Abstract
Influenza A viruses, which are further subtyped on the basis of antigenic differences in external hemagglutinin and neuraminidase glycoproteins, and influenza B viruses are prominent among the viral causes of respiratory diseases and can cause a wide spectrum of illness. Each year these viruses are responsible for recurrent epidemics, frequently in association with genetic variation. There is a requirement for sensitive and rapid diagnostic techniques in order to improve both the diagnosis of infections and the quality of surveillance systems. A new three-dimensional biochip platform (Flow-Thru Chip; MetriGenix) was used to develop a rapid and reliable molecular method for the typing and subtyping of influenza viruses. Oligonucleotide probes immobilized in microchannels of a silicon wafer were selected to recognize multiple fragments of the influenza A virus matrix protein gene; the influenza B virus NS gene; the H1, H3, and H5 hemagglutinin genes; and the N1 and N2 neuraminidase genes. Biotinylated amplicons resulting from either multiplex or random reverse transcription-PCR were hybridized to arrayed oligonucleotides on the influenza virus chip before they were stained with horseradish peroxidase-streptavidin and were imaged by use of a chemiluminescent substrate. The chip analysis procedure, from the time of pipetting of the sample into the chip cartridge to the time of analysis of the results, was performed in less than 5 h. The random PCR exhibited a higher level of performance than the multiplex PCR in terms of the specificity of product hybridization to the influenza virus chip. Analysis of influenza A viruses (H1N1, H3N2, H1N2, and H5N1) and influenza B viruses showed that this microarray-based method is capable of the rapid and unambiguous identification of all types and subtypes of viruses by use of random PCR products. The redundancy of the probes designed for each gene selected yielded an additional criterion of confidence for the subtyping of viruses which are known for antigenic variations in some of their components.
The concept of fabricating miniaturized molecular assay systems that are capable of conducting thousands of analyses simultaneously led to the development of the DNA microarray, or DNA chip, technology in the 1990s (22). This technology found immediate applications in a wide range of fields, including monitoring of gene expression, evaluation of gene function, gene sequencing, drug evaluation, and more recently, the detection of bacterial or viral pathogens. While numerous differences appeared as a function of the chip substrate (nylon membrane, glass, or silicon), probes (cDNA or oligonucleotides), and probe deposition method (photolithography, mechanical microspotting, ink spotting, etc.) used as well as the number of probes arrayed per chip (from the 10s [low-density microarrays] to 10s of thousands to hundreds of thousands [high-density microarrays]), a feature common to all commercialized or in-house DNA chips has been the two-dimensional nature of the format. A novel platform was recently introduced in which molecular interactions occur within the three-dimensional volumes of ordered microchannels rather than at a two-dimensional surface. The technology has been termed the Flow-Thru Chip (FTC) (2) and has primarily been applied to profiling of the expression of genes involved in cancer, inflammation, proliferation, and cell signaling, to name a few.
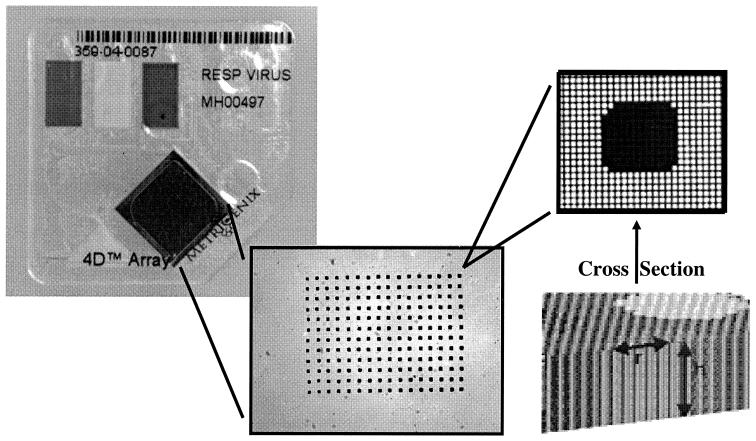
FTC is a microarray substrate that comprises more than 700,000 uniform microchannels per square centimeter. Each microchannel has a diameter of 10 μm and a length of 450 μm. When probes, such as oligonucleotides, cDNA, antibodies, or proteins, are printed on the FTC, the resulting spots include about 100 of the individual microchannels, with a total surface area for probe binding that is 100 times greater than that achieved on an impermeable, two-dimensional surface. Accordingly, FTC provides a greater binding capacity for targets than a flat chip, but the microchannel substrate geometry provides other benefits as well. First, the hybridization reaction is enhanced by the dimensionally favorable microenvironment of the microchannel. The diffusion of targets to probes on the chip substrate wall is rapid, on the order of seconds, in the 10-μm microchannels of the FTC, whereas the diffusion distances on a flat chip can be on the order of millimeters. Second, because the FTC structure is reminiscent of that of a filter, the chip can be embedded in a fluidics system in an analogous manner. The result is a disposable cartridge unit that accepts all of the reagents necessary to complete an assay in an easy-to-use format. Figure 1 provides an image of an FTC in the MetriGenix 4D cartridge, along with a magnified view of a spot.
FIG. 1.
MetriGenix 4D cartridge with a ×10 zoom of the array on the FTC and ×100 zoom on an individual spot. Each spot is roughly 140 μm in diameter on a 350-μm center-to-center spacing. Spots appear dark after printing because the printing solution fills the microchannels.
The FTC technology has displayed advantages in terms of its greater binding capacity, reduced hybridization times, and the reduced sample amounts required when it is applied to gene expression analysis. These advantages distinguish the platform as a promising tool in the development of a new method for the diagnosis of respiratory viral infections. During the last decade, numerous molecular methods for the detection of influenza viruses and other respiratory viruses have been published, and the majority of these have been based on PCR technology (e.g., PCR, PCR-enzyme immunoassay, and multiplex PCR [MPCR]). Due to the difficulties with the design of compatible multiplex primer sets and the confirmation of amplicon identity by conventional methods, the maximum number of viruses detectable in a single assay has been quite limited. In order to solve such a problem, data on the use of microarrays for respiratory virus identification (16, 28) have started to appear.
We developed an Influenza Chip for the typing and subtyping of influenza viruses as a model for analyzing the suitability of use of the FTC array for the further development of a larger respiratory chip; indeed, because symptoms clinically indistinguishable from those associated with true influenza can result from infection with other respiratory viral pathogens, the latter must be tested in parallel, implying the spotting of selected corresponding probes. Several oligonucleotide probes specific for each selected influenza virus gene were designed in order to increase the confidence in viral identification. Two different protocols, an MPCR and a random PCR, were developed for target amplification and are discussed in terms of the specificities of amplicon hybridization to the chip and the ease of use for the screening of larger numbers of respiratory viruses.
MATERIALS AND METHODS
Chip design. (i) Chip substrate.
A uniformly porous silicon substrate (Infineon, Munich, Germany) was used in place of a flat, impenetrable material. The pores of the microchannels connect the upper and the lower faces of the chip in such a manner that the fluid can flow through the chip. The microchannels are roughly 9-μm-diameter-square pores on a 12-μm center-to-center spacing, yielding a porosity of 56% and a total surface area of 100 cm2 per chip (1 by 1 cm). The silicon wafer is coated with a thin film of epoxysilane to promote probe adhesion.
(ii) Probe design.
Twenty-nine oligonucleotide probes (45 to 65 bp) were designed for recognition of seven different genes: the matrix protein (MP) gene of influenza A virus (MP/A); the nonstructural (NS) gene of influenza B virus (NS/B); hemagglutinin genes H1, H3, H5; and neuraminidase genes N1 and N2. Highly conserved viral sequences were obtained from the National Center for Biotechnology Information (NCBI) database; these were each segmented into two to seven overlapping pieces. The length of the overlapping domain between two consecutive gene segments was generally 10 to 15 bp, but exceptionally shorter (5 bp) or longer (25 bp) lengths were designed as a function of the sizes of the specific gene segments. Most of probes exhibited a 40 to 50% G+C content and melting temperatures (Tms) ranging from 65 to 70°C (Table 1).
TABLE 1.
