Abstract
Adenosine deaminase acting on RNA1 (ADAR1) catalyzes the C6 deamination of adenosine (A) to produce inosine (I) in regions of RNA with double-stranded (ds) character. This process is known as A-to-I RNA editing. Alternative promoters drive the expression of the Adar1 gene and alternative splicing gives rise to transcripts that encode 2 ADAR1 protein size isoforms. ADAR1 p150 is an interferon (IFN)-inducible dsRNA adenosine deaminase found in the cytoplasm and nucleus, whereas ADAR1 p110 is constitutively expressed and nuclear in localization. Dependent on the duplex structure of the dsRNA substrate, deamination of adenosine by ADAR can be either highly site-selective or nonspecific. A-to-I editing can alter the stability of RNA structures and the coding of RNA as I is read as G instead of A by ribosomes during mRNA translation and by polymerases during RNA replication. A-to-I editing is of broad physiologic significance. Both the production and the action of IFNs, and hence the subsequent interaction of viruses with their hosts, are among the processes affected by A-to-I editing.
Introduction
Double-stranded RNA (dsRNA) is a potent inducer of interferon (IFN) production. Both synthetic dsRNAs and naturally occurring dsRNAs generated during pathogen infection are effective IFN inducers (Stewart 1979). Among the natural inducers, Philip Marcus and Margaret Sekellick demonstrated nearly 40 years ago that a copyback defective interfering viral RNA, vesicular stomatitis virus (VSV) DI-011, was an efficient trigger of the innate immune response and that just a single DI particle was sufficient to induce a quantum yield of IFN (Marcus and Sekellick 1977). We now have considerable insight into the dsRNA signaling mechanisms that lead to the transcriptional activation of IFN genes. IFN system sensors of dsRNA include the RIG-like receptors RIG-I and MDA5 (Yoneyama and Fujita 2010; Ramos and Gale 2011) and the Toll-like receptor TLR3 (Kawai and Akira 2010; Yu and Levine 2011). The IFN-inducible protein kinase regulated by RNA (PKR) also is a sensor of dsRNA (Toth and others 2006; Pindel and Sadler 2011). Likewise, the IFN-inducible RNA adenosine deaminase that utilizes dsRNA as a substrate, adenosine deaminase acting on RNA1 (ADAR1), is a sensor of dsRNA (Toth and others 2006; Hundley and Bass 2010; Nishikura 2010; George and others 2011). ADAR1 catalyzes the deamination of adenosine in dsRNA structures to yield inosine, thereby potentially altering the coding capacity and structure of the substrate RNA. However, in contrast to the activity of PKR, ADAR1 responses often are proviral and antiapoptotic (Pfaller and others 2011; Samuel 2011). ADAR1, although IFN inducible, has emerged as a suppressor of the type I IFN response. What, then, is ADAR1? What are the biochemical mechanisms by which ADAR1 acts, and what is the evidence that ADAR1 impairs the type I IFN response?
Adar1 Gene and Proteins
There are 3 characterized Adar genes in the mouse and human, Adar1, Adar2, and Adar3 (Bass and others 1997; Toth and others 2006; Nishikura 2010; George and others 2011). Among these, Adar1 is IFN inducible (Patterson and Samuel 1995; Patterson and others 1995). The Adar1 gene maps to human chromosome 1q21.1 and mouse chromosome 3F2 (Wang and others 1995; Weier and others 1995, 2000). Transcription of the mammalian Adar1 gene is driven by multiple promoters; one is IFN inducible and the others are constitutively active, both in human and mouse (George and Samuel 1999a, 1999b; Kawakubo and Samuel 2000; George and others 2005). The IFN inducible promoter of Adar1 possesses a consensus IFN-stimulated response element. The human Adar1 gene includes 17 exons and spans ∼40-kbp (Liu and others 1997). The exon–intron organizations of the human and mouse Adar1 genes are highly conserved (Liu and others 1997; Hartner and others 2004; Wang and others 2004; George and others 2008).
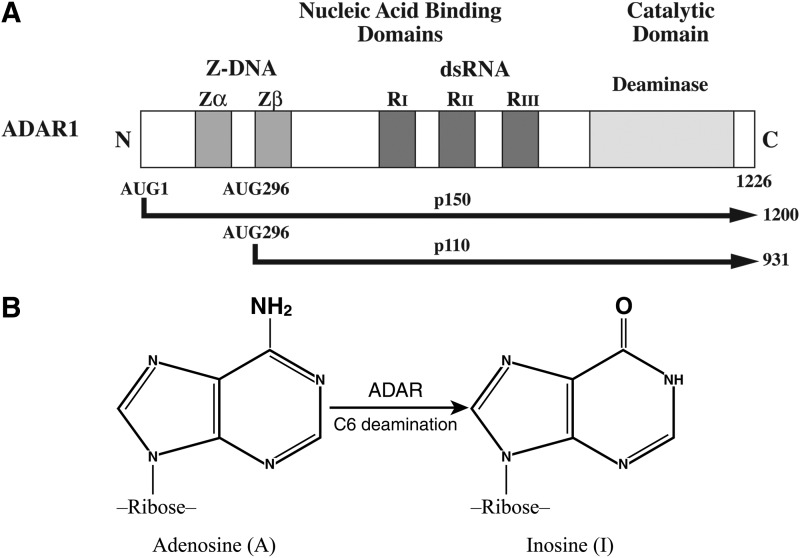
Adar1 transcripts undergo alternative splicing, including exon 1 that occurs in at least 3 alternative forms, to generate the mature transcripts that encode 2 differently sized ADAR1 proteins: an IFN-inducible p150 ADAR1; and, a constitutively expressed p110 ADAR1 (Patterson and Samuel 1995). The predominant IFN-inducible transcript possesses the alternative exon 1A that includes an AUG translation initiation codon and also an alternative form of exon 7; this transcript encodes the p150 protein of 1,200 amino acids in human cells and 1,152 amino acids in mouse cells (Toth and others 2006; George and others 2011). By contrast, the alternative exons 1B and 1C of constitutively expressed Adar1 transcripts lack a functional AUG codon; translation of the ORF of these mRNAs, therefore, begins at AUG296 present in exon 2 to generate the p110 protein that is 931 amino acids in human cells (Toth and others 2006; George and others 2011). The constitutively expressed mouse protein is 903 amino acids.
