Abstract
Several animal viruses encode proteins that bind double-stranded RNA (dsRNA) to counteract host dsRNA-dependent antiviral responses. This article discusses the structure and function of the dsRNA-binding proteins of influenza A virus and Ebola viruses (EBOVs).
Introduction
Several host antiviral defenses are activated by double-stranded RNA (dsRNA), including retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA-5), protein kinase R (PKR), and 2′-5′ oligo(A) synthetase (2′-5′ OAS) (Silverman 1994; Gale and Katze 1998; Jiang and others 2011; Kowalinski and others 2011). As one countermeasure against these dsRNA-activated antiviral responses, several animal viruses encode proteins that bind dsRNA. These viruses include both DNA viruses (vaccinia virus) (Chang and Jacobs 1993) and RNA viruses (influenza viruses, reoviruses, and Ebola viruses [EBOVs]) (Imani and Jacobs 1988; Hatada and Fukuda 1992; Lu and others 1995; Chien and others 1997; Liu and others 1997; Basler and others 2003; Hartman and others 2004; Cardenas and others 2006). In this study, the focus will be on the structure and function of the dsRNA-binding proteins encoded by 2 RNA viruses, influenza viruses and EBOVs. These 2 viruses are important human pathogens, and their dsRNA-binding proteins are potential targets for the development of antivirals.
Influenza A Virus NS1 Protein
Influenza A viruses, which cause an annual highly contagious respiratory disease in humans, infect many avian and mammalian species and are responsible for periodic human pandemics that can result in high mortality rates (Wright and others 2012). Influenza A viruses are enveloped RNA viruses that contain 8 single-stranded, negative-sense genome segments encoding 13 or 14 proteins (Wise and others 2012; Krug and Fodor 2013). The smallest genome segment encodes the nonstructural protein 1 (NS1) protein, which is synthesized at high levels in infected cells, but is not incorporated into virus particles. The NS1 proteins of wild-type influenza A viruses range in size from 215 to 237 amino acids long and are comprised of 2 functional domains connected by a short linker: the N-terminal RNA-binding domain (RBD) (amino acids 1-73), which binds dsRNA, and the C-terminal effector domain (ED) (amino acids 85-end) (Krug and Garcia-Sastre 2013) (Fig. 1A).
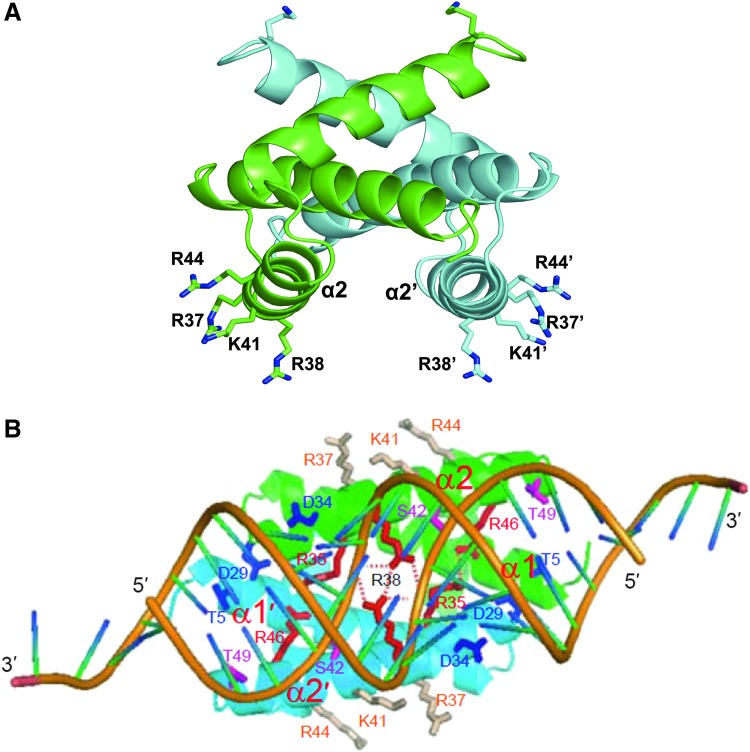
FIG. 1.
