46A (ns2H46A) and H126R (ns2H126R), and the deletion mutant JHM.WU-Δns2 were used to further investigate the role of ns2 in pathogenesis. While both the A59 catalytic site mutants and JHM.WU-Δns2 were able to replicate in the central nervous system, both sets of ns2 mutants were unable to replicate in the liver of infected mice indicating that ns2 was an organ-specific virulence factor (Zhao and others 2011).
We investigated the possibility that ns2 was an IFN antagonist. Previous studies in the Weiss laboratory demonstrated that MHV induced type I IFN only in macrophages and furthermore was sensitive to IFN pretreatment primarily in macrophages (Roth-Cross and others 2009; Zhao and others 2011). Comparison of the replication of wild-type (WT) A59 and the ns2H126R mutant in bone marrow-derived macrophages (BMM) revealed that replication of the ns2 mutant was impaired, yielding a titer 100–1,000-fold less than for the WT virus. However, mutant virus recovered a WT level of replication in BMM derived from type I IFN receptor (IFNAR) gene knockout macrophages, suggesting that ns2 was an IFN antagonist.
Interestingly, in addition to the difference in the requirement of ns2 for virulence in the liver versus the brain, the requirement for ns2 was also cell type-specific. Expression of WT ns2 was essential for replication in several types of myeloid cells in addition to BMM, including brain resident microglia, liver resident Kupffer cells, bone marrow-derived dendritic cells and in liver sinusoidal endothelial cells (unpublished); however, ns2 was not required for efficient replication of MHV in other types of primary murine cells derived from the liver (hepatocytes) or the brain (neurons, astrocytes, and oligodendrocytes) or in several transformed cells lines (Zhao and others 2011, 2012). Consistent with the cell type-specific requirement for ns2, myeloid cells expressed significantly higher basal levels of IFN-stimulated genes including (as mentioned previously) mOas1a, mOas2, and mOas3, than other cell types (Zhao and others 2011, 2013), suggesting that ns2 was needed to overcome the antiviral effects of IFN in these cell types.
ns2 did not inhibit induction of IFN or the JAK-STAT pathway of IFN signaling. Thus, it seemed likely that ns2 would inhibit one or more IFN-induced antiviral activities. We therefore compared replication of ns2 mutant and WT A59 in BMM derived from mouse strains deficient in each of several antiviral activities, IFIT1, IFIT2, ISG15, and PKR; the ns2 mutant was still unable to replicate in the absence of any of these activities (data not shown). Remarkably, however, ns2 mutant completely regained WT ability to replicate in BMM derived from RNase L-deficient mice (Zhao and others 2012). Having established that MHV ns2 enables the virus to evade the antiviral activity of IFN in BMM by antagonizing RNase L, we proceeded to investigate whether ns2 might degrade 2-5A and thus prevent RNase L activation (Fig. 1).
ns2 Is a 2′,5′-Phosphodiesterase That Degrades 2-5A and Prevents RNase L Activation
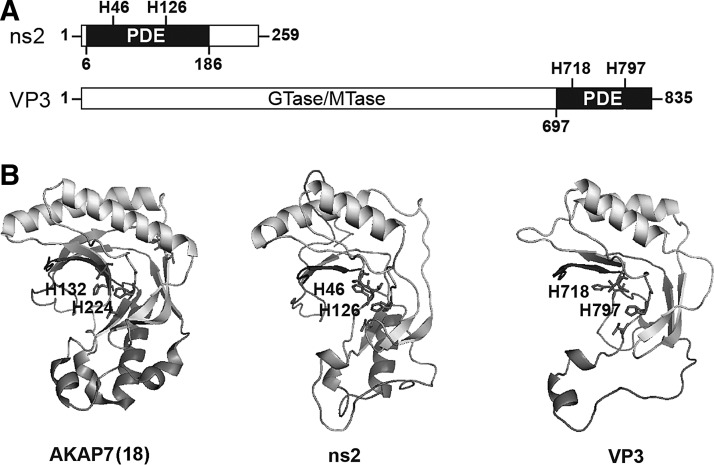
As mentioned above, based on sequence profile analysis methods, ns2 was previously assigned to the eukaryotic-viral LigT-like group (family II) of the 2H phosphoesterase superfamily (Mazumder and others 2002). The positions of the 2 conserved catalytic histidines in ns2 are shown (Fig. 2A). The most ancient members of the 2H phosphoesterase superfamily are archaeo-bacterial tRNA-ligating LigT proteins with 2′-5′ RNA ligase activity for tRNA precursors (Arn and Abelson 1996). Escherichia coli 2′-5′ RNA ligase is also able to perform the reverse 2′,5′-phosphodiesterase (PDE) reaction in vitro. The similarities between ns2 and bacterial LigT suggested to us that perhaps ns2 favors the reverse reaction and degrades 2-5A.
FIG. 2.
Homology between rat AKAP7 (PDB ID code 2VFY), MHV ns2 (strain A59), and rotavirus VP3 (strain SA11) [reprinted from Zhang and others (2013)]. (A) PDE domains in MHV ns2 and rotavirus VP3. VP3 domains involved in RNA capping are GTase, guanylyltransferase, and MTase, methyltransferase. (B) Alignments between the x-ray crystal structure of rat AKAP7 (PDB ID code 2VFY) and predicted structures of SA11 VP3-CTD and MHV A59 ns2. The 2 conserved His residues are shown as sticks. PyMol (http://pymol.org) was used for molecular graphics. CTD, C-terminal domain; MHV, mouse hepatitis virus.