ADAR1 proteins possess 2 kinds of nucleic acid binding domains (Fig. 1A). Three copies of a double-stranded RNA binding motif are present in the central region of the p110 and p150 proteins (Patterson and Samuel 1995; Liu and Samuel 1996; Fierro-Monti and Mathews 2000). The dsRNA binding motifs found in ADAR1 proteins are similar to the prototypical dsRNA binding motif discovered initially in PKR (Toth and others 2006). Either 1 (p110) or 2 (p150) copies of a Z-DNA binding motif (Zα, Zβ) are present in the N-terminal region of ADAR1 (Herbert and others 1997). The Z-DNA binding motifs found in ADAR1 share homology with the N-terminal region of the poxvirus E3L protein (Patterson and Samuel 1995), also a Z-DNA binding protein (Schwartz and others 1999; Kim and others 2003). Because the A-form of dsRNA with purine-pyrimidine repeats can be transformed to generate a left-handed Z-RNA helix and because the Zα domain of p150 can bind Z-RNA (Placido and others 2007), the precise nature of nucleic acid (Z-DNA or Z-RNA) bound by the Z-domain of ADAR1 p150 under physiologic conditions remains unclear. The C-terminal region of ADAR1 p110 and p150 proteins includes the deaminase catalytic domain (Kim and others 1994; O'Connell and others 1995; Patterson and Samuel 1995; Liu and Samuel 1996). The biochemical activities assigned to the nucleic acid binding and catalytic domains have been verified by mutational analyses (Toth and others 2006; George and others 2011). Both p150 and p110 are active enzymes that catalyze the C6 deamination of adenosine in duplex RNA structures; the product is inosine (Fig. 1B). The p150 protein possesses a nuclear export signal and p150 is present in both the cytoplasm and nucleus, whereas the p110 form of ADAR1 is predominantly if not exclusively a nuclear protein (Patterson and Samuel 1995; Poulsen and others 2001). Genetic disruption of the mouse Adar1 gene in a manner that either knocks out expression of both p150 and p110 ADAR1 proteins (Hartner and others 2004; Wang and others 2004) or only the p150 protein (Ward and others 2011) results in embryonic lethality.
FIG. 1.
Domain organization and reaction catalyzed by adenosine deaminase acting on RNA1 (ADAR1). (A) Domain organization of human ADAR1. Alternative promoters and alternative splicing generate 2 size-isoforms of ADAR1, an interferon (IFN)-inducible p150 protein and a constitutively expressed p110 protein. The 2 Z-DNA binding domains (Zα, Zβ), the 3 double-stranded RNA (dsRNA) binding domains (RI, RII, and RIII), and the deaminase catalytic domain are shown. Alternative exon 1 and exon 7 structures give rise to the 1,200 amino acid p150 protein and 931 aa p110 protein in human cells. (B) Both p150 and p110 ADAR1 catalyze the hydrolytic C6 deamination of adenosine (A) to yield inosine (I) in dsRNA. Adapted from George and others (2011).
RNA Substrates of ADAR1
Both cellular and viral RNAs are edited by ADAR1 and, dependent upon the duplex structure and sequence of the substrate RNA, the editing can be either highly site-selective with one or very few adenosine residues edited or nonselective with multiple sites edited (Samuel 2011). A-to-I editing is effectively an RNA nucleotide substitution process. Hence, A-to-I editing of RNA has the capacity to alter RNA coding and RNA structure (Hundley and Bass 2010; Nishikura 2010; George and others 2011; Maas 2012). The purine I is decoded as G instead of A by ribosomes during mRNA translation and also by viral polymerases during RNA-dependent RNA replication, thereby potentially altering the coding of genetic information. A-to-I editing can also alter the stability of RNA structures, either by decreasing stability as I:U mismatches are less stable than A:U base pairs or by increasing stability as I:C base pairs are more stable than A:C mismatch pairs (Bass and Weintraub 1988; Wagner and others 1989; Levanon and others 2004).
A-to-I editing was discovered during antisense RNA studies because of destablization of dsRNA within cells. An activity described initially as a dsRNA unwinding activity present in Xenopus and mammalian cells was later shown instead to covalently modify dsRNA substrates by adenosine deamination, thereby changing A:U base pairs to less stable I:U mismatches (Bass and Weintraub 1988; Wagner and others 1989). Two Adar genes are now known that specify catalytically active dsRNA adenosine deaminases, Adar1 and Adar2 (Bass and others 1997; Samuel 2011). The ADAR1 and 2 enzymes act with overlapping specificity on duplex RNA, without sequence specificity for binding dsRNA, but showing a 3′-neighbor preference of a purine for adenosine deamination and with selectivity of the deamination conferred by bulges and mismatches and presumably a higher order structure within the dsRNA substrates (Lehmann and Bass 1999; Dawson and others 2004; Riedmann and others 2008).
Among the first biologically relevant and still best characterized substrates of ADAR1 that result in amino acid coding changes following editing are transcripts for the GluR-B and 5-HT2c-R neurotransmitter receptors for glutamate and serotonin, respectively (Sommer and others 1991; Higuchi and others 1993; Burns and others 1997; Liu and Samuel 1999; Liu and others 1999). In the cases of these RNA substrates, the high selectivity of the editing reaction is conferred by imperfect duplex structures that form between exonic and adjacent intronic sequences. This results in site-specific deamination by ADAR1 and ADAR2 of specific adenosine residues present in ORFs of GluR-B and 5-HT2c-R transcripts that subsequently lead to amino acid substitutions to produce receptor proteins with altered functional activity (Seeburg and Hartner 2003; Hood and Emeson 2012). A-to-I editing of hepatitis delta virus (HDV) agent antigenome RNA also provides another early and well-characterized example of a highly selective adenosine deamination reaction (Casey and others 1992; Casey and Gerin 1995). In the case of HDV, the editing involves the selective conversion by ADAR1 of an amber UAG stop codon within a dsRNA duplex rod-like structure to a UIG codon that then is decoded as tryptophan (UGG), thereby allowing for synthesis of large delta antigen (Casey 2012). By contrast, the hyperediting of measles virus RNA during persistent infection is nonspecific (Oldstone 2009; Samuel 2011).
Deep sequencing and bioinformatic approaches have identified additional candidate A-to-I editing sites in the human transcriptome, some present in nonrepetitive coding sequences, but most occurring in repetitive elements present in noncoding regions of the RNA, including Alu elements (Athanasiadis and others 2004; Kim and others 2004; Levanon and others 2004; Paz-Yaacov and others 2010; Garncarz and others 2013; Ramaswami and others 2013). However, the functional significance of editing of Alu elements is not well established. Among the newly validated A-to-I editing targets in coding regions identified by transcriptome sequencing is that for the cellular protein AZIN1 that encodes antizyme inhibitor 1. Selective editing of AZIN1 RNA by ADAR1, resulting in a Ser to Gly amino acid substitution, confers phenotypes manifested by augmented tumor-initiating potential and a more aggressive behavior that predisposes to hepatocellular carcinoma (Chen and others 2013).