Structure of the nonstructural protein 1 (NS1) RNA-binding domain (RBD). (A) Structure of the NS1 RBD in the absence of double-stranded RNA (dsRNA). The α2 and α2' helicases contain the amino acids that bind dsRNA. Generated from PBD accession number 1NS1. (B) Structure of the NS1 RBD in complex with a 19-bp dsRNA. From Cheng and others (2009) with permission.
The 3-dimensional structure of the RBD shows that it forms a unique 6-helical head-to-tail homodimer (Chien and others 1997; Liu and others 1997) (Fig. 1B, top). Only one amino acid (R38/R38’) in the RBD is absolutely required for dsRNA binding (Wang and others 1999). The binding site for dsRNA is in the pocket at the bottom of this structure and involves amino acids in helices α2 and α2′. As shown by the crystal structure of the RBD in complex with a 19-bp dsRNA (Cheng and others 2009), the RBD binds the major groove of A form dsRNA, and recognition is entirely with the dsRNA backbone and not with specific base pairs (Fig. 1B, bottom). The R38/R38′ residues from the 2 monomers form hydrogen bonds with each other, as well as with the 2 RNA strands, thereby anchoring the dsRNA in the protein-binding site. In addition to R38, positively charged residues in the middle of the dsRNA-binding surface, such as R35, R37, and K41 make hydrogen bonds and electrostatic interactions with both strands of the dsRNA. As measured using a 16-bp dsRNA, the isolated RBD has a low affinity for dsRNA (Kd of ∼1 μM) (Chien and others 2004). Higher affinity was observed with longer dsRNAs (Aramini and others 2011), indicating some cooperativity in the binding of NS1 molecules along the lengths of longer dsRNAs. In addition, the full-length NS1 protein was shown to have a 5-fold higher affinity for longer dsRNAs than the isolated RBD, indicating that the ED increases cooperative binding, possibly by enhancing oligomerization of NS1 molecules along the dsRNA chains (Aramini and others 2011; Kerry and others 2011).
Several early studies to determine the roles of NS1-mediated dsRNA binding used transient transfection experiments to overexpress the NS1 protein. These experiments showed that the overexpressed NS1 protein inhibited many cellular dsRNA-dependent activities, including RIG-I-mediated activation of interferon regulatory factor 3 (IRF-3) and NF-kappa B that results in interferon-β (IFN-β) production (Talon and others 2000; Wang and others 2000). In addition, in vitro experiments using purified proteins showed that the NS1 protein inhibited the dsRNA activation of PKR (Lu and others 1995). However, it was subsequently established that these functions do not correspond to the actual function of NS1 protein-mediated dsRNA binding in influenza A virus-infected cells. To determine the role of NS1 dsRNA binding in influenza A virus-infected cells, a recombinant influenza A/Udorn/72 (Ud) virus was generated that expresses an NS1 protein in which R38 was changed to alanine (Min and others 2006). This amino acid substitution also abolishes the nuclear localization signal (NLS) in the RBD (Greenspan and others 1988; Melen and others 2007). However, because the Ud NS1 protein has a second NLS in the ED (Melen and others 2007), the R38A mutant NS1 protein is imported into the nucleus (Min and others 2006). Consequently, defects in the replication of this R38A mutant virus can be attributed largely to the loss of dsRNA-binding activity of the NS1 protein. Neither the PKR activation nor an increase in the IFN-β mRNA production was detected in R38 mutant virus-infected cells. The primary defect was shown to be the inability to block activation of the IFN-induced 2′-5′ OAS/RNAse L pathway (Min and others 2006). Because 2′-5′ OAS has to be activated by dsRNA to activate RNase L (Silverman 2007), this result demonstrated that the primary role of dsRNA binding by the NS1 protein in infected cells is to sequester dsRNA away from 2′-5′ OAS. Interestingly, 2′-5′ OAS has a very lower affinity for dsRNA (Hartmann and others 2003), indicating that the NS1 protein can effectively compete for dsRNA only with cellular proteins with low affinity for dsRNA. The amino acids of the NS1 RBD that participate in dsRNA binding also participate in the binding of several proteins, including α-importin, TRIM25, and the viral nucleoprotein (Melen and others 2007; Gack and others 2009; Robb and others 2011). Because this region of the NS1 RBD is crucial for multiple functions that are required for virus infection, it should be an excellent target for the development of new influenza virus antivirals.