Design of oligonucleotide probes
| Probe | Standard virus | Accession no. | Gene location | Probe length (mer) | G+C content (%) | Tm (°C) |
|---|---|---|---|---|---|---|
| MP/A-2 | A/PR/8/34 | ISDN13425 | 141-200 | 60 | 47 | 68 |
| MP/A-3 | A/PR/8/34 | 191-250 | 60 | 53 | 70 | |
| MP/A-4 | A/PR/8/34 | 241-300 | 60 | 48 | 68 | |
| MP/A-5 | A/PR/8/34 | 291-350 | 60 | 37 | 67 | |
| MP/A-6 | A/PR/8/34 | 341-400 | 60 | 47 | 68 | |
| NS/B-1 | B/Yamanashi/166/98 | AF100410 | 24-80 | 57 | 47 | 77 |
| NS/B-2 | B/Yamanashi/166/98 | 56-106 | 51 | 41 | 64 | |
| NS/B-3 | B/Yamanashi/166/98 | 81-132 | 52 | 50 | 67 | |
| H1-1 | A/Taiwan/1/86 | D00407 | 490-548 | 59 | 45 | 67 |
| H1-2 | A/Taiwan/1/86 | 539-598 | 60 | 43 | 66 | |
| H1-3 | A/Taiwan/1/86 | 894-655 | 62 | 45 | 67 | |
| H3-1 | A/Sydney/05/97 | ISDNASYD97 | 272-330 | 59 | 49 | 69 |
| H3-2 | A/Sydney/05/97 | 321-380 | 60 | 47 | 68 | |
| H3-3 | A/Sydney/05/97 | 366-425 | 65 | 43 | 67 | |
| H3-4 | A/Sydney/05/97 | 416-465 | 50 | 32 | 60 | |
| H3-5 | A/Sydney/05/97 | 455-503 | 49 | 35 | 60 | |
| H5-1 | A/Hong Kong/156/97 | AF046096 | 894-950 | 57 | 47 | 68 |
| H5-2 | A/Hong Kong/156/97 | 941-1000 | 60 | 50 | 69 | |
| H5-3 | A/Hong Kong/156/97 | 991-1050 | 60 | 47 | 68 | |
| H5-4 | A/Hong Kong/156/97 | 1041-1100 | 60 | 45 | 67 | |
| H5-5 | A/Hong Kong/156/97 | 1091-1150 | 60 | 52 | 70 | |
| H5-6 | A/Hong Kong/156/97 | 1141-1200 | 60 | 45 | 67 | |
| H5-7 | A/Hong Kong/156/97 | 1191-1245 | 55 | 40 | 64 | |
| N1-1 | A/Chile/1/83 | X15281 | 1059-1118 | 62 | 40 | 65 |
| N1-2 | A/Chile/1/83 | 1111-1165 | 55 | 44 | 65 | |
| N2-1 | A/Johannesburg/33/94 | U43425 | 779-839 | 62 | 48 | 69 |
| N2-2 | A/Johannesburg/33/94 | 821-880 | 60 | 49 | 69 | |
| N2-3 | A/Johannesburg/33/94 | 861-920 | 60 | 44 | 67 | |
| N2-4 | A/Johannesburg/33/94 | 911-955 | 45 | 29 | 58 |
(iii) Synthesis of probes and spotting of the probes onto chips.
Oligonucleotide probes were synthesized by using standard phosphoramidite chemistry (Glen Research, Sterling, Va.) in a 96-well format (PolyPlex; Gene Machines, San Carlos, Calif.). Oligonucleotides were eluted in 5% Tris-EDTA-acetate, dried, and resuspended in 250 μl of molecular biology-grade water (Quality Biological, Gaithersburg, Md.). Probe quality was determined by capillary electrophoresis (P/ACE MDQ; Beckman Coulter, Fullerton, Calif.) and determination of the concentration by measurement of the optical density at 260 nm (Saffire; Tecan, Reading, United Kingdom). Probes at a concentration of 3 μM each in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) were printed on the inner walls of the microchannels with an Omnigrid Array (Gene Machines) and a Stealth 4B pin (Telechem, Sunnyvale, Calif.). Spots were printed on individual chips in an 8-by-12 format at a 350-μm pitch. Each spot was roughly 120 μm in diameter and included about 80 discrete microchannels.
(iv) Microarray platform.
Printed FTC Influenza Chips (MetriGenix) were sealed inside a polystyrene cartridge with a biocompatible adhesive (MedTec, Chapel Hill, N.C.). The FTC cartridge is an integrated part of the MetriGenix 4D system for microarray analysis. The cartridge interfaces with both an automated assay station (MGX 2000; MetriGenix) and a chip reader (MGX 1200CL; MetriGenix). The cartridge contains reservoirs for three assay reagents (sample, stain, and blocker) and ports that accept 100- to 300-μl pipette tips. All assay reagents either are added to the cartridge reservoirs or are available from the assay controller, and all experimental parameters (operation times, temperatures, and fluid flow rates) are controlled by the automated assay station (MGX 2000). In-depth technical information as well as a process animation can be found on the MetriGenix website.
(v) Reagents.
The reagents necessary for sample hybridization included hybridization and wash buffers, a sample dilution buffer, stain, blocker, and a spike-in mixture. Buffer 1 (washing buffer) consisted of a buffered solution containing NaCl, NaH2PO4, Triton X-100, and EDTA. Buffer 2 (hybridization buffer) was a buffered solution containing morpholineethanesulfonic acid, formamide, EDTA, sarcosine, and NaCl. The sample dilution buffer was buffer 2, to which sheared herring sperm DNA (Promega, Madison, Wis.) was added at 1 μg/μl. The stain was streptavidin-horseradish peroxidase (HRP; Pierce, Rockford, Ill.), which was used at 5 μg/ml in 1× SSC. The blocker was 8% (by mass) goat serum in 1× SSC. The spike-in mixture was a buffered solution containing three synthetic oligonucleotides (Arabidopsis thalianca cDNA) at different concentrations and herring and salmon sperm DNA. On chips, as a function of their low, medium, and high concentrations in spike-in control, these three different nucleotides give low, medium, and high signal intensities, respectively, with matched probes. The three biotinylated synthetic targets in the spike-in mixture were complementary to probes present on the array and were included in the hybridization buffer at 20 nM. The reagents used for image capture included enhanced luminol and peroxide solutions (MetriGenix).
Influenza virus strains and isolates.
Four prototype human influenza virus strains were used in the study: Sydney/05/97 (A/H3/N2), PR/8/34 (A/H1N1), B/Yamanashi/166/98, and Hong Kong/156/97 (A/H5/N1). All of them were propagated in MDCK cells adapted to grow in serum-free medium (Ultra-MDCK; Cambrex Bioscience, Walkersville, Md.).
Clinical samples were received at the World Health Organization National Influenza Centre. They were used for amplification of virus isolates after inoculation of clinical samples into MDCK cells.
RNA isolation.
Viral RNA was extracted by the single-step method of Chomczynski and Sacchi (8), subsequently modified by replacing the guanidinium thiocyanate-phenol-chloroform mixture by the Tri-Reagent (Sigma Aldrich, St. Louis, Mo.). Briefly, 100 μl of an infected cell culture supernatant was pooled with 500 μl of Tri-Reagent and then mixed with 200 μl of chloroform. After 15 min of centrifugation at 12,000 × g, the aqueous phase was transferred to a tube containing isopropyl alcohol and the tube was incubated overnight at −20°C. After 30 min of centrifugation at 12,000 × g, the pellet containing RNA was washed with 1 ml of 75% ethanol and was then dissolved in 20 μl of nuclease-free water. The RNA yield obtained from 100 μl of cell culture supernatant ranged from 360 to 600 ng when a prototype strain was used and from 150 to 1,400 ng when a clinical isolate was used.
Viral RNA amplification.
Two different protocols were used for RNA transcription and amplification, based on the use of either specific or random primers.
(i) One-step RT-MPCR.
The primers used for amplification of conserved regions on the different genes were those mentioned by Poddar (19). The amplicon sizes for our prototype strains were 311 bp for MP/A (A/PR/8/34), 109 bp for NS (B/Yamanashi/166/98), 166 bp for H1 (A/Taiwan/118/96), 230 bp for H3 (A/Sydney/05/97), 352 bp for H5 (A/HongKong/481/97), 106 bp for N1 (A/Chile/1/83), and 176 bp for N2 (A/Johannesburg/33/94). Forward primers were unlabeled, while reverse primers were biotinylated (MetriGenix).