To determine whether ns2 prevented RNase L activation during viral infections, BMM were infected with MHV WT A59 or mutant ns2H126R virus. Following infection with the ns2 mutant virus, rRNA degradation products highly characteristic of RNase L activity (Wreschner and others 1981a; Silverman and others 1983) were observed (Zhao and others 2012). In contrast, rRNA remained intact in A59 virus-infected BMM, indicating that ns2 was blocking or otherwise preventing RNase L activity. Furthermore, cellular levels of 2-5A oligonucleotides were greatly elevated in ns2 mutant virus-infected BMM compared with A59 virus-infected BMM. As mentioned above, replication of the ns2 mutant virus was restored in RNase L−/− BMM. As a direct test for 2′,5′-PDE activity, recombinant ns2 and ns2-H126R were expressed in bacteria and purified. Upon incubation, WT ns2 degraded trimeric 2-5A [(2′-5′)p3A3] to (2′-5′)p3A2 and 5′AMP, and eventually to 5′AMP and 5′ATP, as determined with high-performance liquid chromatography. The mutant ns2 failed to degrade (2′-5′)p3A3. Those studies established that ns2 has 2′,5′-PDE activity dependent on a conserved catalytic histidine residue.
Mutant ns2 virus replicated near the lower limit of detection, if at all, in the liver, spleen, lung, and kidney of WT B6 mice. In contrast, replication of the mutant virus was restored in all of these organs in RNase L−/− mice. Remarkably, at 5 days postinfection the titer in liver was 5 log10 units higher in RNase L−/− mice than in WT mice. As a result, the mutant ns2 virus caused hepatitis in RNase L−/− mice but not in WT mice. Evidence of virus-mediated pathology, including necrosis, apoptosis, and inflammation (with induction of TNF-α and IFN-γ) was observed after infection with mutant ns2 virus in RNase L−/− mice but not in WT mice. These studies demonstrate that the 2′,5′-PDE function of ns2 is required for efficient replication of MHV in the mouse liver and for pathogenesis.
The C-Terminal Domain of the VP3 Protein of a Group A Rotavirus Has 2′,5′-PDE Activity
Group A rotaviruses in the family Reoviridae have segmented dsRNA genomes requiring a replication strategy very different from that of positive RNA strand coronaviruses. Rotaviruses are important human pathogens that cause severe diarrhea in children worldwide (Patton 2012). The rotavirus minor core protein VP3 has both methyltransferease and guanylyltransferase activities involved in mRNA capping (Fig. 2A). In addition, the sequence of the VP3 C-terminal domain (CTD), characterized by 2 conserved His-X-Thr/Ser motifs in the catalytic ligand-binding cleft, places it in the same family of 2H phosphoesterases as MHV ns2 (Mazumder and others 2002; Zhang and others 2013). Both rotavirus VP3 CTD and ns2 were also predicted to be similar to an X-ray crystal structure of the rat A-kinase (PKA) anchoring protein AKAP15/18δ (AKAP7δ) complexed with 5′-AMP (Fig. 2B) (Gold and others 2008; Roth-Cross and others 2009; Zhang and others 2013). VP3-CTD was confirmed to be a 2′,5′-PDE by incubation of purified recombinant protein with (2′-5′)p3A3 as a substrate, which resulted in sequential release of 5′-AMP moieties from the 2′,3′-terminus until only 5′-ATP and 5′-AMP remained (Zhang and others 2013). Mutant VP3-CTD (VP3H718A;H797R) in which the catalytic histidines were mutated was enzymatically inactive. VP3-CTD was also able to prevent accumulation of 2-5A in cells treated with the synthetic dsRNA, poly(rI):poly(rC), an activator of OAS enzymatic activity. Due to the lack of a robust rotavirus reverse genetic system, we used chimeric MHV to determine whether VP3-CTD might enable virus to evade the antiviral activity of RNase L. MHV(A59) chimeras were constructed in which the endogeneous ns2 gene was mutated to abrogate PDE activity and the VP3-CTD cDNA (either WT or mutated) was inserted into the nonessential gene ns4. Remarkably, chimeric MHV encoding mutant ns2 and WT VP3-CTD replicated nearly as well as the parental MHV strain in BMM in vitro, and to a 1,000-fold higher titer than virus harboring mutant ns2 and mutant VP3-CTD, in the livers of WT mice. However, replication of the chimeric virus in which the conserved histidines were mutated in both ns2 and VP3-CTD was not impaired in RNase L−/− mice. Further experiments that examined the integrity of rRNA in infected BMM directly showed that rotavirus VP3-CTD prevented RNase L activation. Therefore, the rate of 2-5A cleavage by VP3-CTD or ns2 apparently exceeded its rate of synthesis by OAS resulting in an insufficient level of 2-5A to activate RNase L and thus inhibit virus replication (Fig. 1).