EBOV VP35 Protein
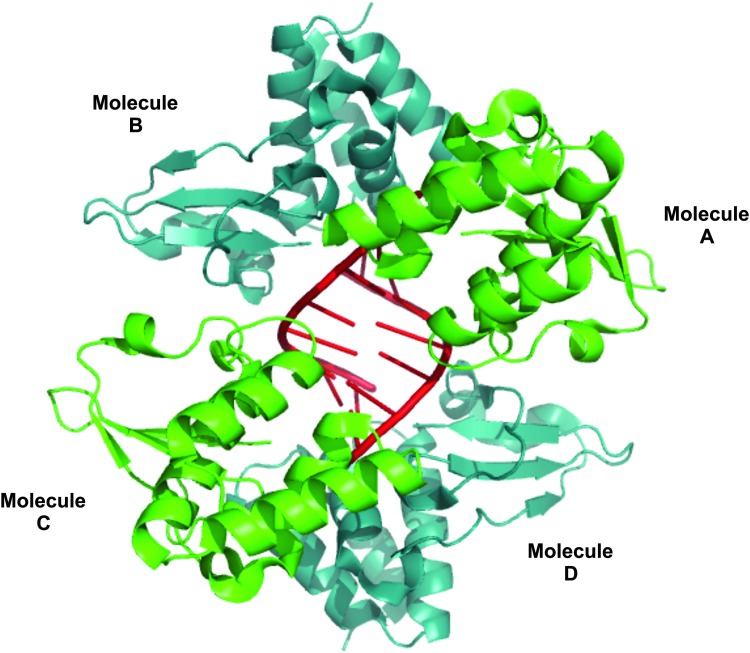
EBOVs are enveloped viruses that contain a nonsegmented, negative-sense single-stranded RNA genome. These viruses cause hemorrhagic fever in humans and nonhuman primates with very high mortality rates (Geisbert and Hensley 2004). The VP35 structural protein, which contains 340 amino acids, is comprised of 2 domains: the N-terminal oligomerization domain (amino acids 1–220); and the C-terminal RBD (amino acids 221–340) (Basler and others 2003; Hartman and others 2004; Cardenas and others 2006). There are 2 different modes of binding of the EBOV VP5 protein and its RBD to dsRNA: binding to the blunt end of a dsRNA, termed end-capping; and binding to the backbone of dsRNA (Kimberlin and others 2010; Leung and others 2010; Hastie and others 2012). In contrast, the influenza A virus NS1 protein binds only to the dsRNA backbone (Fig. 1). The determination of the affinity of EBOV VP35 for dsRNA is complicated because of the 2 modes of binding. Nonetheless, the affinity of the full-length VP35 protein for a 500-bp dsRNA appears to be high (∼3–4 nM) (Zinzula and others 2012), which is probably significantly higher than the affinity of the influenza NS1 protein for dsRNA. Several X-ray crystal structures of VP35 RBDs bound to small dsRNAs have been published (Kimberlin and others 2010; Leung and others 2010). Figure 2 shows the structure of VP35 RBDs bound to an 8-bp blunt end dsRNA (Leung and others 2010). Assymetric RBD dimers wrap around the 2 ends of the dsRNA. Because molecules A and B are equivalent to molecules C and D, respectively, it is appropriate to focus on only one of these 2 dimers, for example, the A-B dimer. Central basic patches in molecules A and B, particularly Arg312 and Arg322, mediate 2 different intermolecular interactions. Molecules A and B interact through hydrogen bonds that include Arg312 and Arg322 of molecule A. These 2 Args in molecule B, the end-capping RBD, have a different function: they form direct contacts with the phosphodiester backbone near the end of dsRNA. Molecule B also interacts with the end of the dsRNA through hydrophobic residues. In contrast to molecule B, amino acids in molecule A other than those centered around Arg312 and Arg322 directly contact the dsRNA backbone. Another study indicates that other VP35 molecules coat the backbone of longer dsRNAs (Bale and others 2013). Recognition of dsRNA molecules by the V35 RBD is entirely with the dsRNA backbone and not with specific base pairs (Kimberlin and others 2010; Leung and others 2010), as is also the case for the influenza A virus NS1 protein.