Reverse transcription (RT)-MPCR was performed with the RT-PCR one-step kit (Qiagen, Valencia, Calif.). Ten microliters of extracted RNA at a 1/10 dilution (prototype strains) or undiluted (isolates and clinical samples) was mixed with 25 μl of a mixture of seven forward primers and seven reverse primers in sterile nuclease-free water. The primers specific for MP/A, H5, and N2 were used at a final concentration of 0.1 μM each; the primers specific for NS/B, H1, and N1 were used at final concentration of 0.2 μM each; and the primers specific for H3 were used at a final at concentration of 0.225 μM each. After 5 min at 90°C and subsequent cooling in ice, the samples were mixed with 15 μl of a mixture containing 10 μl of 1× RT-PCR buffer, 2 μl of deoxynucleoside triphosphates (final concentration, 400 μM each), 2 μl of an enzyme mixture (Omniscript and Sensiscript RNA polymerase plus HotStar Taq DNA polymerase), and 1 μl of RNase inhibitor. After a precycle of 30 min at 50°C for RT, followed by 15 min at 90°C (for inhibition of RNA polymerase and activation of the HotStar polymerase), samples were submitted to 30 (prototype strains) or 40 (isolates) cycles of amplification in an I-Cycler (Bio-Rad Laboratories, Richmond, Calif.). Each cycle consisted of 94°C for 15 s, 56°C for 25 s, and 72°C for 60 s. Finally, an additional cycle of 72°C for 7 min was performed to achieve good extension of all DNA strands.
(ii) Random RT-PCR.
Viral samples were amplified by a modified version of a random PCR protocol (28). At the time that this report was prepared, similar modifications to this protocol were reported by Rota et al. (20) for severe acute respiratory syndrome coronavirus RNA amplification.
Ten microliters (75 to 300 ng of RNA) of total RNA was reverse transcribed by using primer A (5′-GTT-TCC-CCA-GTC-ACG-ATC-NNN-NNN-NNN; 40 pmol/μl; Eurogentec, Liege, Belgium) and avian myeloblastosis virus reverse transcriptase (10 U/μl; Promega). After 60 min at 37°C and 5 min at 95°C, the product was chilled to 4°C and was then amplified by PCR. Thirty-five cycles of amplification in the thermocycler were performed with primer B (5′GTT-TCC-CAG-TCA-CGA-TC; 100 pM/μl; Eurogentec) in a biotinylated form and Taq polymerase (5 U/μl; Perkin-Elmer, Inc., Norwalk, Conn.). Each cycle consisted of 94°C for 30 s, 40°C for 30 s, 50°C for 30 s, and 72°C for 160 s. After an additional 7 min at 72°C, the amplified product was chilled to 4°C.
Purification and gel analysis of amplicons.
Both RT-MPCR and random RT-PCR products were purified, unless mentioned otherwise, with the DNA Clean & Concentrator 5 kit (Zymo Research Inc., Orange, Calif.) prior to hybridization on an Influenza Chip. For specified experiments, the RT-MPCR product was run on agarose gels; specific bands were cut, pooled, and extracted with the QIAQuick Gel Extraction kit (Qiagen); and the resulting product was then used for hybridization to the arrays.
Crude and/or purified amplicons derived from either RT-MPCR or random RT-PCR were separated on agarose gels (2%) containing 0.5 μg of ethidium bromide per ml. After migration, the DNA bands or DNA smears were visualized by UV transillumination.
Microarray hybridization and image analysis.
Hybridization of the amplified products to the Influenza Chips was performed according to the recommendations of the manufacturer. Prior to hybridization, the product from the amplified sample was mixed with spike-in controls (for sample normalization) in sample dilution buffer and denatured for 5 min at 90°C. The blocking reagent, the sample, and HRP-streptavidin staining were distributed at a volume of 68 μl each into the corresponding reservoirs of the chip cartridge. The hybridization fluid station (MGX 2000) automatically controlled all subsequent steps (blocking and buffer flushes, hybridization time, and temperature). Four hours after the start of the hybridization process (2 h for hybridization to the corresponding probes and 2 h for the different steps before and after hybridization, including blocking, washing, and staining of the reactive spots with HRP-streptavidin), the cartridge was removed from the MGX 2000 instrument and placed in the MGX 1200CL detection unit for chemiluminescence (CL) detection (5). During detection, a continuous flow of the luminol and peroxide mixture was delivered with a syringe pump at a rate of 600 μl min−1, with exposure times usually ranging from 2 to 5 s. The CL substrate was directed to flow through the chip by the detection unit for 15 s prior to image capture to ensure stable luminescence intensity. An image was captured with a charge-coupled device camera (Hamamatsu, Hamamatsu City, Japan), and subsequent image analysis was performed with custom software (MetriSoft; MetriGenix). Normalization of the spot signal values was performed automatically by comparison with the values obtained with spike-in control. For each probe-target hybridization, the normalized signal value was the ratio between the number of relative light units obtained with the probe and the number obtained with the oligonucleotide from spike-in control giving the medium signal intensity value.
RESULTS
Hybridization of DNA derived from single-step RT-MPCR for influenza virus.
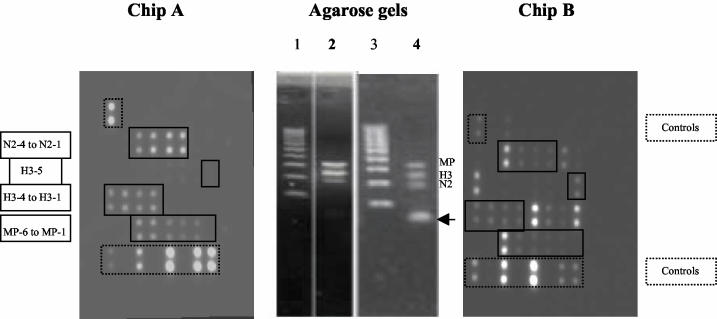
Our first approach for evaluating the microarray for the typing and subtyping of influenza viruses was to amplify viral RNA by RT-MPCR. For this aim, A/Sydney/05/97 (H3N2) virus was amplified under two different experimental conditions: (i) multiplex subtyping conditions with three primer pairs (primer pairs specific for H3, N2, and MP/A) and (ii) multiplex typing and subtyping conditions with seven primer pairs (primer pairs specific for H3, H1, H5, N1, N2, MP/A, and NS/B). As shown in Fig. 2, both multiplex conditions resulted in three DNA bands of 311, 206, and 176 bp on agarose gels, corresponding to the expected amplification products of the MP/A, H3, and N2 genes, respectively. Hybridization to the microarrays of nonpurified influenza virus derived from RT-MPCR with three primer pairs (Fig. 2, chip A) resulted in specific hybridization to the expected spots corresponding to the probes specific for H3, N2, and MP/A. On the contrary, hybridization of nonpurified products derived from RT-MPCR with seven primer pairs (Fig. 2, chip B) resulted in specific hybridization to the expected spots and nonspecific hybridization to heterologous probes. Agarose gels revealed that the amplicons resulting from RT-MPCR with the seven primers showed the presence of primers-dimers (Fig. 2). Primers-dimers were shown to be highly dependent on the number of primers used during gene amplification and are thus believed to be responsible for the nonspecific hybridization observed on chip B. In order to solve this problem, the RT-MPCR products were purified with different systems, such as the Qiaquick PCR purification kit (100 bp to 10 kb) from Qiagen or the DNA Clean & Concentrator 5 kit from Zymo Research Inc. The purified RT-MPCR products on agarose gels (Fig. 3) confirmed the rather complete elimination of primers-dimers, especially when the Zymo Research kit was used; but nonspecific spots still persisted after hybridization of the purified products to the Influenza Chips, although they were reduced in number (Fig. 3, chip B). In order to confirm the role of primers-dimers in such a heterospecific hybridization, the three bands corresponding to the MP/A, H3, and N2 gene amplification products were pooled and extracted from the agarose gel with a Qiaquick Gel Extraction kit (Qiagen). As shown in Fig. 3, the extracted DNA was completely devoid of primers-dimers when it was analyzed on agarose gels and showed specific hybridization to the expected A/H3N2-specific probes (probes specific for MP/A, H3, and N2) on the Influenza Chip (Fig. 3, chip A).
FIG. 2.