Biological Activities Mediated by Alterations in ADAR1 and A-to-I Editing
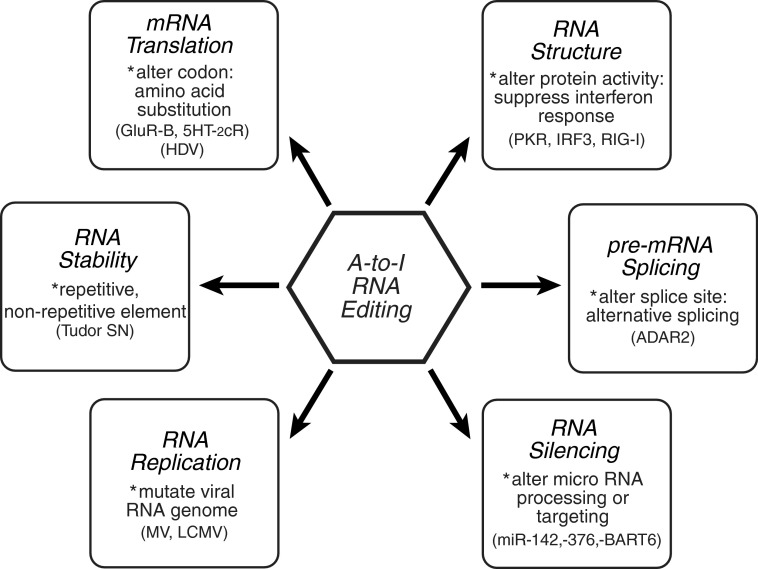
Studies with cell culture and intact animal systems have revealed a range of physiological changes caused by altered expression of ADAR1 and subsequent perturbation of A-to-I editing homeostasis. ADAR1-mediated RNA editing has been shown to affect a number of biologic responses including virus growth and persistence, cell proliferation, neurotransmitter function, and innate immune responses. Changing the sequence of an RNA by deamination can affect multiple biochemical processes (Fig. 2), thereby impacting gene expression and function.
FIG. 2.
A-to-I RNA editing affects multiple biochemical processes, thereby altering gene expression and function. Because I base pairs as G instead of A, nucleotide substitution of an A with an I may affect mRNA translation by altering a codon potentially leading to an amino acid substitution; RNA structure-dependent activities that trigger IFN responses may be suppressed if dsRNA structures are destabilized; the pre-mRNA splicing pattern of an RNA may be altered by editing a conserved A involved in splice site selection; RNA silencing may be altered by affecting dsRNA structures involved in either micro RNA processing or targeting; RNA virus genome stability may be altered by changing template and therefore complementary product sequences during viral RNA synthesis leading to A-to-G (U-to-C) transitions; and, A-to-I editing of noncoding repetitive (Alu) or nonrepetitive RNA elements may potentially affect RNA stability by altering cellular localization or nuclease recognition. Adapted from Samuel (2011).
Virus growth and persistence
Sequence changes consistent with A-to-I editing have been described for viral RNAs of a number of viruses including measles virus, influenza virus, parainfluenza virus, VSV, hepatitis C virus (HCV), hepatitis D virus, lymphocytic choriomeningitis virus, and polyoma virus (Toth and others 2006; Gelinas and others 2011; Samuel 2011). How the observed sequence changes attributed to A-to-I RNA editing affect virus replication appears to differ for different virus–host combinations. For example, with HDV, the selective editing of viral RNA has a proviral effect by permitting synthesis of large delta antigen required for RNA replication (Casey 2012). A long history also exists for the involvement of A-to-I editing in measles virus–host interactions. In subacute sclerosing panencephalitis, a rare but often fatal disease in which persistent measles virus infection occurs in the brain, clustered A-to-G mutations are described mostly in the M gene and less so in other genes (Cattaneo and others 1988; Oldstone 2009). The mutations found in the M gene RNA would prevent synthesis of the matrix protein that is required for virion assembly and release, and therefore are believed to contribute to the viral persistence phenotype. A combination of approaches including overexpression, genetic knockout, and stable knockdown established ADAR1 as a proviral host factor in the context of measles virus acute infection (Toth and others 2009; Okonski and Samuel 2013), VSV (Nie and others 2007; Li and others 2010), and human immunodeficiency virus (HIV) (Phuphuakrat and others 2008; Clerzius and others 2009; Doria and others 2009; Schoggins and others 2011). However, under some conditions, ADAR1 also displays an antiviral role with measles virus (Ward and others 2011) and HIV (Biswas and others 2012). The emerging picture is one of a finely balanced interplay between ADAR1 and other IFN-stimulated gene products including PKR that together contribute to determining the outcome of an infection (Pfaller and others 2011).
IFN is a therapeutic presently used for treatment of HCV infection, which can lead to chronic liver disease including hepatocellular carcinoma. In addition to HCV virus genotype, among the cellular genes identified that affect HCV responsiveness to IFN therapy is ADAR1 (Welzel and others 2009). Interestingly, overexpression of ADAR1 in cell culture was found to be proviral for a number of medically important RNA viruses (Gelinas and others 2011; Samuel 2011; Schoggins and others 2011), and an increased A-to-I editing of the cellular AZIN1 transcript recently has been reported to predispose to hepatocellular carcinoma (Chen and others 2013). ADAR1 and PKR differentially affect the formation of stress granules, cytosolic aggregates of stalled translation preinitiation complexes (Okonski and Samuel 2013). In combination with IFN, HCV infection induces dynamic oscillation of stress granules that HCV may exploit to establish persistence (Ruggieri and others 2012).
Among the biochemical mechanisms suggested to account for the apparent pan-viral enhancement of virus growth by ADAR1 seen with some virus-cell combinations but not others is the antagonism of PKR activation by ADAR1 (Gelinas and others 2011; Samuel 2011). This mechanism is exemplified by measles virus. Studies with human cells stably knocked down for ADAR1 show increased activation of PKR, increased apoptosis, increased stress granule formation, and reduced viral growth following infection with measles virus (Toth and others 2009; Li and others 2012; Okonski and Samuel 2013). Complementation of ADAR1-deficient cells with ADAR1 p150 wild type but not the p150 catalytic mutant enhances virus growth and suppresses both PKR activation and stress granule formation (Okonski and Samuel 2013). Likewise, with mouse cells genetically knocked out for ADAR1 p150 expression, increased virus-induced cytotoxicity is observed compared with ADAR1-sufficient wild type cells following infection with negative-strand RNA viruses including measles virus, Newcastle disease virus, Sendai virus, canine distemper virus, and influenza virus (Ward and others 2011). These results are consistent with a protective function of the ADAR1 p150 cytoplasmic protein against RNA virus-induced cellular stress.