FIG. 2.
Structure of the Ebola virus (EBOV) VP35 RBD in complex with an 8-bp dsRNA. Generated from PBD accession number 3L25.
The strong binding of VP35 to the blunt end and backbone of dsRNA affords protection against cellular dsRNA sensors like RIG-I and MDA-5 that recognize both the terminus and backbone of dsRNA (Kimberlin and others 2010; Leung and others 2010; Jiang and others 2011; Kowalinski and others 2011; Hastie and others 2012). This protection was documented in EBOV-infected cells. EBOV recombinant viruses were generated to encode a VP35 protein containing a mutation in the basic stretch of the RBD that eliminates dsRNA binding, for example, Arg312 mutated to alanine (Hartman and others 2008a, 2008b; Prins and others 2010). These viruses were attenuated and did not block RIG-I-mediated activation of IRF-3 and IFN production. In fact, these recombinant viruses were avirulent in mice and guinea pigs (Hartman and others 2008a; Prins and others 2010), indicating that the inhibition of this antiviral response is required for virulence. Consequently, small molecules that inhibit VP35 binding to dsRNA, including the VP35 dimerization that is required for the 2 modes of dsRNA binding, would be expected to be potent antivirals against lethal EBOV infections.
Concluding Remarks
Influenza A viruses and EBOVs encode dsRNA-binding proteins, namely, the NS1 and VP50 proteins, respectively. These 2 viral dsRNA-binding proteins differ in the mode of dsRNA binding, and in the role of their dsRNA binding in combating host antiviral defenses in infected cells. The influenza A virus NS1 protein binds with relatively low affinity to the backbone of dsRNA and apparently inhibits the function of only one dsRNA-binding protein in infected cells, the IFN-induced 2′-5′ OAS enzyme, which has a low affinity for dsRNA (Min and others 2006). In contrast, the EBOV VP35 binds with higher affinity to both the blunt end and backbone of dsRNA, and this dsRNA binding is responsible for the suppression of RIG-I-mediated activation of IRF-3 and IFN production in infected cells (Hartman and others 2008a, 2008b; Prins and others 2010).
Acknowledgment
Research in the author's laboratory is supported by NIH grant AI 11772.
Author Disclosure Statement
No competing financial interests exist.
References
- Aramini JM, Ma LC, Zhou L, Schauder CM, Hamilton K, Amer BR, Mack TR, Lee HW, Ciccosanti CT, Zhao L, et al. 2011. Dimer interface of the effector domain of non-structural protein 1 from influenza A virus: an interface with multiple functions. J Biol Chem 286:26050–26060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S, Julien JP, Bornholdt ZA, Krois AS, Wilson IA, Saphire EO. 2013. Ebolavirus VP35 coats the backbone of double-stranded RNA for interferon antagonism. J Virol 87:10385–10388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Muhlberger E, Bray M, Klenk HD, Palese P, Garcia-Sastre A. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol 77:7945–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas WB, Loo YM, Gale M, Jr., Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol 80:5168–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW, Jacobs BL. 1993. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology 194:537–547 [DOI] [PubMed] [Google Scholar]
- Cheng A, Wong SM, Yuan YA. 2009. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res 19:187–195 [DOI] [PubMed] [Google Scholar]
- Chien CY, Tejero R, Huang Y, Zimmerman DE, Rios CB, Krug RM, Montelione GT. 1997. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat Struct Biol 4:891–895 [DOI] [PubMed] [Google Scholar]
- Chien CY, Xu Y, Xiao R, Aramini JM, Sahasrabudhe PV, Krug RM, Montelione GT. 2004. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry 43:1950–1962 [DOI] [PubMed] [Google Scholar]
- Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. (2009). Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M, Jr., Katze MG. 1998. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol Ther 78:29–46 [DOI] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE. 2004. Ebola virus: new insights into disease aetiopathology and possible therapeutic interventions. Expert Rev Mol Med 6:1–24 [DOI] [PubMed] [Google Scholar]
- Greenspan D, Palese P, Krystal M. 1988. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J Virol 62:3020–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Bird BH, Towner JS, Antoniadou ZA, Zaki SR, Nichol ST. 2008a. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of ebola virus. J Virol 82:2699–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Ling L, Nichol ST, Hibberd ML. 2008b. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J Virol 82:5348–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Towner JS, Nichol ST. 2004. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology 328:177–184 [DOI] [PubMed] [Google Scholar]
- Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. 2003. Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase. Mol Cell 12:1173–1185 [DOI] [PubMed] [Google Scholar]
- Hastie KM, Bale S, Kimberlin CR, Saphire EO. 2012. Hiding the evidence: two strategies for innate immune evasion by hemorrhagic fever viruses. Curr Opin Virol 2:151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatada E, Fukuda R. 1992. Binding of influenza A virus NS1 protein to dsRNA in vitro. J Gen Virol 73:3325–3329 [DOI] [PubMed] [Google Scholar]
- Imani F, Jacobs BL. 1988. Inhibitory activity of the interferon-induced protein kinase is associated with the reovirus serotype 1 s3 protein. Proc Natl Acad Sci U S A 88:7887–7891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr., Patel SS, Marcotrigiano J. 2011. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479:423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerry PS, Ayllon J, Taylor MA, Hass C, Lewis A, Garcia-Sastre A, Randall RE, Hale BG, Russell RJ. 2011. A transient homotypic interaction model for the influenza A virus NS1 protein effector domain. PLoS One 6:e17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin CR, Bornholdt ZA, Li S, Woods VL, Jr., MacRae IJ, Saphire EO. 2010. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc Natl Acad Sci U S A 107:314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. 2011. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147:423–435 [DOI] [PubMed] [Google Scholar]
- Krug RM, Fodor E. 2013. The virus genome and its replication. In: Webster RG, Braciale TJ, Lamb RA, editors. Influenza textbook, 2nd ed. Hoboken (NJ): Wiley-Blackwell, p 57–66 [Google Scholar]
- Krug RM, Garcia-Sastre A. 2013. The NS1 protein: a master regulator of host and viral functions. In: Webster RG, Braciale TJ, Lamb RA, editors. Influenza textbook, 2nd ed. Hoboken (NJ): Wiley-Blackwell, p 114–132 [Google Scholar]
- Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, Nix JC, Helgeson LA, Otwinowski Z, Honzatko RB, et al. 2010. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol 17:165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lynch PA, Chien CY, Montelione GT, Krug RM, Berman HM. 1997. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol 4:896–899 [DOI] [PubMed] [Google Scholar]
- Lu Y, Wambach M, Katze MG, Krug RM. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222–228 [DOI] [PubMed] [Google Scholar]
- Melen K, Kinnunen L, Fagerlund R, Ikonen N, Twu KY, Krug RM, Julkunen I. 2007. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J Virol 81:5995–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J-Y, Krug RM. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2'-5' OAS/RNase L pathway. Proc Natl Acad Sci U S A 103:7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins KC, Delpeut S, Leung DW, Reynard O, Volchkova VA, Reid SP, Ramanan P, Cardenas WB, Amarasinghe GK, Volchkov VE, et al. 2010. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J Virol 84:3004–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb NC, Chase G, Bier K, Vreede FT, Shaw PC, Naffakh N, Schwemmle M, Fodor E. 2011. The influenza A virus NS1 protein interacts with the nucleoprotein of viral ribonucleoprotein complexes. J Virol 85:5228–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RH. 1994. Fascination with 2-5A-dependent RNase: a unique enzyme that functions in interferon action. J Interferon Res 14:101–104 [DOI] [PubMed] [Google Scholar]
- Silverman RH. 2007. Viral encounters with 2',5'-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 81:12720–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol 74:7989–7996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Riedel K, Lynch K, Chien C, Montelione GT, Krug RM. 1999. RNA-binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, Garcia-Sastre A. 2000. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol 74:11566–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, et al. 2012. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog 8:e1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P, Neumann G, Kawaoka 2012. Orthomyxoviruses. In: Knipe DM, Howley PM, editors. Fields virology, 6th ed. Philadelphia (PA): Lippincott Williams & Wilkins, p 1186–1243 [Google Scholar]
- Zinzula L, Esposito F, Pala D, Tramontano E. 2012. dsRNA binding characterization of full length recombinant wild type and mutants Zaire ebolavirus VP35. Antiviral Res 93:354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]