Chip A was processed with amplified, nonpurified products derived from RT-MPCR of A/Sydney/1/97 with three primer pairs (primers specific for MP/A, H3, and N2); as shown in the agarose gel control lane (lane 2), no band of primers-dimers was present for the amplicons, and only the spots expected for hybridization with MP/A, H3, and N2 were found on the chip. Chip B was processed with amplified, nonpurified product derived from RT-MPCR of A/Sydney/1/97 with seven primer pairs (primers specific for MP/A, NS/B, H1, H3, H5, N1, and N2); as shown on the agarose gel control lane (lane 4), a strong band of primers-dimers (arrow) was present in amplicons and numerous nonhomologous probes hybridized on the chip. Lanes 1 and 3, Hi-Lo DNA size markers (100-bp DNA ladder; Cambrex Bioscience).
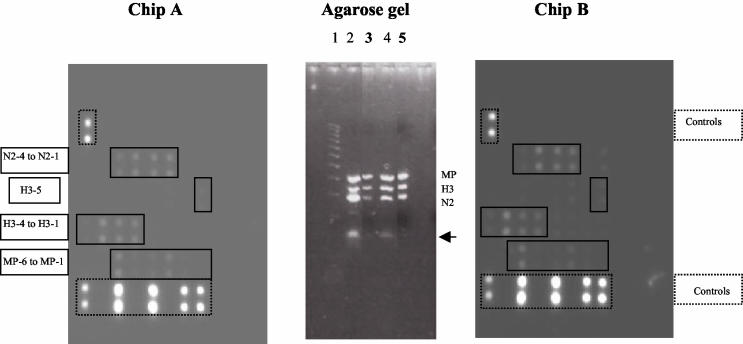
FIG. 3.
Chip A was processed with amplified products derived from RT-MPCR of A/Sydney/1/97 (H3N2) with seven primer pairs (primers specific for MP/A, NS/B, H1, H3, H5, N1, and N2) and then purified after gel extraction. As shown on the agarose gel, the extracted product (lane 3) was devoid of primers-dimers, whereas the nonpurified product was not (lane 2). Only the probes specific for the expected MP/A, H3, and N2 genes were hybridized. Chip B was processed with amplified product derived from RT-MPCR with A/Sydney/1/97 (H3N2) with seven primer pairs (primers specific for MP/A, NS/B, H1, H3, H5, N1, and N2) and then purified with the Zymo Research purification kit. Agarose gel electrophoresis of the purified product (lane 5) showed limited primers-dimers compared to the numbers of primers-dimers for the nonpurified product (lane 4); as a consequence, chip B showed greater specificity. The arrow indicates bands corresponding to primers-dimers. Lanes 1 and 3, Hi-Lo DNA size markers (100-bp DNA ladder; Cambrex Bioscience).
Three different hybridization temperatures (37, 42, and 55°C) were tested for comparative evaluation of both specific and nonspecific hybridization to oligonucleotide probes. Hybridization at 42°C was found to be optimal when both the numbers (Table 2), and the normalized signal values of spots (data not shown) were considered. Indeed, an increase in the hybridization temperature to 55°C resulted in a reduced number of spots with nonspecific labeling compared with that at 42°C (two and four probes, respectively), but the signal values with homologous probes were found to be 5 to 10 times lower (data not shown).
TABLE 2.
Influence of hybridization temperature on recognition of specific complementary probes by A/Sydney/05/97 DNA from RT-MPCR
| Probe | No. of chips probe positive/no. of chips processed at the following hybridization temp (°C):
|
||
|---|---|---|---|
| 37 | 42 | 55 | |
| N2-1 | 2/2 | 4/4 | 4/6 |
| N2-2 | 2/2 | 4/4 | 4/6 |
| N2-3 | 2/2 | 4/4 | 6/6 |
| N2-4 | 2/2 | 4/4 | 6/6 |
| H3-1 | 2/3 | 4/4 | 4/6 |
| H3-2 | 3/3 | 4/4 | 4/6 |
| H3-3 | 2/3 | 4/4 | 5/6 |
| H3-4 | 3/3 | 4/4 | 2/6 |
| H3-5 | 3/3 | 4/4 | 5/6 |
| MP-1 | 1/2 | 1/4 | 0/6 |
| MP-2 | 0/2 | 1/4 | 0/6 |
| MP-3 | 0/2 | 3/4 | 5/6 |
| MP-4 | 1/2 | 4/4 | 0/6 |
| MP-5 | 2/2 | 4/4 | 2/6 |
| MP-6 | 2/2 | 4/4 | 6/6 |
The demonstration of both the residual presence of primers-dimers in purified RT-MPCR products and their role in nonspecific hybridization to the Influenza Chip prompted a switch to a random RT-PCR protocol for amplification of influenza virus from samples prior to analysis. The random RT-PCR protocol also has the advantage of requiring only a single primer pair for all of the strains on the Influenza Chip and may be suitable for use with a more comprehensive respiratory virus panel as well.
Hybridization of DNA derived from random RT-PCR. (i) Evaluation of random RT-PCR protocol.
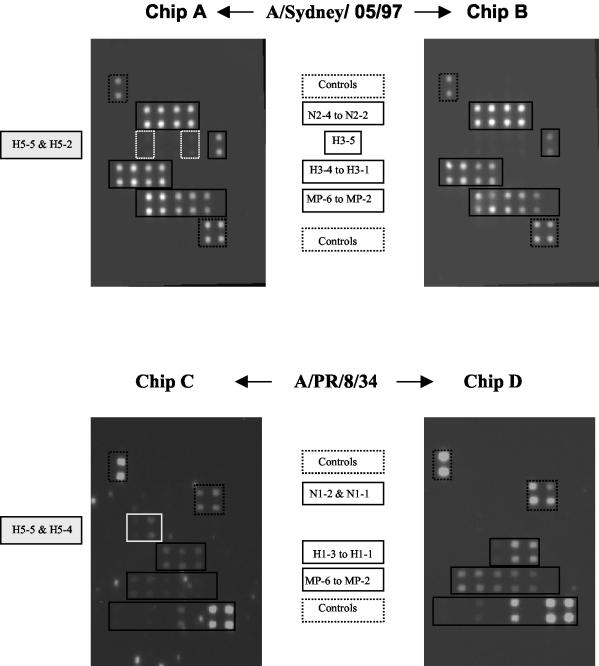
Influenza viruses A/PR/8/34 (H1N1) and A/Sydney/05/97 (H3N2) were used to validate the random RT-PCR as the protocol selected for amplification of influenza virus RNA and subsequent analysis on the microarrays. From the previous observations with amplified products from RT-MPCR, hybridization to the oligonucleotide probes was performed at 42°C. Purified and nonpurified products from the amplicons of both A/PR/8/34 (H1N1) and A/Sydney/05/97 (H3N2) were hybridized to the chips in parallel. As shown in Fig. 4, purified products from random RT-PCR amplification of RNA from both A/Sydney/05/97 (H3N2) (chip B) and A/PR/8/34 (H1N1) (chip D) hybridized specifically to the expected probes: MP/A, H3, and N2 for the former and MP/A, H1, and N1 for the latter. All probes specific for the H1, H3, N1, N2, and MP/A genes were recognized by the corresponding amplicons. When the amplified products were used in the nonpurified form, products amplified from A/H3N2 (chip A) and A/H1N1 (chip C) reacted with homologous immobilized probes in a manner similar to that for the purified products, but in addition, they both exhibited low degrees of cross-reactivity with some H5-specific probes (probes H5-2 and H5-5 with the A/H3N2 amplicons, probes H5-5 and H5-4 with the A/H1N1 amplicons). As a function of the storage conditions of the amplified products (4°C or frozen) and the ages of the reagents, some bright “specks” could be observed after the images of chips processed with nonpurified products were captured. Such bright contaminants could easily be distinguished from hybridized spots due to differences in size and shape, but they were shown to be capable of interfering with the quantitative analysis of the signal if they were located on the spotted area of the chip. As a consequence, we favored the use of purified random RT-PCR products for further experiments.
FIG. 4.
Chips A and B were processed with amplified products derived from random RT-PCR of A/Sydney/05/97, and nonpurified (chip A) and purified (chip B) products were used. Purified amplicons exhibited specific reactivities with the expected probes (H3-, N2-, and MP/A-specific probes), while nonpurified products showed additional reactivities with probes H5-5 and H5-2. Chips C and D were processed with amplified products derived from random RT-PCR of A/PR/8/34, and both nonpurified (chip C) and purified (chip D) products were used. Purified amplicons exhibited specific reactivities with the expected probes (H1-, N1-, and MP/A-specific probes), while nonpurified products showed additional reactivities with probes H5-5 and H5-4.