Neurotransmitter function
A-to-I RNA editing represents an important mechanism by which neuronal activity is modulated (Seeburg and Hartner 2003; George and others 2011; Hood and Emeson 2012). Some of the best characterized substrates of ADAR enzymes are neurotransmitter receptor and channel mRNAs. Among the most extensively studied are transcripts coding for AMPA and kainate glutamate-gated ion channel receptors, the serotonin 5-HT2c receptor, the α3 subunit of the GABAA receptor and the Kv1.1 potassium ion channel. Editing of these RNAs results in the expression of proteins with altered amino acid sequence and altered physiologic function. For example, the ionotrophic glutamate receptors, GluRs, regulate excitatory synaptic neurotransmission (Seeburg and Hartner 2003). A-to-I editing of GluR mRNAs regulate calcium permeability of the channels (Burnashev and others 1992), assembly and stoichiometry of the channel subunits (Greger and others 2003), and the rate of recovery from receptor desensitization (Lomeli and others 1994). Of the 2 editing sites present in GluR-B mRNA, the Q/R and R/G sites, ADAR1 contributes to the editing of the R/G site. ADAR1 also efficiently edits the GluR-B pre-mRNA at the intron 11 hotspot site +60 (Liu and Samuel 1999). The 5-HT2c serotonin receptor is a 7-transmembrane, phospholipase C-linked receptor encoded by mRNA transcripts that undergo editing at 5 sites termed A, B, C, D (Burns and others 1997), and E (Fitzgerald and others 1999; Niswender and others 1999; Wang and others 2000). Editing of the 5-HT2c transcript at all sites results in 3 amino acid substitutions in the rat and human receptor proteins, which cause a reduction in G-protein signaling compared with unedited RNA. Sites A and B are primarily edited by ADAR1 (Liu and Samuel 1999; Hartner and others 2004; Wang and others 2004).
Alterations in editing of RNAs encoding glutamate and serotonin neurotransmitter receptors have been implicated with neurologic and behavioral disorders (Morabito and Emeson 2009; O'Neil and Emeson 2012; Slotkin and Nishikura 2013). Altered editing patterns of 5-HT2c receptor transcripts, including at the A site primarily edited by ADAR1, have been observed in suicide victims with a history of major depression and in response to antidepressant treatment (Niswender and others 2001; Gurevich and others 2002). Increased cortical expression of ADAR1 but not ADAR2 also has been described in depressive suicide victims compared with patients who do not commit suicide (Simmons and others 2010). Increased expression of ADAR1 and ADAR2 and increased editing of 5-HT2c receptor mRNAs also has been implicated with addictive substance abuse behavior in model organisms (Dracheva and others 2009; Watanabe and others 2013). Depression can be a negative effect of IFNα therapy that adversely affects quality of life with potentially serious complications, and while the molecular mechanism has yet to be established, serotonin-mediated effects have been implicated, which could have as their basis altered RNA editing mediated by IFN-inducible ADAR1 p150 (Menkes and MacDonald 2000; Hood and Emeson 2012; O'Neil and Emeson 2012). Finally, mutations in ADAR1 resulting in its dysfunction has been shown to cause Aicardi-Goutieres (AG) syndrome, which is a genetically determined inflammatory disorder that affects newborns and infants resulting in severe mental and physical handicaps (Rice and others 2012). AG syndrome disease is associated with an upregulation of type I IFN in the absence of normal ADAR1 function (Rice and others 2012; Livingston and others 2013).
Cell proliferation and cancer
A growing body of evidence has revealed that disruption of the normal balance of ADAR1- and ADAR2-mediated A-to-I RNA editing influences the growth and progression of some human tumors (Galeano and others 2012; Chan and others 2013; Chen and others 2013; Qin and others 2013). Disruption of ADAR1 function also influences apoptotic responses both in virus-infected cells (Toth and others 2009; Ward and others 2011) and during embryonic development (Hartner and others 2004; Wang and others 2004; XuFeng and others 2009).
During embryonic development, ADAR1 is essential for hematopoiesis in mice (Hartner and others 2009; XuFeng and others 2009). ADAR1 is necessary for maintenance of both fetal and adult hematopoietic stem cells; genetic disruption of Adar1 in hematopoietic stem cells leads to rapid apoptosis (Hartner and others 2009). ADAR1 is required for survival of differentiating hematopoietic progenitor cells in adult mice via an RNA-editing dependent mechanism (XuFeng and others 2009). ADAR1 also suppresses measles virus-induced apoptosis and cytotoxicity in cell culture. HeLa cells stably knocked down for ADAR1 and mouse MEFs genetically deficient in ADAR1 show enhanced measles virus-induced apoptosis measured by PARP cleavage, reduced cell viability measured by colorimetric MTS assay, and enhanced cytotoxicity assessed microscopically (Toth and others 2009; Ward and others 2011).
Dysregulation of A-to-I RNA editing by ADAR1 also has been implicated in the functional alteration of proteins relevant to cancer biology, including AZIN1 and FLNB in hepatocellular carcinoma (Chen and others 2013) and esophageal squamous cell carcinoma (ESCC) (Qin and others 2013), respectively. ADAR1-mediated editing of AZIN1 RNA (antizyme inhibitor 1) has been demonstrated to associate with increased risk of liver cirrhosis and predisposition to liver carcinoma. The AZIN1 protein blocks the effects of an antizyme on ornithine decarboxylase (ODC). ADAR1-mediated A-to-I editing causes a Ser367Gly substitution that generates an AZIN1 protein that is more stable and hence binds antizyme in a manner that impairs its interaction with ODC, compared with the wild type AZIN1. Hence, the edited form of AZIN1 inhibits the antizyme tumor suppressor function and promotes cell proliferation (Chen and others 2013). Large-scale genome sequencing of hepatocellular carcinoma samples also identified, in addition to AZIN1, FLNB (filamin B) as a major target of ADAR1. ADAR1, but not ADAR2 or 3, is overexpressed in primary ESCC, a major form of esophageal cancer (Qin and others 2013). Functional analysis revealed hyperediting of FLNB RNA in addition to AZIN1 RNA in primary ESCC. In vitro studies also support the oncogenic potential of AZIN1 and FLNB associated with unbalanced editing by ADAR1 (Qin and others 2013).