These data suggest that the VP3-CTD, by virtue of its ability to block the OAS/RNase L pathway, may be the virulence factor previously mapped to the VP3 gene of group A rotaviruses (Hoshino and others 1995; Wang and others 2011; Zhang and others 2013). Because VP3 is found in the viroplasm, it was suggested a non-particle-associated VP3 might be responsible for antagonizing RNase L function (Zhang and others 2013). Thus, VP3-CTD may be a second host IFN antagonist encoded by rotaviruses in addition to the previously described rotavirus NSP1 protein that inhibits induction of IFNs by causing proteosomal degradation of IRFs and βTrCP [reviewed in Arnold and others (2013)].
Additional Viruses That Are Likely to Have 2′,5′-PDEs
Homology analysis suggests that many additional RNA viruses encode 2′,5′-PDEs of the eukaryotic-viral LigT-like group of 2H phosphoesterases (Table 1). Coronaviruses with ns2 genes predicted to encode a functional 2′,5′-PDE include human coronaviruses OC43 and HEC4408, which cause respiratory and enteric infections, respectively, and bovine coronavirus (BCV), which causes respiratory and enteric infections in cattle and wild ruminants (Saif 2010). Another member of the coronavirus superfamily, the equine torovirus, Berne virus, which causes gastrointestinal disease, expresses a predicted 2′,5′-PDE in its pp1a C-terminal protein (CTP) (Mazumder and others 2002; Snijder and others 2003). SARS-CoV and MERS-CoV, both human coronaviruses causing severe respiratory infections, lack an ns2 homolog (Snijder and others 2003; Weiss and Navas-Martin 2005; Scobey and others 2013). However, based on the finding that porcine coronavirus transmissible gastroenteritis virus (TGEV) also lack an ns2 homolog and inhibits RNase L by a mechanism distinct from PDE (Cruz and others 2011), as discussed below, it is tempting to speculate that these pathogenic human coronaviruses may have alternative strategies for evading RNase L. Alignments of VP3 from different human and animal group A rotaviruses showed that all encode a similar PDE domain (Zhang and others 2013). In addition, the group B and G rotaviruses, but not group C, F, and H strains, also appear to encode a PDE in the CTD of VP3 (Zhang and others 2013).
Table 1.
Viral 2′,5′-Phosphodiesterases
| Viral (2′,5′-PDE) | Pathogenesis |
|---|---|
| Coronavirus (murine MHV A59 ns2) (Zhao and others 2012) (confirmed) | Hepatitis, encephalitis, gastrointestinal (strain dependent) |
| Coronavirus (human OC43 ns2) (Mazumder and others 2002; Snijder and others 2003) (predicted) | Respiratory |
| Coronavirus (human HEC4408 ns2) (Han and others 2006) (predicted) | Gastrointestinal |
| Coronavirus (bovine BCV ns2) (Mazumder and others 2002; Snijder and others 2003) (predicted) | Respiratory, gastrointestinal |
| Torovirus (equine) (pp1a CTP) (Mazumder and others 2002; Snijder and others 2003) (predicted) | Gastrointestinal |
| Rotavirus, group A (human, simian, avian VP3 CTD) (Zhang and others 2013) (confirmed) | Gastrointestinal |
| Rotavirus, groups B & G (VP3-CTD) (predicted) | Gastrointestinal |
BCV, bovine coronavirus; CTD, C-terminal domain; CTP, C-terminal protein; PDE, phosphodiesterase; MHV, mouse hepatitis virus.
Alternative Viral Mechanisms for Antagonizing OAS/RNase L
OAS/RNase L poses a threat to a wide range of viruses. Accordingly, many viruses have evolved mechanisms for blocking this pathway. These viral evasion strategies include, but are not limited to, viral proteins with 2′,5′-PDE activity. For example, the alphacoronavirus TGEV encodes the gene 7 protein that suppresses RNase L activity, possibly by inducing dephosphorylation of 2-5A through its interaction with protein phosphatase 1 (PP1) (Cruz and others 2011). The L* protein of the neurotropic picornavirus, Theiler's virus, promotes viral persistence and demyelinating disease in mice (van Eyll and Michiels 2002). L* directly binds to the ankyrin repeat domain of mouse RNase L inhibiting its activity in a species-specific manner (Sorgeloos and others 2013). A phylogenetically conserved region in the RNA of poliovirus and other group C enteroviruses, called ciRNA, binds to and competitively inhibits RNase L (Townsend and others 2008a, 2008b; Keel and others 2012). Viral proteins containing dsRNA-binding domains, including influenza A virus ns1 and vaccinia virus E3L, sequester dsRNA from OAS thus preventing 2-5A synthesis and subsequent RNase L activation (Beattie and others 1995; Rivas and others 1998; Xiang and others 2002; Min and Krug 2006). Some DNA viruses (vaccinia virus, herpes simplex virus I and II, and SV40) induce the synthesis of alternative 2-5A-like molecules that either inhibit or fail to activate RNase L (Cayley and others 1984; Hersh and others 1984; Rice and others 1984, 1985). There can be little doubt that other viral mechanisms for antagonizing the OAS/RNase L pathway remain to be discovered.