(ii) Typing-subtyping of influenza viruses on MetriGenix 4D influenza virus microarrays.
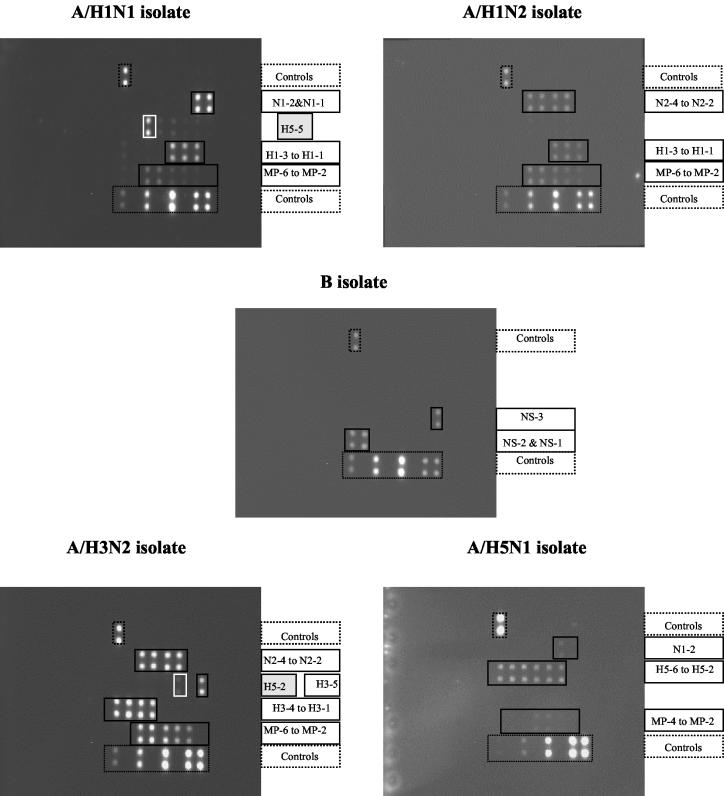
The random RT-PCR protocol was applied to amplification of viral RNA purified from influenza virus isolates identified in our laboratory as A/H1N1 (two isolates), A/H3N2 (two isolates), A/H1N2 (two isolates), and B (two isolates) viruses by hemagglutination and neuraminidase inhibition tests as well as RT-MPCR; in addition, A/Hong Kong/156/97 (H5N1) virus was analyzed in this experiment. As shown in Fig. 5, specific hybridizations between biotin-labeled viral DNAs and the corresponding probes spotted on the chip were observed; the reactivities of all the different spotted probes with virus amplicons are shown in Table 3. No nonspecific spots resulting from hybridization of viral amplicons with heterologous probes were observed, with the exception that low cross-reactivity between some isolates and an H5-specific probe(s) was observed. However, the heterologous binding to only one of the six H5-specific probes did not complicate identification of a viral type and subtype because identifications were based on a positive response by a majority of the probes. The identities of all influenza virus types and subtypes identified previously were confirmed without any ambiguity by hybridization to the MetriGenix 4D influenza virus microarray.
FIG. 5.
Influenza Chips were processed with DNA obtained from A/H1N1, A/H1N2, B, A/H3N2, and A/H5N1 isolates by the random RT-PCR protocol. Specific hybridization was observed for the product from each isolate. Nonspecific hybridization of H5-specific heterologous probes with A/H1N1and A/H3N2 was observed: probe H5-5 for the former and probe H5-2 for the latter.
TABLE 3.
Reactivities of DNAs from influenza virus isolates with homologous and heterologous probes
| Probe | Reactivity of the following influenza isolates with the indicated probea:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| A/H5N1 HK/156/97 | A/H1N1
|
A/H1N2
|
A/H3N2
|
B
|
|||||
| Ly/168/03 | Ly/210/03 | Ly/1723/02 | Ly/26613/03 | Ly/826/03 | Ly/841/03 | Ly/1760/02 | Ly/372/03 | ||
| H1-1 | − | + | +++ | ++ | ++ | − | − | − | − |
| H1-2 | − | + | +++ | ++ | ++ | − | − | − | − |
| H1-3 | − | + | +++ | ++ | ++ | − | − | − | − |
| H3-1 | − | − | − | − | − | +++ | ++++ | − | − |
| H3-2 | − | − | − | − | − | ++ | ++++ | − | − |
| H3-3 | − | − | − | − | − | +++ | ++++ | − | − |
| H3-4 | − | − | − | − | − | ++ | ++++ | − | − |
| H3-5 | − | − | − | − | − | − | +++ | − | − |
| H5-1 | ++++ | − | − | − | − | − | − | − | − |
| H5-2 | ++++ | − | − | + | − | − | + | − | − |
| H5-3 | ++++ | − | + | − | − | − | − | − | − |
| H5-4 | ++++ | − | + | − | − | − | − | − | + |
| H5-5 | ++++ | + | +++ | − | − | − | − | − | − |
| H5-6 | +++ | − | |||||||
| N1-1 | ++ | + | ++++ | − | − | − | − | − | − |
| N1-2 | − | + | ++++ | − | − | − | − | − | − |
| N2-1 | − | − | − | +++ | ++ | ++++ | ++ | − | − |
| N2-2 | − | − | − | +++ | ++ | ++++ | ++ | − | − |
| N2-3 | − | − | − | +++ | ++ | +++ | ++ | − | − |
| N2-4 | − | − | − | ++ | ++ | +++ | ++ | − | − |
| MP-2 | + | − | − | − | + | + | ++ | − | − |
| MP-3 | ++ | + | + | + | + | + | +++ | − | − |
| MP-4 | ++ | + | + | + | ++ | ++ | +++ | − | − |
| MP-5 | − | − | ++ | + | ++ | +++ | ++++ | − | − |
| MP-6 | − | + | ++ | + | ++ | +++ | ++++ | − | − |
| NS-1 | − | − | − | − | − | − | − | ++++ | ++ |
| NS-2 | − | − | − | − | − | − | − | ++++ | ++ |
| NS-3 | − | − | − | − | − | − | − | ++++ | ++ |
++++, signal value > I; +++, 0.5 < signal value < 1; ++, 0.25 < signal value < 0.5; +, 0.05 < signal value < 0.25; −, signal value < 0.05; boldface, heterologous binding.
(iii) Sensitivity of MetriGenix 4D influenza virus microarray.
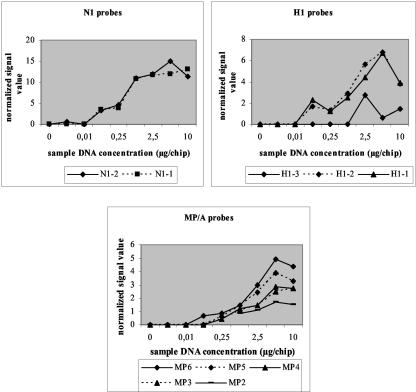
The sensitivity of the MetriGenix 4D influenza virus microarray for typing and subtyping of influenza viruses was tested with A/Pr/8/34 DNA at concentrations ranging from 0.001 to 10 μg/chip. Random RT-PCR allowed the amplification of all genes in the sample (100 to 300 μg of total DNA per reaction mixture), and it was not possible to estimate the number of copies of individual targets in the DNA sample loaded on the chips. All signal values on the chips were normalized to the mean value obtained with the two spots hybridized with the oligonucleotide in spike-in control chosen to get the medium effect. With a threshold factor of 3 (blank value × 3), the mean threshold value for the chips was 2,750 ± 234 relative light units. The normalized signal values obtained with the H1-, N1-, and MP/A-specific probes as a function of the DNA concentration of the sample that hybridized to the chips are plotted in Fig. 6. A number of conclusions emerged from this study: (i) the smallest amount of DNA capable of producing a positive signal ranged from 0.01 to 0.1 μg/chip (equivalent to 102 to 103 infectious particles); (ii) differences in probe reactivities were observed; some probes, like N1-specific probes, exhibited high reactivities, as demonstrated by the high signal values observed with low target DNA concentrations (0.1 μg), while others (probes H1-3 and MP-2) exhibited low reactivities; and (iii) the optimal amount of amplified DNA product for hybridization to the Influenza Chip was 5 μg/chip, independent of the probe considered; an almost linear increase in signal was observed for DNA concentrations ranging from 0.01 to 5 μg/chip.