In case of chronic myeloid leukemia (CML) patients, whole transcriptome sequencing revealed an enhanced expression of ADAR1 p150 isoform and increased A-to-I editing during CML progression. Overexpression of ADAR1 p150 promotes the expression of myeloid transcription factor pu.1, which induces extension of myeloid progenitors and also generates an alternatively spliced form of glycogen synthase kinase involved in self-renewal of leukemia stem cells (Jiang and others 2013). A-to-I editing by ADAR1 also occurs within the 3′-untranslated region (UTR) of the F11R gene transcript. The editing is increased upon hypoxic conditions, thereby altering nuclear retention of the RNA; F11R is associated with cell adhesion and spreading (Ben-Zvi and others 2013). Finally, the Hedgehog signaling pathway implicated in tumor progression and embryonic development is deregulated by A-to-I editing of the glioma-associated oncogene 1 by ADAR1 and ADAR2 (Shimokawa and others 2013). Glioblastoma, an aggressive, malignant brain tumor involving astrocyte glial cells also is associated with hypoediting of glutamate receptor RNA and decreased ADAR2 activity in astrocytoma tumor tissue (Galeano and others 2012).
The biochemical mechanism whereby editing affects tumor growth in most instances is not yet clear. A-to-I editing by ADARs is known to affect both the processing and targeting of micro RNAs (Nishikura 2010). Editing by ADAR1 in invasive ductal breast cancer and glioblastoma specimens inhibits the binding of miR-30b-3p and miR-573 within the 3′-UTR of Rho GTPase activating protein 26 (ARHGAP26) transcripts. ARHGAP26 is a regulator of the Rho family and a putative tumor suppressor, negatively regulating RhoA and Cdc42 in human cancers. miR-47 and miR-432 down-regulate the expression of ADAR1 in cancer cells (Wang and others 2013). Attenuated A-to-I editing of microRNA miR-376a has been found to promote invasiveness of glioblastoma cells (Choudhury and others 2012).
Type I IFN production
The ADAR1 cDNA was initially identified and characterized as an IFN-stimulated gene (Patterson and Samuel 1995; Patterson and others 1995). ADAR1 then was shown through loss of function and complementation studies to suppress innate immune responses and IFN induction in virus-infected cells in culture in a manner dependent upon ADAR1 catalytic activity (Toth and others 2009; Li and others 2012; Okonski and Samuel 2013). Mutation of ADAR1 was also established to enhance IFN production and to trigger a type I IFN signature in mice (Hartner and others 2009) and humans (Rice and others 2012; Livingston and others 2013). Thus, one emerging role of ADAR1 is the downregulation of dsRNA-triggered innate immune responses through destablization of the activator dsRNA and hence suppression of its IFN inducing capacity.
The IFN system is a cornerstone of antiviral innate immunity (Borden and others 2007). Among the pathogen-associated molecular patterns of viruses that trigger the transcriptional induction of type I IFN is dsRNA, which is recognized as foreign by host cell nucleic acid sensors. In the case of measles virus, RIG-I senses infection and signals an IPS-dependent induction of IFN (Randall and Goodbourn 2008). PKR enhances IFNβ induction by measles virus in a manner that depends upon PKR catalytic activity and is prevented by viral C protein expression (McAllister and others 2010, 2012). C protein impairs production of defective copyback viral dsRNA and PKR activation (Pfaller and others 2014). ADAR1 suppresses the activation of PKR (Toth and others 2009), suppresses the activation of IRF3 (Li and others 2012), suppresses the formation of cytoplasmic stress granules (Okonski and Samuel 2013), and suppresses the induction of IFNβ (Li and others 2012; Okonski and Samuel 2013). In human cells stably knocked down for both the p110 and p150 ADAR1 proteins, the transcriptional induction of IFNβ is enhanced by wild type and V mutant measles viruses, whereas in the presence of ADAR1 these viruses are very poor IFNβ inducers (Li and others 2012). The enhanced induction of IFNβ seen with ADAR1-deficient cells correlates with enhanced activation of both IRF3 and PKR (Li and others 2012); activation of PKR leads to enhanced activation of NFkB (McAllister and others 2010, 2012). Complementation of ADAR1-deficient cells reveals that the p150 ADAR1 isoform suppresses both PKR activation and IFN induction (Okonski and Samuel 2013).
The elevated production of type I IFN observed in human cells in culture stably knocked down for ADAR1 also has been observed in Adar1−/− mice genetically knocked out for ADAR1 protein expression (Hartner and others 2009) and in human AG syndrome patients with mutations in ADAR1(Rice and others 2012; Livingston and others 2013). Genetic disruption of Adar1 causes embryonic lethality in mice and results in the upregulation of type I IFN and of IFN-stimulated gene expression in hematopoietic stem cells in the absence of infection (Hartner and others 2009). Mutations that cause AG syndrome mapped to Adar1, which likewise are associated with a type I IFN signature and elevated levels of IFN in the absence of detectable infection (Rice and others 2012; La Piana and others 2013; Livingston and others 2013). These results suggest that ADAR1 is essential for the suppression of type I IFN responses in cultured cells, mice and humans. The biochemical mechanism responsible for the enhanced type I IFN response seen in ADAR1-deficient cells, based on measles virus-cell culture system studies, appears to involve an increased accumulation of dsRNA (Pfaller and others 2014) that subsequently increases the activation of both IRF3 and PKR (Li and others 2012; Okonski and Samuel 2013). Whether a similar mechanism occurs in mice and humans with mutations in the Adar1 gene, which conceivably might permit accumulation of increased levels of cytoplasmic cellular RNA with stable dsRNA structure sufficient to trigger the observed type I IFN signature, remains to be established.
ADAR1 deficiency in cultured cells, mice and humans all lead to enhanced type I IFN production (Hartner and others 2009; Rice and others 2012; Okonski and Samuel 2013). Whether this is due to an anticipated reduction in inosine-containing RNA present in ADAR1-deficient cells remains to be established. Synthetic inosine-containing dsRNA exemplified by poly rI:poly rC was identified long ago as a potent inducer of IFN in a variety of cultured cells (Field and others 1967). Recently, inosine incorporation into synthetic single-stranded RNA (ssRNA) also has been demonstrated to potentiate production of IFNα and tumor necrosis factor α in human peripheral blood mononuclear cells by TLR7/8 sensing of the ssRNA (Sarvestani and others 2014). A-to-I editing of influenza virus ssRNA also enhanced TLR7 sensing. The biological effect of the presence of inosine within an RNA, presumably generated under physiological conditions by the action of an ADAR, on the IFN-inducing capacity of a given ssRNA or dsRNA is likely determined by the combination of factors including size, overall structure and concentration of the I-containing RNA, the subcellular localization of the RNA, and the kind of cell in which the I-containing RNA is present and hence the innate immune response triggered.