Candidate Cellular Enzymes That Degrade 2-5A and Limit Damage to the Host RNA Caused by RNase L
Cellular enzymes that degrade 2-5A perform an important function by preventing perpetual activation of RNase L that would otherwise lead to extensive RNA damage and tissue injury. 2-5A has long been known to be unstable in cell extracts, and its half-life is typically measured in minutes (Williams and others 1978; Torrence and others 1983). Two types of enzymes are implicated: (1) 2′,5′ PDEs (similar to viral ns2 and VP3) that sequentially release 5′-AMP moieties from the 2′,3′-terminus of 2-5A, and (2) phosphatases that remove the 5′-phosphoryl groups that are required for RNase L activation (Schmidt and others 1979; Johnston and Hearl 1987; Trujillo and others 1988). While PDE, and not phosphatase, activity have been suggested to be the predominant mechanism for degrading 2-5A (Williams and others 1978; Trujillo and others 1988), their relative importance might vary according to the cell type. Three PDEs are candidates for the regulation of 2-5A turnover in cells: PDE12 (2′-PDE) (Kubota and others 2004), ENPP1 (Poulsen and others 2012), and AKAP7 (also known as AKAP15 or 18) (Zhang and others 2013). Of these, only AKAP7 is a member of the 2H phosphoesterase family with conserved sequences and predicted structural homology with both ns2 and VP3 (Fig. 2B) (Gold and others 2008; Roth-Cross and others 2009; Zhang and others 2013). PDE12, a member of the endonuclease/exonuclease/phosphatase family of deadenylases, has both 3′,5′- and 2′,5′-PDE activities and localizes to the mitochondria matrix where it removes 3′,5′-poly(rA) tails from and modulates stability of some mitochondrial mRNAs (Kubota and others 2004; Poulsen and others 2011; Rorbach and others 2011). Downregulation of PDE12 (using siRNA), or inhibition with a small molecule (A-74528a) had only a modest (2-fold) antiviral effect against vaccinia virus (Kubota and others 2004). ENPP1 degrades 3′,5′-linked RNA, DNA, 2′,3′-cAMP, and pyrophosphate linkages of nucleotides and nucleotide sugars, in addition to 2-5A, and its catalytic domain is extracellular (Poulsen and others 2012). Therefore, neither PDE12 nor ENPP1 co-localize with replicating viruses.
The AKAPs are a family of 43 proteins that bind the R subunits of PKA and function as scaffolds that localize and regulate cAMP signaling during a range of physiological processes, including cardiac excitation–contraction, renal homeostasis, neuronal synaptic plasticity, and sperm motility [reviewed in Mauban and others (2009), McConnachie and others (2006), Welch and others (2010)]. However, only AKAP7 has a central domain with predicted structural and sequence homology, including the 2 conserved histidine motifs, with the 2H phosphoesterase family (Mazumder and others 2002; Gold and others 2008). There are 4 splice variants of AKAP7, 2 short forms of 15 and 18 kDa (α and β), which lack the central domain but contain a membrane targeting region and the PKA-binding motif, and 2 long forms of 37 and 42 kDa (γ and δ), which contain from the N- to the C-teminal a nuclear localization signal, the central domain with 2H phosphoesterase homology, and the AKAP helix and leucine zipper domain responsible for binding the R subunits of PKA and also Ca+2 and Na+ channels (Gold and others 2008). Mice homozygous for a deletion of AKAP7 exon 7, encoding the leucine zipper domain and the 3′-UTR, were phenotypically normal (Jones and others 2012). Moreover, β-adrenergic stimulation of the calcium current in adult ventricular cardiomyocytes was also normal, and therefore, it was suggested that another AKAP isoforms might be responsible for this function in mouse cardiomyocytes (Jones and others 2012). We recently determined that AKAP7γ has 2′,5′-PDE activity that degrades 2-5A in vitro (Gusho, E., Zhang, R., Jha, B.K., Dong, B., Thornbrough, J.M., Gaughan, C., Elliott, R., S.R.W., and R.H.S., unpublished). AKAP7 resides in the nucleus (Brown and others 2003) and would presumably have access to 2-5A synthesized either there or in the cytoplasm where it could diffuse into the nucleus through the nuclear pores. However, 2′,5′-PDE activity in the nucleus may be insufficient to reduce 2-5A levels in the cytoplasm where many RNA viruses replicate. Direct evidence that these 3 enzymes (PDE12, ENPP1, and AKAP7) degrade 2-5A during viral infections has yet to be reported and alternative or additional functions are likely. A possible function of host enzymes that degrade 2-5A is to limit the damage to cellular RNA following viral clearance. In addition, some studies have suggested a tumor suppressor function for RNase L, making it possible that enzymes that degrade 2-5A might regulate tumorigenesis (Carpten and others 2002; Kruger and others 2005; Madsen and others 2008).
Concluding Remarks
Coronavirus ns2 and rotavirus VP3 (and possibly related proteins in other RNA viruses) block dsRNA signaling to RNase L by degrading 2-5A and represent a newly recognized viral strategy for evading host innate immunity. Coronaviruses and rotaviruses are unrelated RNA viruses suggesting that these viruses independently acquired a 2′,5′-PDE gene from the host, possibly AKAP7. 2-5A stability is a critical control point in the OAS/RNase L antiviral pathway that these viruses have exploited (Fig. 1). Activation of RNase L can depend on basal levels of OAS expression, which vary among cell and tissue types, and virus encoded PDEs can antagonize this activation. The balance between activation and antagonism of RNase L can determine the extent of viral success. In this regard, in the case of MHV-induced hepatitis viral antagonism of RNase L is a striking determinant of virulence. Future studies will be directed at investigating activation/antagonism of RNase L in other organs systems including the lung and the gastrointestinal tract, 2 targets of human coronavirus, and rotavirus infections. Further understanding of the competition between virus and the OAS/RNase L pathway may also aid in the development or refinement of therapeutic strategies against viral diseases. For example, specific inhibitors of the viral 2′,5′-PDEs could function as antiviral agents by restoring the virus-induced accumulation of 2-5A specifically in virus-infected cells with little effect on uninfected cells.