FIG. 6.
Sensitivity of the Influenza Chip established by using A/PR/8/34 DNA concentrations ranging from 0.001 to 10 μg/chip.
(iv) Specificity of MetriGenix 4D influenza virus microarray.
The specificity of influenza virus detection by the approach that combines random RT-PCR and the Influenza Chip was evaluated by analyzing the results in terms of the hybridization of both homologous (specific) and heterologous (nonspecific) probes to viral DNA. The results in Table 4 were derived from the analysis of 37 chips processed with DNA from different influenza virus types and subtypes (H1N1, H3N2, H1N2, H5N1, and B). Two groups of chips were analyzed separately due to differences in the washing conditions during the last step of the hybridization program. The volume and flow of buffer 1 were 2,000 μl and 500 μl/min, respectively, for chips in series I and 3,000 μl and 750 μl/min, respectively, for chips in series II. Two points must be emphasized: (i) increases in both the volume and the flow of the final wash significantly increased, by up to 60%, the number of chips on which specific hybridization to the expected spots was shown due to hybridization of viral DNA to the corresponding homologous probes, and (ii) probes showing cross-reactivity with nonmatched DNAs were exclusively H5-specific probes (probes H5-2, H5-3, H5-4, and H5-5), with probe H5-4 most often or exclusively reacting as a function of the viral target and the washing conditions. Indeed, increased flushing of the chips with washing buffer (series II experiments) was shown to decrease both the number of chips with unrelated spots and the number of unrelated spots per chip. The question of a potential relationship between the amount of sample DNA and the reactivity with heterologous probes arose. To try to answer this question, chips from series II were processed with different concentrations of A/PR/8/34 DNA ranging from 0.1 to 0.6 and 2.5 to 10 μg/chip and analyzed in terms of homologous signal with H1 probes and heterologous signal with H5 probes. As shown in Table 5, an increase in the A/PR/8/34 DNA concentration from 0.1 to 10 μg/chip did not increase the level of heterologous hybridization to probe H5-4, although it did increase the level of homologous binding to H1-specific probes, when the number of chips with heterologous spots (35%) and when the normalized signal intensities of heterologous spots are considered. Due to the number of different genes and probes spotted on the Influenza Chip, the potential cross-reactivity of influenza A and B viruses with probe H5-4 does not interfere with the typing and subtyping results. Another control for the specificity of the protocol with the Influenza Chip and random RT-PCR was performed by mixing virus A/PR/8/34 and coronavirus 229-E prior to RNA extraction. Subsequent amplification and processing on the Influenza Chip showed no heterologous hybridization, with the exception of the previously noted hybridization with probe H5-4 (data not shown).
TABLE 4.
Specificity of MetriGenix 4D microarray for influenza virus
| Chip series and virusa | No. (%) of processed chips:
|
No. of chips with the following no. of heterologous probes/chip:
|
No. of chips with reactivity with heterologous probe:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | With homologous spots only | With homologous and heterologous spots | |||||||
| 1 | >1 | H5-2 | H5-3 | H5-4 | H5-5 | ||||
| Series I | |||||||||
| A/H1N1 | 10 | 3 | 7 | 5 | 2 | 0 | 1 | 6 | 3 |
| A/H1N2 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 |
| A/H3N2 | 5 | 2 | 3 | 2 | 1 | 3 | 0 | 0 | 1 |
| A/H5N1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| B | 3 | 2 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| Total | 21 | 9 (43) | 12 (57) | 8 | 4 | 4 | 2 | 7 | 5 |
| Series II | |||||||||
| A/H1N1 | 14 | 9 (65) | 5 (35) | 5 | 0 | 0 | 0 | 5 | 0 |
The washing conditions for the chips are described in the text (see Results).
TABLE 5.
DNA concentrations of samples and generation of heterologous hybridization in chips
| Homologous H1-specific reactivitya | No. (%) of chips with heterologous probe H5-4 reactivity
|
||
|---|---|---|---|
| No signal | Low signal (≤1) | High signal (>3) | |
| Medium signal | 5 (62) | 3 (38) | 0 (0) |
| High signal | 4 (66) | 1 (17) | 1 (17) |
Reactivity with H1-specific probes H1-1, H1-2, and H1-3. A medium signal was >1 but <3 (normalized signal value). A high signal was >3. The ranges of DNA concentrations were 0.1 to 0.6 μg of DNA/chip for the medium signal and 2.5 to 10 μg of DNA/chip for the high signal.
(v) Interchip reproducibility.
Interchip reproducibility was tested by using virus A/PR/8/34 DNA at a concentration of 0.5 μg/chip. Four chips were processed, and the mean signal values were calculated for each probe. As shown in Table 6, the coefficients of variation for each probe except the N1-specific probes routinely ranged from 10 to 20%; the N1-specific probes exhibited higher normalized signal values and higher coefficients of variation. In addition, the results in Table 6 obtained from hybridization of A/PR/8/34 amplicons showed that perfect probe-target matching (100% homology with MP-specific probes derived from the virus A/PR/8/34 MP gene sequence) and nonperfect probe-target matching (86 to 92% homology with H1-specific probes derived from the virus A/Taiwan/1/86 hemagglutinin gene sequence and 92% homology with N1-specific probes derived from the virus A/Chile/1/83 NA gene sequence) gave similar ranges of normalized signal values. Variability in signal intensity was also noted as a function of the different probes selected from the same gene; for example, probes MP-5 and MP-6 consistently showed higher signals than probes MP-2, MP-3, and MP-4, while the interchip coefficients of variation were similar for all five probes.
TABLE 6.
Interchip reproducibility tested with A/PR/8/34 DNAa
| Probe | Normalized spot signal valueb | SD | CVc (%) |
|---|---|---|---|
| N1-1 | 9.6 | 3.2 | 33.0 |
| N1-2 | 10 | 2.7 | 27.0 |
| H1-1 | 4.8 | 0.8 | 16.8 |
| H1-2 | 5 | 0.8 | 16.0 |
| H1-3 | 2.3 | 0.45 | 19.5 |
| MP-2 | 2.7 | 0.55 | 20.3 |
| MP-3 | 4 | 0.6 | 15.2 |
| MP-4 | 4.8 | 0.75 | 15.6 |
| MP-5 | 8.6 | 0.6 | 10.9 |
| MP-6 | 7.8 | 1.6 | 20.4 |
The DNA concentration was 0.5 μg/chip.
Mean of results from four assays, with two spots per assay.
CV, coefficient of variation.
DISCUSSION
At present there is considerable interest in the development of a rapid and reliable method for the identification of viral respiratory pathogens. Regarding influenza viruses, the segmented nature of their genomes makes reassortment among viruses an important mechanism for generating genetic diversity. Reassortment among influenza A virus subtypes is of particular importance because of their role in the generation of new pandemic strains in humans and generates a need for the development of new assays which are capable of supplying information on the hemagglutinins and neuraminidases of influenza A viruses (10). For this aim, we have developed a genomic strategy for parallel screening of respiratory viruses by focusing presently on the typing and subtyping of influenza viruses.