Acknowledgments
This work was supported in part by the National Institute of Allergy and Infectious Diseases, NIH, U.S. Public Health Service.
Author Disclosure Statement
No competing financial interests exist.
References
- Athanasiadis A, Rich A, Maas S. 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2(12):e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL, Nishikura K, Keller W, Seeburg PH, Emeson RB, O'Connell MA, Samuel CE, Herbert A. 1997. A standardized nomenclature for adenosine deaminases that act on RNA. RNA 3(9):947–949 [PMC free article] [PubMed] [Google Scholar]
- Bass BL, Weintraub H. 1988. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55(6):1089–1098 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi M, Amariglio N, Paret G, Nevo-Caspi Y. 2013. F11R expression upon hypoxia is regulated by RNA editing. PLoS One 8(10):e77702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas N, Wang T, Ding M, Tumne A, Chen Y, Wang Q, Gupta P. 2012. ADAR1 is a novel multi targeted anti-HIV-1 cellular protein. Virology 422(2):265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov 6(12):975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnashev N, Monyer H, Seeburg PH, Sakmann B. 1992. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron 8(1):189–198 [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387(6630):303–308 [DOI] [PubMed] [Google Scholar]
- Casey JL. 2012. Control of ADAR1 editing of hepatitis delta virus RNAs. Curr Top Microbiol Immunol 353:123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JL, Bergmann KF, Brown TL, Gerin JL. 1992. Structural requirements for RNA editing in hepatitis delta virus: evidence for a uridine-to-cytidine editing mechanism. Proc Natl Acad Sci U S A 89(15):7149–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JL, Gerin JL. 1995. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J Virol 69(12):7593–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, Billeter MA. 1988. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55(2):255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TH, Lin CH, Qi L, Fei J, Li Y, Yong KJ, Liu M, Song Y, Chow RK, Ng VH, Yuan YF, Tenen DG, Guan XY, Chen L. 2013. A disrupted RNA editing balance mediated by ADARs (adenosine deaminases that act on RNA) in human hepatocellular carcinoma. Gut [Epub ahead of print];DOI: 10.1136/gutjnl-2012-304037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, Liu M, Yuan YF, Fu L, Kong KL, Qi L, Li Y, Zhang N, Tong AH, Kwong DL, Man K, Lo CM, Lok S, Tenen DG, Guan XY. 2013. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med 19(2):209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury Y, Tay FC, Lam DH, Sandanaraj E, Tang C, Ang BT, Wang S. 2012. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J Clin Invest 122(11):4059–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerzius G, Gelinas JF, Daher A, Bonnet M, Meurs EF, Gatignol A. 2009. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J Virol 83(19):10119–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TR, Sansam CL, Emeson RB. 2004. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J Biol Chem 279(6):4941–4951 [DOI] [PubMed] [Google Scholar]
- Doria M, Neri F, Gallo A, Farace MG, Michienzi A. 2009. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res 37(17):5848–5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracheva S, Lyddon R, Barley K, Marcus SM, Hurd YL, Byne WM. 2009. Editing of serotonin 2C receptor mRNA in the prefrontal cortex characterizes high-novelty locomotor response behavioral trait. Neuropsychopharmacology 34(10):2237–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field AK, Tytell AA, Lampson GP, Hilleman MR. 1967. Inducers of interferon and host resistance. II. Multistranded synthetic polynucleotide complexes. Proc Natl Acad Sci U S A 58(3):1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro-Monti I, Mathews MB. 2000. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem Sci 25(5):241–246 [DOI] [PubMed] [Google Scholar]
- Fitzgerald LW, Iyer G, Conklin DS, Krause CM, Marshall A, Patterson JP, Tran DP, Jonak GJ, Hartig PR. 1999. Messenger RNA editing of the human serotonin 5-HT2C receptor. Neuropsychopharmacology 21(2 Suppl):82S–90S [DOI] [PubMed] [Google Scholar]
- Galeano F, Tomaselli S, Locatelli F, Gallo A. 2012. A-to-I RNA editing: the “ADAR” side of human cancer. Semin Cell Dev Biol 23(3):244–250 [DOI] [PubMed] [Google Scholar]
- Garncarz W, Tariq A, Handl C, Pusch O, Jantsch MF. 2013. A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol 10(2):192–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelinas JF, Clerzius G, Shaw E, Gatignol A. 2011. Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase. J Virol 85(17):8460–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Das S, Samuel CE. 2008. Organization of the mouse RNA-specific adenosine deaminase Adar1 gene 5′-region and demonstration of STAT1-independent, STAT2-dependent transcriptional activation by interferon. Virology 380(2):338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Gan Z, Liu Y, Samuel CE. 2011. Adenosine deaminases acting on RNA, RNA editing, and interferon action. J Interferon Cytokine Res 31(1):99–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Samuel CE. 1999a. Characterization of the 5′-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene 229(1–2):203–213 [DOI] [PubMed] [Google Scholar]
- George CX, Samuel CE. 1999b. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci U S A 96(8):4621–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Wagner MV, Samuel CE. 2005. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J Biol Chem 280(15):15020–15028 [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Kong X, Ziff EB. 2003. AMPA receptor tetramerization is mediated by Q/R editing. Neuron 40(4):763–774 [DOI] [PubMed] [Google Scholar]
- Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. 2002. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 34(3):349–356 [DOI] [PubMed] [Google Scholar]
- Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. 2004. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 279(6):4894–4902 [DOI] [PubMed] [Google Scholar]
- Hartner JC, Walkley CR, Lu J, Orkin SH. 2009. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol 10(1):109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A, Alfken J, Kim YG, Mian IS, Nishikura K, Rich A. 1997. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc Natl Acad Sci U S A 94(16):8421–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. 1993. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell 75(7):1361–1370 [DOI] [PubMed] [Google Scholar]
- Hood JL, Emeson RB. 2012. Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr Top Microbiol Immunol 353:61–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Bass BL. 2010. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci 35(7):377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Crews LA, Barrett CL, Chun HJ, Court AC, Isquith JM, Zipeto MA, Goff DJ, Minden M, Sadarangani A, Rusert JM, Dao KH, Morris SR, Goldstein LS, Marra MA, Frazer KA, Jamieson CH. 2013. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci U S A 110(3):1041–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11(5):373–384 [DOI] [PubMed] [Google Scholar]