Acknowledgments
We thank Carolina B. Lopez, Judith M. Phillips, and Joshua Thornbrough (Philadelphia) and Babal Kant Jha and Elona Gusho (Cleveland) for comments made during writing of the article, and to David Schumick, Center for Medical Art and Photography, Cleveland Clinic for expert art design. This work was supported by National Institutes of Health Grants RO1 AI104887 (to R.H.S. and S.R.W.), RO1 CA44059 (to R.H.S.), and RO1 NS081001 (to S.R.W.).
Author Disclosure Statement
No competing financial interests exist.
References
- Arn EA, Abelson JN. 1996. The 2′-5′ RNA ligase of Escherichia coli. Purification, cloning, and genomic disruption. J Biol Chem 271:31145–31153 [DOI] [PubMed] [Google Scholar]
- Arnold MM, Sen A, Greenberg HB, Patton JT. 2013. The battle between rotavirus and its host for control of the interferon signaling pathway. PLoS Pathog 9:e1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Chakrabarti A, Jha BK, Weiss SR, Silverman RH. 2014. Cell-type specific effects of RNase L on viral induction of beta interferon. Mbio 5:e00856-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie E, Denzler KL, Tartaglia J, Perkus ME, Paoletti E, Jacobs BL. 1995. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J Virol 69:499–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benech P, Mory Y, Revel M, Chebath J. 1985. Structure of two forms of the interferon-induced (2′-5′) oligo A synthetase of human cells based on cDNAs and gene sequences. EMBO J 4:2249–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek PJ, Pachuk CJ, Noten AF, Charite J, Luytjes W, Weiss SR, Spaan WJ. 1990. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res 18:1825–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, August SL, Williams CJ, Moss SB. 2003. AKAP7gamma is a nuclear RI-binding AKAP. Biochem Biophys Res Commun 306:394–401 [DOI] [PubMed] [Google Scholar]
- Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, Faruque M, Moses T, Ewing C, Gillanders E, Hu P, Bujnovszky P, Makalowska I, Baffoe-Bonnie A, Faith D, Smith J, Stephan D, Wiley K, Brownstein M, Gildea D, Kelly B, Jenkins R, Hostetter G, Matikainen M, Schleutker J, Klinger K, Connors T, Xiang Y, Wang Z, De Marzo A, Papadopoulos N, Kallioniemi OP, Burk R, Meyers D, Gronberg H, Meltzer P, Silverman R, Bailey-Wilson J, Walsh P, Isaacs W, Trent J. 2002. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet 30:181–184 [DOI] [PubMed] [Google Scholar]
- Castelli JC, Hassel BA, Maran A, Paranjape J, Hewitt JA, Li XL, Hsu YT, Silverman RH, Youle RJ. 1998. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ 5:313–320 [DOI] [PubMed] [Google Scholar]
- Castelli JC, Hassel BA, Wood KA, Li XL, Amemiya K, Dalakas MC, Torrence PF, Youle RJ. 1997. A study of the interferon antiviral mechanism: apoptosis activation by the 2-5A system. J Exp Med 186:967–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayley PJ, Davies JA, McCullagh KG, Kerr IM. 1984. Activation of the ppp(A2′p)nA system in interferon-treated, herpes simplex virus-infected cells and evidence for novel inhibitors of the ppp(A2′p)nA-dependent RNase. Eur J Biochem 143:165–174 [DOI] [PubMed] [Google Scholar]
- Chakrabarti A, Ghosh PK, Banerjee S, Gaughan C, Silverman RH. 2012. RNase L triggers autophagy in response to viral infections. J Virol 86:11311–11321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby C, Duesberg PH. 1969. Double-stranded RNA in vaccinia virus infected cells. Nature 222:940–944 [DOI] [PubMed] [Google Scholar]
- Courtney SC, Di H, Stockman BM, Liu H, Scherbik SV, Brinton MA. 2012. Identification of novel host cell binding partners of Oas1b, the protein conferring resistance to flavivirus-induced disease in mice. J Virol 86:7953–7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JL, Sola I, Becares M, Alberca B, Plana J, Enjuanes L, Zuniga S. 2011. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog 7:e1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Silverman RH. 1995. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem 270:4133–4137 [DOI] [PubMed] [Google Scholar]
- Donovan J, Dufner M, Korennykh A. 2013. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci U S A 110:1652–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbahesh H, Jha B., Silverman RH, Scherbik SV, Brinton MA. 2010. The Flvr-encoded murine oligoadenylate synthetase 1b (Oas1b) suppresses 2-5A synthesis in intact cells. Virology 409(2):262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MG, Smith FD, Scott JD, Barford D. 2008. AKAP18 contains a phosphoesterase domain that binds AMP. J Mol Biol 375:1329–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JQ, Barton DJ. 2002. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA 8:512–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MG, Cheon DS, Zhang X, Saif LJ. 2006. Cross-protection against a human enteric coronavirus and a virulent bovine enteric coronavirus in gnotobiotic calves. J Virol 80:12350–12356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R, Olsen HS, Widder S, Jorgensen R, Justesen J. 1998. p59OASL, a 2′-5′ oligoadenylate synthetase like protein: a novel human gene related to the 2′-5′ oligoadenylate synthetase family. Nucleic Acids Res 26:4121–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassel BA, Zhou A, Sotomayor C, Maran A, Silverman RH. 1993. A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J 12:3297–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh CL, Brown RE, Roberts WK, Swyryd EA, Kerr IM, Stark GR. 1984. Simian virus 40-infected, interferon-treated cells contain 2′,5′-oligoadenylates which do not activate cleavage of RNA. J Biol Chem 259:1731–1737 [PubMed] [Google Scholar]