While DNA arrays are considered promising tools for the genome-based detection of pathogens, they have still found only a very few applications for virus analysis and identification. The DNA array technology has been reported to have been used for the genotyping of human immunodeficiency virus (26, 29), influenza virus (16), respiratory viruses (28), rotavirus (7), and human papillomavirus (1, 12, 15) and for the detection of hepatitis B virus DNA in the sera of patients with chronic hepatitis (14). A common feature of the various types of arrays used by other investigators, whether they are commercially available, such as the human immunodeficiency virus type 1 GeneChip (Affymetrix) or the human papillomavirus DNA chip (Biomedlab), or homemade, is the flat, two-dimensional chip substrate used for probe binding. We have chosen a novel three-dimensional FTC due to its potential for enhanced performance over planar substrates. When FTC is applied to gene expression analysis, it has shown an improved responsiveness and an improved dynamic range due to the increased surface area relative to that for a flat-surface geometry, as well as reduced assay times due to dynamic fluid delivery to the chip (3, 6, 23). Enhanced mass transport leading to shorter hybridization times is the primary advantage of the FTC system in gene expression applications with RNA products transcribed in vitro, so it may be possible that such an advantage was mitigated in the present application by the use of PCR for amplification. Nevertheless, it is necessary to keep in mind that the random RT-PCR protocol is a linear PCR (4) because only the forward primer is available, resulting in a rather moderate level of amplification. The assay time reported in this paper is similar to (21) or less than (17, 28) those applied for influenza virus cDNA detection on two-dimensional microarrays. According to recent results obtained with a later version of the Influenza Chip, which was expanded to include additional respiratory viruses, there is no doubt that the hybridization time can be reduced significantly when prototype strains or isolates obtained from cell culture are studied (data not included here). We have not yet optimized the protocol to shorten the assay time, because our principal concern was to apply such an RT-PCR and microarray technology directly to clinical samples (nasal swabs or throat washings) that might have viral loads lower than those in infected cell culture supernatants. Oligonucleotide sequences of 45 to 65 nucleotides were chosen for spotting on the microarray since it has been shown that no compromise in sensitivity occurs from the use of 50-mer oligonucleotide probes compared to that from the use of PCR-derived probe species (13). Oligonucleotide sense probes were all synthesized with a C-6 amino acid linker modification at the 5′ end to enhance covalent immobilization to the activated silicon surface (6), and their Tms, with a few exceptions, ranged from 65 to 70°C. Several probes specific for each gene were selected in order to increase the chance that they would hybridize to viruses with small variations in the corresponding gene sequence.
Chemiluminescence (CL) detection for hybridization assays on the FTC was chosen due to its inherently low background. CL results in no illumination in the volume of the chip, as is the case with fluorescence; rather, the CL reagents are pumped into the volume of the chip, where they are converted into emitting species by the enzyme-target conjugates on the inner walls of the individual microchannels. Unlike fluorescence, for which an excitation source is needed and nonspecific radiation can be produced, photons are generated only when converted enzymes are present. In addition, the microchannels that traverse the chip and house the capture probes behave much like fiberoptic cables, channeling the light generated within their volumes to the surface of the chip. Finally, enhanced spatial resolution and enhanced image analysis are obtained with flash CL reagents that minimize the diffusion effects associated with glowing CL reagents (6).
Our first approach to the amplification of viral RNA was RT-MPCR. MPCR, which amplifies multiple DNA fragments in one reaction, has found increasing applications for influenza virus RNA amplification (19, 24, 30), with the identification of amplified target gene fragments by agarose gel electrophoresis. It is interesting that for the typing and subtyping of influenza viruses, none of the investigators introduced a complete set of primers (six to seven pairs) in one PCR mixture but divided the primer pairs into two to three sets and performed as many reactions as there were primer sets. In order to optimize our protocol, we performed RT-MPCR by introducing all primer pairs in one reaction mixture. Both subsequent hybridization of the amplicons to the Influenza Chips and migration of the amplicons on agarose gels showed significant differences as a function of the number of primers used in the MPCR. When three primer pairs were used for amplification of virus A/H3N2, the amplicons hybridized exclusively to the expected MP/A-, H3-, and N2-specific probes and only the three corresponding bands were detected on the gel. Unfortunately, when the MPCR was performed with seven primer pairs in order to mimic the analysis of an unknown sample, numerous nonspecific hybridizations were observed on the chips, while an additional low-molecular-weight band, identified as a primer-dimer, appeared on the agarose gels. When we checked for possible primers-dimers using Primo Multiplex (version 3.2) online software, we found that numerous possible dimers could occur when seven primer pairs are introduced into the reaction mixture. Nine potential dimer combinations exhibited a high probability of structure formation on the basis of the high negative values of the free energies (−3 to −6 kcal/mol) of these combinations. Purification of amplicons with different commercial kits was shown to be incapable of completely eliminating primers-dimers and the generation of heterologous specific spots on the chips. In agreement with the data of Kane et al. (13), who worked with 50-mer arrayed probes, an increase in the hybridization temperature to 55°C resulted in a slight improvement in the specificities and a simultaneous decrease in the sensitivities of numerous probes.
In view of the primer-dimer situation and in consideration of the fact that typing and subtyping of influenza virus is the first step in the development of a larger Respiratory Virus Chip, we moved to random RT-PCR for viral RNA amplification. Experiments performed with RNA isolated from prototype strains A/PR/8/34 (an ancient H1N1 strain) and A/Sydney/05/97 (a recent H3N2 strain) confirmed the reactivities and specificities of the probes selected to be arrayed on the flowthrough influenza virus array. Indeed, the gene segments which were chosen for use in the design of the probes belong to virus strains isolated from 1934 (MP/A) to 1997 (H3 and H5). These probes were shown to be very efficient for the hybridization of complementary labeled targets, independent of the differences in the years of isolation of both the probe and the target viral strains. For example, A/Sydney/05/97 amplicons hybridized perfectly to the MP/A-specific probes derived from A/PR/8/34, and A/PR/8/34 amplicons hybridized perfectly to the H1-specific probes derived from A/Taiwan/1/86 and the N1-specific probes derived from A/Chile/1/83. Hybridization to any of the other probes on the array was not detected.
The results obtained by hybridization of the A/H5N1 amplicons were very interesting. A/HongKong/156/97 virus, which was isolated for the first time from a 3-year-old child who died from respiratory failure, was shown to contain gene segments of avian origin exclusively (25). Hemagglutinin gene sequence analysis showed features that are associated with highly pathogenic H5 avian viruses, and neuraminidase gene sequence analysis confirmed the presence of the N1 subtype, whose neuraminidase gene is closely related to that of A/Parrot/Ulster/73 (H7N1). Nucleic acid sequence analysis of genes coding for internal proteins showed that they were closely related to known genes of avian origin. The potential corresponding probes arrayed on the Influenza Chips were of avian origin when it is considered that the H5-specific probe was designed from the hemagglutinin gene sequence of virus A/Hong Kong/156/97 and were of human origin when it is considered that the H1- and N1-specific probes were derived from the sequences of human virus strains A/Taiwan/1/86 and A/Chile/1/83, respectively. A very strong hybridization of A/H5N1 amplicons was observed with all seven probes designed from the virus A/Hong Kong/156/97 hemagglutinin. Failures of hybridization (false-negative results) to N1- and MP/A-specific probes were observed with probe N1-2 and probes MP-5 and MP-6. Alignment of the N1 sequence from virus A/Hong Kong/156/97 and that of probe N1-2 (A/Che/1/83) showed the existence of a break after the first 4 nucleotides of the probe, which aligned with positions 999 to 1003 of the neuraminidase gene of virus A/H5N1, while the remaining part of the probe sequence aligned to positions 1058 to 1108. In addition, 10 substitutions appeared between these two regions. Such a break of 55 nucleotides could be responsible for the absence of hybridization or for the instability of the structure resulting from hybridization of neuraminidase amplicons of virus H5N1 to probe N1-2. The absence of binding of A/H5N1 MP amplicons to probes MP-5 and MP-6 (A/PR/8/34) could be explained by the high number of substitutions on these two segments of the MP gene of A/Hong Kong/156/97. Sequence alignment showed 9 and 12 nucleotide substitutions in the A/H5N1 MP gene segment corresponding to probes MP-5 and MP-6, respectively. A low reactivity of probe N1-1 was also detected after hybridization of the H5N1 amplicons; this could be explained by the high number of base substitutions in the A/Hong Kong/156/97 N1 neuraminidase compared to the sequence of N1 from A/Chile/1/83 in the array. In view of these results, it would be interesting to enlarge the number of influenza virus-specific probes on chips by adding additional hemagglutinin-specific (H9, H7) and neuraminidase-specific (N7) probes, particularly since transmission of subtypes H9N2 and H7N7 from avian species to humans was observed in Hong Kong in 1999 (17, 18) and The Netherlands in 2003 (11), respectively. In all these cases, except for the very limited spread of A/Hong Kong/156/97 (H5N1), there was no evidence of human-to-human spread of virus. The main danger for humans would be the emergence of reassortant viruses and simultaneous infection of humans with both a “human” virus and an “avian” virus, which could result in a true influenza pandemic (5).