- Kawakubo K, Samuel CE. 2000. Human RNA-specific adenosine deaminase (ADAR1) gene specifies transcripts that initiate from a constitutively active alternative promoter. Gene 258(1–2):165–172 [DOI] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. 2004. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res 14(9):1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. 1994. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci U S A 91(24):11457–11461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Muralinath M, Brandt T, Pearcy M, Hauns K, Lowenhaupt K, Jacobs BL, Rich A. 2003. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc Natl Acad Sci U S A 100(12):6974–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Piana A, Uggetti C, Olivieri I, Tonduti D, Balottin U, Fazzi E, Orcesi S. 2013. Bilateral striatal necrosis in two subjects with Aicardi-Goutieres syndrome due to mutations in ADAR1 (AGS6). Am J Med Genet part A 9999:1–5 [DOI] [PubMed] [Google Scholar]
- Lehmann KA, Bass BL. 1999. The importance of internal loops within RNA substrates of ADAR1. J Mol Biol 291(1):1–13 [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, Olshansky M, Rechavi G, Jantsch MF. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22(8):1001–1005 [DOI] [PubMed] [Google Scholar]
- Li Z, Okonski KM, Samuel CE. 2012. Adenosine deaminase acting on RNA 1 (ADAR1) suppresses the induction of interferon by measles virus. J Virol 86(7):3787–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wolff KC, Samuel CE. 2010. RNA adenosine deaminase ADAR1 deficiency leads to increased activation of protein kinase PKR and reduced vesicular stomatitis virus growth following interferon treatment. Virology 396(2):316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Emeson RB, Samuel CE. 1999. Serotonin-2C receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J Biol Chem 274(26):18351–18358 [DOI] [PubMed] [Google Scholar]
- Liu Y, George CX, Patterson JB, Samuel CE. 1997. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J Biol Chem 272(7):4419–4428 [DOI] [PubMed] [Google Scholar]
- Liu Y, Samuel CE. 1996. Mechanism of interferon action: functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J Virol 70(3):1961–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Samuel CE. 1999. Editing of glutamate receptor subunit B pre-mRNA by splice-site variants of interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J Biol Chem 274(8):5070–5077 [DOI] [PubMed] [Google Scholar]
- Livingston JH, Lin JP, Dale RC, Gill D, Brogan P, Munnich A, Kurian MA, Gonzalez-Martinez V, De Goede CG, Falconer A, Forte G, Jenkinson EM, Kasher PR, Szynkiewicz M, Rice GI, Crow YJ. 2013. A type I interferon signature identifies bilateral striatal necrosis due to mutations in ADAR1. J Med Genet 51(2):76–82 [DOI] [PubMed] [Google Scholar]
- Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266(5191):1709–1713 [DOI] [PubMed] [Google Scholar]
- Maas S. 2012. Posttranscriptional recoding by RNA editing. Adv Protein Chem Struct Biol 86:193–224 [DOI] [PubMed] [Google Scholar]
- Marcus PI, Sekellick MJ. 1977. Defective interfering particles with covalently linked [+/−]RNA induce interferon. Nature 266(5605):815–819 [DOI] [PubMed] [Google Scholar]
- McAllister CS, Taghavi N, Samuel CE. 2012. Protein kinase PKR amplification of interferon beta induction occurs through initiation factor eIF-2alpha-mediated translational control. J Biol Chem 287(43):36384–36392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister CS, Toth AM, Zhang P, Devaux P, Cattaneo R, Samuel CE. 2010. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. J Virol 84(1):380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkes DB, MacDonald JA. 2000. Interferons, serotonin and neurotoxicity. Psychol Med 30(2):259–268 [DOI] [PubMed] [Google Scholar]
- Morabito MV, Emeson RB. 2009. RNA editing as a therapeutic target for CNS disorders. Neuropsychopharmacology 34(1):246. [DOI] [PubMed] [Google Scholar]
- Nie Y, Hammond GL, Yang JH. 2007. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J Virol 81(2):917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem 79:321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender CM, Copeland SC, Herrick-Davis K, Emeson RB, Sanders-Bush E. 1999. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J Biol Chem 274(14):9472–9478 [DOI] [PubMed] [Google Scholar]
- Niswender CM, Herrick-Davis K, Dilley GE, Meltzer HY, Overholser JC, Stockmeier CA, Emeson RB, Sanders-Bush E. 2001. RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology 24(5):478–491 [DOI] [PubMed] [Google Scholar]
- O'Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A, Keller W. 1995. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol 15(3):1389–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil RT, Emeson RB. 2012. Quantitative analysis of 5HT(2C) receptor RNA editing patterns in psychiatric disorders. Neurobiol Dis 45(1):8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonski KM, Samuel CE. 2013. Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J Virol 87(2):756–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone MB. 2009. Modeling subacute sclerosing panencephalitis in a transgenic mouse system: uncoding pathogenesis of disease and illuminating components of immune control. Curr Top Microbiol Immunol 330:31–54 [DOI] [PubMed] [Google Scholar]
- Patterson JB, Samuel CE. 1995. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol 15(10):5376–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JB, Thomis DC, Hans SL, Samuel CE. 1995. Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology 210(2):508–511 [DOI] [PubMed] [Google Scholar]
- Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, Amariglio N, Eisenberg E, Rechavi G. 2010. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci U S A 107(27):12174–12179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller CK, Li Z, George CX, Samuel CE. 2011. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr Opin Immunol 23(5):573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller CK, Radeke MJ, Cattaneo R, Samuel CE. 2014. Measles virus C protein impairs production of defective copyback double-stranded viral RNA and activation of protein kinase R. J Virol 88(1):456–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuphuakrat A, Kraiwong R, Boonarkart C, Lauhakirti D, Lee TH, Auewarakul P. 2008. Double-stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. J Virol 82(21):10864–10872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pindel A, Sadler A. 2011. The role of protein kinase R in the interferon response. J Interferon Cytokine Res 31(1):59–70 [DOI] [PubMed] [Google Scholar]