- Hoshino Y, Saif LJ, Kang SY, Sereno MM, Chen WK, Kapikian AZ. 1995. Identification of group A rotavirus genes associated with virulence of a porcine rotavirus and host range restriction of a human rotavirus in the gnotobiotic piglet model. Virology 209:274–280 [DOI] [PubMed] [Google Scholar]
- Huang H, Zeqiraj E, Dong B, Jha BK, Duffy N, Orlicky S, Thevakumaran M, Pillon MC, Ceccarelli DF, Wan L, Juang YC, Mao DYL, Gaughan C, Brinton MA, Perelygin AA, Kourinov I, Guarne A, Silverman RH, Sicheri F. 2014. Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon induced antiviral activity. Mol Cell 53(2):221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen H, Czarniecki CW, Krause D, Friedman RM, Silverman RH. 1983. Interferon-induced synthesis of 2-5A-dependent RNase in mouse JLS-V9R cells. Virology 125:496–501 [DOI] [PubMed] [Google Scholar]
- Johnston MI, Hearl WG. 1987. Purification and characterization of a 2′-phosphodiesterase from bovine spleen. J Biol Chem 262:8377–8382 [PubMed] [Google Scholar]
- Jones BW, Brunet S, Gilbert ML, Nichols CB, Su T, Westenbroek RE, Scott JD, Catterall WA, McKnight GS. 2012. Cardiomyocytes from AKAP7 knockout mice respond normally to adrenergic stimulation. Proc Natl Acad Sci U S A 109:17099–17104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justesen J, Hartmann R, Kjeldgaard NO. 2000. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci 57:1593–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta S, Shibata S, Iwakura Y. 2002. Genomic structure of the mouse 2′,5′-oligoadenylate synthetase gene family. J Interferon Cytokine Res 22:981–993 [DOI] [PubMed] [Google Scholar]
- Keel AY, Jha BK, Kieft JS. 2012. Structural architecture of an RNA that competitively inhibits RNase L. RNA 18:88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr IM, Brown RE. 1978. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci U S A 75:256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe DM, Howley PM. (eds). 2007. Fields virology, 5th ed. Philadelphia, PA: Lippincott, Williams & Wilkins [Google Scholar]
- Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. 2011. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res 31:41–47 [DOI] [PubMed] [Google Scholar]
- Kruger S, Silber AS, Engel C, Gorgens H, Mangold E, Pagenstecher C, Holinski-Feder E, von Knebel Doeberitz M, Moeslein G, Dietmaier W, Stemmler S, Friedl W, Ruschoff J, Schackert HK. 2005. Arg462Gln sequence variation in the prostate-cancer-susceptibility gene RNASEL and age of onset of hereditary non-polyposis colorectal cancer: a case-control study. Lancet Oncol 6:566–572 [DOI] [PubMed] [Google Scholar]
- Kubota K, Nakahara K, Ohtsuka T, Yoshida S, Kawaguchi J, Fujita Y, Ozeki Y, Hara A, Yoshimura C, Furukawa H, Haruyama H, Ichikawa K, Yamashita M, Matsuoka T, Iijima Y. 2004. Identification of 2′-phosphodiesterase, which plays a role in the 2-5A system regulated by interferon. J Biol Chem 279:37832–37841 [DOI] [PubMed] [Google Scholar]
- Lee MS, Kim B, Oh GT, Kim YJ. 2013. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat Immunol 14:346–355 [DOI] [PubMed] [Google Scholar]
- Madsen BE, Ramos EM, Boulard M, Duda K, Overgaard J, Nordsmark M, Wiuf C, Hansen LL. 2008. Germline mutation in RNASEL predicts increased risk of head and neck, uterine cervix and breast cancer. PLoS One 3:e2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra RK, McMillan NA, Desai S, McSwiggen J, Hovanessian AG, Sen G, Williams BR, Silverman RH. 1994. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology 204:823–827 [DOI] [PubMed] [Google Scholar]
- Malathi K, Dong B, Gale M, Jr., Silverman RH. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448:816–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Saito T, Crochet N, Barton DJ, Gale M, Jr., Silverman RH. 2010. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA 16:2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie I, Blanco J, Rebouillat D, Hovanessian AG. 1997. 69-kDa and 100-kDa isoforms of interferon-induced (2′-5′)oligoadenylate synthetase exhibit differential catalytic parameters. Eur J Biochem 248:558–566 [DOI] [PubMed] [Google Scholar]
- Marie I, Hovanessian AG. 1992. The 69-kDa 2-5A synthetase is composed of two homologous and adjacent functional domains. J Biol Chem 267:9933–9939 [PubMed] [Google Scholar]
- Marques J, Anwar J, Eskildsen-Larsen S, Rebouillat D, Paludan SR, Sen G, Williams BR, Hartmann R. 2008. The p59 oligoadenylate synthetase-like protein possesses antiviral activity that requires the C-terminal ubiquitin-like domain. J Gen Virol 89:2767–2772 [DOI] [PubMed] [Google Scholar]