- Placido D, Brown BA, 2nd, Lowenhaupt K, Rich A, Athanasiadis A. 2007. A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure 15(4):395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen H, Nilsson J, Damgaard CK, Egebjerg J, Kjems J. 2001. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol Cell Biol 21(22):7862–7871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YR, Qiao JJ, Chan TH, Zhu YH, Li FF, Liu H, Fei J, Li Y, Guan XY, Chen L. 2013. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma (ESCC). Cancer Res 74(3):840–851 [DOI] [PubMed] [Google Scholar]
- Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O'Connell MA, Li JB. 2013. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods 10(2):128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos HJ, Gale M., Jr.2011. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol 1(3):167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89(Pt 1):1–47 [DOI] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, Battini R, Bertini E, Brogan PA, Brueton LA, Carpanelli M, De Laet C, de Lonlay P, del Toro M, Desguerre I, Fazzi E, Garcia-Cazorla A, Heiberg A, Kawaguchi M, Kumar R, Lin JP, Lourenco CM, Male AM, Marques W, Jr., Mignot C, Olivieri I, Orcesi S, Prabhakar P, Rasmussen M, Robinson RA, Rozenberg F, Schmidt JL, Steindl K, Tan TY, van der Merwe WG, Vanderver A, Vassallo G, Wakeling EL, Wassmer E, Whittaker E, Livingston JH, Lebon P, Suzuki T, McLaughlin PJ, Keegan LP, O'Connell MA, Lovell SC, Crow YJ. 2012. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genet 44(11):1243–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedmann EM, Schopoff S, Hartner JC, Jantsch MF. 2008. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA 14(6):1110–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri A, Dazert E, Metz P, Hofmann S, Bergeest JP, Mazur J, Bankhead P, Hiet MS, Kallis S, Alvisi G, Samuel CE, Lohmann V, Kaderali L, Rohr K, Frese M, Stoecklin G, Bartenschlager R. 2012. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe 12(1):71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. 2011. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 411(2):180–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvestani ST, Tate MD, Moffat JM, Jacobi AM, Behlke MA, Miller AR, Beckham SA, McCoy CE, Chen W, Mintern JD, O'Keeffe M, John M, Williams BR, Gantier MP. 2014. Inosine-mediated modulation of RNA sensing by TLR7/8. J Virol 88(2):799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472(7344):481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. 1999. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 284(5421):1841–1845 [DOI] [PubMed] [Google Scholar]
- Seeburg PH, Hartner J. 2003. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr Opin Neurobiol 13(3):279–283 [DOI] [PubMed] [Google Scholar]
- Shimokawa T, Rahman MF, Tostar U, Sonkoly E, Stahle M, Pivarcsi A, Palaniswamy R, Zaphiropoulos PG. 2013. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA Biol 10(2):321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M, Meador-Woodruff JH, Sodhi MS. 2010. Increased cortical expression of an RNA editing enzyme occurs in major depressive suicide victims. Neuroreport 21(15):993–997 [DOI] [PubMed] [Google Scholar]
- Slotkin W, Nishikura K. 2013. Adenosine-to-inosine RNA editing and human disease. Genome Med 5(11):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Kohler M, Sprengel R, Seeburg PH. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67(1):11–19 [DOI] [PubMed] [Google Scholar]
- Stewart WE. 1979. The interferon system. New York: Springer-Verlag [Google Scholar]
- Toth AM, Li Z, Cattaneo R, Samuel CE. 2009. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J Biol Chem 284(43):29350–29356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AM, Zhang P, Das S, George CX, Samuel CE. 2006. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol 81:369–434 [DOI] [PubMed] [Google Scholar]
- Wagner RW, Smith JE, Cooperman BS, Nishikura K. 1989. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci U S A 86(8):2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hui H, Guo Z, Zhang W, Hu Y, He T, Tai Y, Peng P, Wang L. 2013. ADAR1 regulates ARHGAP26 gene expression through RNA editing by disrupting miR-30b-3p and miR-573 binding. RNA 19(11):1525–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. 2004. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 279(6):4952–4961 [DOI] [PubMed] [Google Scholar]
- Wang Q, O'Brien PJ, Chen CX, Cho DS, Murray JM, Nishikura K. 2000. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J Neurochem 74(3):1290–1300 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zeng Y, Murray JM, Nishikura K. 1995. Genomic organization and chromosomal location of the human dsRNA adenosine deaminase gene: the enzyme for glutamate-activated ion channel RNA editing. J Mol Biol 254(2):184–195 [DOI] [PubMed] [Google Scholar]
- Ward SV, George CX, Welch MJ, Liou LY, Hahm B, Lewicki H, de la Torre JC, Samuel CE, Oldstone MB. 2011. RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc Natl Acad Sci U S A 108(1):331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Yoshimoto K, Tatebe H, Kita M, Nishikura K, Kimura M, Tanaka M. 2013. Enhancement of alcohol drinking in mice depends on alterations in RNA editing of serotonin 2C receptors. Int J Neuropsychopharmacol 17:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weier HU, George CX, Greulich KM, Samuel CE. 1995. The interferon-inducible, double-stranded RNA-specific adenosine deaminase gene (DSRAD) maps to human chromosome 1q21.1–21.2. Genomics 30(2):372–375 [DOI] [PubMed] [Google Scholar]
- Weier HU, George CX, Lersch RA, Breitweser S, Cheng JF, Samuel CE. 2000. Assignment of the RNA-specific adenosine deaminase gene (Adar) to mouse chromosome 3F2 by in situ hybridization. Cytogenet Cell Genet 89(3–4):214–215 [DOI] [PubMed] [Google Scholar]
- Welzel TM, Morgan TR, Bonkovsky HL, Naishadham D, Pfeiffer RM, Wright EC, Hutchinson AA, Crenshaw AT, Bashirova A, Carrington M, Dotrang M, Sterling RK, Lindsay KL, Fontana RJ, Lee WM, Di Bisceglie AM, Ghany MG, Gretch DR, Chanock SJ, Chung RT, O'Brien TR. 2009. Variants in interferon-alpha pathway genes and response to pegylated interferon-Alpha2a plus ribavirin for treatment of chronic hepatitis C virus infection in the hepatitis C antiviral long-term treatment against cirrhosis trial. Hepatology 49(6):1847–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- XuFeng R, Boyer MJ, Shen H, Li Y, Yu H, Gao Y, Yang Q, Wang Q, Cheng T. 2009. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc Natl Acad Sci U S A 106(42):17763–17768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. 2010. Recognition of viral nucleic acids in innate immunity. Rev Med Virol 20(1):4–22 [DOI] [PubMed] [Google Scholar]
- Yu M, Levine SJ. 2011. Toll-like receptor, RIG-I-like receptors and the NLRP3 inflammasome: key modulators of innate immune responses to double-stranded RNA viruses. Cytokine Growth Factor Rev 22(2):63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]