- Mashimo T, Glaser P, Lucas M, Simon-Chazottes D, Ceccaldi PE, Montagutelli X, Despres P, Guenet JL. 2003. Structural and functional genomics and evolutionary relationships in the cluster of genes encoding murine 2′,5′-oligoadenylate synthetases. Genomics 82:537–552 [DOI] [PubMed] [Google Scholar]
- Mashimo T, Lucas M, Simon-Chazottes D, Frenkiel MP, Montagutelli X, Ceccaldi PE, Deubel V, Guenet JL, Despres P. 2002. A nonsense mutation in the gene encoding 2′-5′-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc Natl Acad Sci U S A 99:11311–11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauban JR, O'Donnell M, Warrier S, Manni S, Bond M. 2009. AKAP-scaffolding proteins and regulation of cardiac physiology. Physiology (Bethesda) 24:78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder R, Iyer LM, Vasudevan S, Aravind L. 2002. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res 30:5229–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnachie G, Langeberg LK, Scott JD. 2006. AKAP signaling complexes: getting to the heart of the matter. Trends Mol Med 12:317–323 [DOI] [PubMed] [Google Scholar]
- Melchjorsen J, Kristiansen H, Christiansen R, Rintahaka J, Matikainen S, Paludan SR, Hartmann R. 2009. Differential regulation of the OASL and OAS1 genes in response to viral infections. J Interferon Cytokine Res 29:199–207 [DOI] [PubMed] [Google Scholar]
- Min JY, Krug RM. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci U S A 103:7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JT. 2012. Rotavirus diversity and evolution in the post-vaccine world. Discov Med 13:85–97 [PMC free article] [PubMed] [Google Scholar]
- Perelygin AA, Scherbik SV, Zhulin IB, Stockman BM, Li Y, Brinton MA. 2002. Positional cloning of the murine flavivirus resistance gene. Proc Natl Acad Sci U S A 99:9322–9327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelygin AA, Zharkikh AA, Scherbik SV, Brinton MA. 2006. The mammalian 2′-5′ oligoadenylate synthetase gene family: evidence for concerted evolution of paralogous Oas1 genes in Rodentia and Artiodactyla. J Mol Evol 63:562–576 [DOI] [PubMed] [Google Scholar]
- Poulsen JB, Andersen KR, Kjaer KH, Durand F, Faou P, Vestergaard AL, Talbo GH, Hoogenraad N, Brodersen DE, Justesen J, Martensen PM. 2011. Human 2′-phosphodiesterase localizes to the mitochondrial matrix with a putative function in mitochondrial RNA turnover. Nucleic Acids Res 39:3754–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen JB, Andersen KR, Kjaer KH, Vestergaard AL, Justesen J, Martensen PM. 2012. Characterization of human phosphodiesterase 12 and identification of a novel 2′-5′ oligoadenylate nuclease—the ectonucleotide pyrophosphatase/phosphodiesterase 1. Biochimie 94:1098–1107 [DOI] [PubMed] [Google Scholar]
- Rebouillat D, Hovnanian A, Marie I, Hovanessian AG. 1999. The 100-kDa 2′,5′-oligoadenylate synthetase catalyzing preferentially the synthesis of dimeric pppA2′p5′A molecules is composed of three homologous domains. J Biol Chem 274:1557–1565 [DOI] [PubMed] [Google Scholar]
- Rebouillat D, Marie I, Hovanessian AG. 1998. Molecular cloning and characterization of two related and interferon-induced 56-kDa and 30-kDa proteins highly similar to 2′-5′ oligoadenylate synthetase. Eur J Biochem 257:319–330 [DOI] [PubMed] [Google Scholar]
- Rice AP, Kerr SM, Roberts WK, Brown RE, Kerr IM. 1985. Novel 2′,5′-oligoadenylates synthesized in interferon-treated, vaccinia virus-infected cells. J Virol 56:1041–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice AP, Roberts WK, Kerr IM. 1984. 2-5A accumulates to high levels in interferon-treated, vaccinia virus-infected cells in the absence of any inhibition of virus replication. J Virol 50:220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas C, Gil J, Melkova Z, Esteban M, Diaz-Guerra M. 1998. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2-5A synthetase enzyme. Virology 243:406–414 [DOI] [PubMed] [Google Scholar]
- Rorbach J, Nicholls TJ, Minczuk M. 2011. PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucleic Acids Res 39:7750–7763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross JK, Stokes H, Chang G, Chua MM, Thiel V, Weiss SR, Gorbalenya AE, Siddell SG. 2009. Organ-specific attenuation of murine hepatitis virus strain A59 by replacement of catalytic residues in the putative viral cyclic phosphodiesterase ns2. J Virol 83:3743–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif LJ. 2010. Bovine respiratory coronavirus. The Veterinary clinics of North America. Food Anim Pract 26:349–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar SN, Bandyopadhyay S, Ghosh A, Sen GC. 1999. Enzymatic characteristics of recombinant medium isozyme of 2′-5′ oligoadenylate synthetase. J Biol Chem 274:1848–1855 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Chernajovsky Y, Shulman L, Federman P, Berissi H, Revel M. 1979. An interferon-induced phosphodiesterase degrading (2′-5′) oligoisoadenylate and the C-C-A terminus of tRNA. Proc Natl Acad Sci U S A 76:4788–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz B, Routledge E, Siddell SG. 1990. Murine coronavirus nonstructural protein ns2 is not essential for virus replication in transformed cells. J Virol 64:4784–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, Graham RL, Swanstrom J, Bove PF, Kim JD, Grego S, Randell SH, Baric RS. 2013. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 110:16157–16162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui MA, Malathi K. 2012. RNase L induces autophagy via c-Jun N-terminal kinase and double-stranded RNA-dependent protein kinase signaling pathways. J Biol Chem 287:43651–43664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RH. 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 81:12720–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM. 1983. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J Virol 46:1051–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331:991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgeloos F, Jha BK, Silverman RH, Michiels T. 2013. Evasion of antiviral innate immunity by Theiler's virus L* protein through direct inhibition of RNase L. PLoS Pathog 9:e1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrence PF, Imai J, Johnston MI. 1983. Assay of 2′,5′-oligoadenylate phosphodiesterase activity in mouse L-cell extracts. Anal Biochem 129:103–110 [DOI] [PubMed] [Google Scholar]
- Townsend HL, Jha BK, Han JQ, Maluf NK, Silverman RH, Barton DJ. 2008a. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA 14:1026–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend HL, Jha BK, Silverman RH, Barton DJ. 2008b. A putative loop E motif and an H-H kissing loop interaction are conserved and functional features in a group C enterovirus RNA that inhibits ribonuclease L. RNA Biol 5:263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo MA, Barbet J, Cailla HL. 1988. Mechanisms of degradation of 2′-5′ oligoadenylates. Biochimie 70:1733–1744 [DOI] [PubMed] [Google Scholar]
- van Eyll O, Michiels T. 2002. Non-AUG-initiated internal translation of the L* protein of Theiler's virus and importance of this protein for viral persistence. J Virol 76:10665–10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Donnelly B, Bondoc A, Mohanty SK, McNeal M, Ward R, Sestak K, Zheng S, Tiao G. 2011. The rhesus rotavirus gene encoding VP4 is a major determinant in the pathogenesis of biliary atresia in newborn mice. J Virol 85:9069–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SR, Navas-Martin S. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev 69:635–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch EJ, Jones BW, Scott JD. 2010. Networking with AKAPs: context-dependent regulation of anchored enzymes. Mol Interv 10:86–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C, Gale M., Jr.2010. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol 22:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Kerr IM, Gilbert CS, White CN, Ball LA. 1978. Synthesis and breakdown of pppA2′p5′A2′p5′A and transient inhibiton of protein synthesis in extracts from interferon-treated and control cells. Eur J Biochem 92:455–462 [DOI] [PubMed] [Google Scholar]
- Wreschner DH, James TC, Silverman RH, Kerr IM. 1981a. Ribosomal RNA cleavage, nuclease activation and 2-5A(ppp(A2′p)nA) in interferon-treated cells. Nucleic Acids Res 9:1571–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. 1981b. Interferon action—sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature 289:414–417 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Condit RC, Vijaysri S, Jacobs B, Williams BR, Silverman RH. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J Virol 76:5251–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Ma L, Stein P, Pangas SA, Burns KH, Bai Y, Schultz RM, Matzuk MM. 2005. Mice deficient in oocyte-specific oligoadenylate synthetase-like protein OAS1D display reduced fertility. Mol Cell Biol 25:4615–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Jha BK, Ogden KM, Dong B, Zhao L, Elliott R, Patton JT, Silverman RH, Weiss SR. 2013. Homologous 2′,5′-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc Natl Acad Sci U S A 110:13114–13119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Birdwell LD, Wu A, Elliott R, Rose KM, Phillips JM, Li Y, Grinspan J, Silverman RH, Weiss SR. 2013. Cell-type-specific activation of the oligoadenylate synthetase-RNase L pathway by a murine coronavirus. J Virol 87:8408–8418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. 2012. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11:607–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Rose KM, Elliott R, Van Rooijen N, Weiss SR. 2011. Cell-type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J Virol 85:10058–10068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Hassel BA, Silverman RH. 1993. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72:753–765 [DOI] [PubMed] [Google Scholar]
- Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman RH. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J 16:6355–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltick PW, Leibowitz JL, Oleszak EL, Weiss SR. 1990. Mouse hepatitis virus ORF 2a is expressed in the cytosol of infected mouse fibroblasts. Virology 174:605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]