Among the influenza virus isolates, the A/H1N1 viruses isolated during 2003 hybridized perfectly to the three H1-specific probes and the two N1-specific probes and hybridized to all MP-specific probes except MP-2. Since the sequences of the MP genes from these isolates were not available, we aligned the MP-2 probe sequence with that of the MP gene of a recent H1N1 strain, A/Saudi Arabia/7971/2000, which was available in the Influenza Sequence Database. A single nucleotide substitution was found; however, a single substitution should not be sufficient to explain the hybridization failure. Two hypotheses could explain such a phenomenon. First, the isolates tested could have larger numbers of substitutions in the MP gene than the isolate whose sequence was available, or second, there could be a problem with the reactivity of probe MP-2. Products amplified from additional A/H3N2 and B isolates hybridized perfectly to the appropriate probes and showed no significant false-positive reactions with the other probes.
Improvement of Influenza Chip specificity was observed when the volume and flow rate of the wash buffer used to wash the arrays after staining were increased, but it is interesting that the specific probes which exhibited cross hybridization with nontarget amplicons independently of the hybridization parameters were H5-specific probes, specifically, probes H5-4 and H5-5. Probe H5-4 frequently hybridized to H1N1 amplicons, and the ratio of the heterologous signal to the homologous signal was shown to be dependent on the target DNA concentration. Ratios of <0.5 were usually observed with target concentrations of less than 1 μg of total DNA, while ratios of ≥1 signaled the use of target DNA in amounts greater than 1 μg. The probe H5-4 and H1 sequences exhibited 84% identity (from a search of the NCBI database with the BLAST2 algorithm) and 11 continuous oligonucleotides, which could explain the heterologous binding of A/H1N1 amplicons to probe H5-4. Very few chips of series I showed heterologous reactivities of H1 and probe H5-5, and no such reactivity was observed with chips from series II. No significant similarity was found between the H1 sequence and probe H5-5 when the NCBI database was searched by use of the BLAST2 algorithm, but comparison of both sequences by use of the ClustalW algorithm (Infobiogen) showed a homology of 45 bases with 8 substitutions. Such a homology could be responsible for the partial and unstable hybridization between the target and nonmatched probe H5-5. Additional modification of the stringency of the assay may eliminate such heterologous reactivity between H1 and probe H5-5; however, on the basis of the possible sequence overlap, redesign of probes H5-4 and H5-5 would most likely provide the best alternative. Two-thirds of the Influenza Chips processed were devoid of any cross-reactivity, but even when false hybridization to an H5-specific probe was observed, there was no doubt regarding the identity of the virus due to probe redundancy for each target gene. The influenza virus array can be extremely specific for types and subtypes because of the probe redundancy and because relative intensity thresholds can be established.
On the basis of the measured titer (the 50% tissue culture infective dose) of A/PR/8/34 that we used to study the sensitivity of the array, the Influenza Chip was shown to detect as few as 1 × 102 to 5 × 102 influenza virus particles, a value similar to that previously reported by Wang et al. (28) for microarray-based detection of rhinoviruses. Such sensitivity is similar to or slightly lower than that reported for influenza virus detection by MPCR and subsequent agarose gel electrophoresis analysis (9, 24, 27, 30), but as emphasized by Wang et al. (28), coupling of random amplification and microarray technology bypasses the limitations usually associated with conventional PCR. The use of amplification with random primers will allow the development of an enlarged Respiratory Virus Chip whose viral type and subtype specificities will be strictly dependent on the probes selected for use in the array.
At the time that this paper was revised, Sengupta et al. (21) published interesting data on the detection and identification of influenza viruses by a two-dimensional microarray hybridization. The approach of Sengupta et al. (21) was completely different from that used in the present study in terms of the methodology (the oligonucleotide probes and PCR protocol chosen) and the subsequent application of such a microarray. As mentioned above, our ultimate purpose was to develop a tool that could be applied to the detection and identification of a larger panel of potential causative agents of viral respiratory diseases directly from the amplification of the viral RNA in clinical samples. To answer this need, and starting with influenza viruses, we used a random RT-PCR protocol and chose to print on FTCs probes that are highly specific for individual selected genes in order to generate images which could allow the immediate and easy identification of a causative agent in a clinical sample.
In summary, the assay with the Influenza Chip proved to be a sensitive and reliable method for the identification of influenza viruses. The whole process from hybridization to image capture can be completed in several hours and provides a rapid means of identification of the types and subtypes of influenza viruses. The Influenza Chip, as described in this paper, is preliminary, but recent data obtained with a second generation of Respiratory Virus Chip, which was developed with probes specific for major respiratory viruses and for which the amplification protocol and hybridization conditions were improved, show promise that such a chip could be beneficial for the identification and surveillance of influenza viruses, as well as viruses other than influenza viruses which are known to induce symptoms clinically indistinguishable from those of true influenza.
Acknowledgments
We thank the “Groupement d'Etudes Immunologiques” for financial support of O. Ferraris during this work.
We gratefully acknowledge the efforts of Helen Schiltz and Ana Munoz at MetriGenix in assisting with Influenza Chip development and Mridula Iyer and Eric Eastman for critical review of the manuscript.
REFERENCES
- 1.An, H. J., N. H. Cho, S. Y. Lee, I. H. Kim, C. Lee, S. J. Kim, M. S. Mun, S. H. Kim, and J. K Jeong. 2003. Correlation of cervical carcinoma and precancerous lesions with human papillomavirus (HPV) genotypes detected with HPV DNA chip microarray method. Cancer 97:1672-1680. [DOI] [PubMed] [Google Scholar]
- 2.Beattie, K. L. December 1998. Microfabricated, flowthrough porous apparatus for discrete detection of binding reactions. U.S. patent 5,843,767.
- 3.Benoit, V., A. Steel, M. Torres, Y. Y. Yu, H. Yang, and J. Cooper. 2001. Evaluation of three-dimensional microchannel glass biochip for multiplexed nucleic acid fluorescence hybridization assays. Anal. Chem. 73:2412-2420. [DOI] [PubMed] [Google Scholar]
- 4.Bohlander, S. K., R. E. Espinosa III, M. M. Le Beau, J. D. Rowley, and M. O. Diaz. 1992. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics 13:1322-1324. [DOI] [PubMed] [Google Scholar]
- 5.Capua, I., and D. J. Alexander. 2002. Avian influenza and human health. Acta Trop. 83:1-6. [DOI] [PubMed] [Google Scholar]
- 6.Cheek, B. D., A. Steel, M. P. Torres, Y. Y. Yu, and H. Yang. 2001. Chemiluminescence detection for hybridization assays on the Flow-Thru chip, a three-dimensional micro channel biochip. Anal. Chem. 73:5777-5783. [DOI] [PubMed] [Google Scholar]
- 7.Chizikov, V., M. Wagner, A. Ivshina, Y. Hoshino, A. Z. Kapikian, and K. Chumakov. 2002. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 40:2398-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 9.Ellis, J., D. Fleming, and M. Zambon. 1997. Multiplex reverse transcription-PCR for surveillance of influenza A and B viruses in England and Wales in 1995 and 1996. J. Clin. Microbiol. 35:2076-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis, J. S., and M. C. Zambon. 2002. Molecular diagnosis of influenza. Rev. Med. Virol. 12:375-389. [DOI] [PubMed] [Google Scholar]
- 11.Galama, J. M. 2003. Avian influenza and oseltamivir; a retrospective view. Ned. Tijdschr. Geneeskd. 147:1100-1102. (In Dutch.) [PubMed] [Google Scholar]
- 12.Hwang, T. S., J. K. Jeong, M. Park, H. S. Han, H. K. Choi, and T. S. Park. 2003. Detection and typing of HPV genotypes in various cervical lesions by HPV oligonucleotide microarray. Gynecol. Oncol. 90:51-56. [DOI] [PubMed] [Google Scholar]
- 13.Kane, M. D., T. A. Jatkoe, C. R. Stumpf, J. Lu, J. D. Thomas, and S. J. Madore. 2000. Assessment of the sensitivity and specificity of oligonucleotide (50 mer) microarrays. Nucleic Acids Res. 28:4552-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawagushi, K., S. Kaneko, M. Honda, H. F. Kawai, Y. Shirota, and K. Kobayashi. 2003. Detection of hepatitis B virus DNA in sera from patients with chronic hepatitis B virus infection by DNA microarray method. J. Clin. Microbiol. 41:1701-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, C. J., J. K. Jeong, M. Park, T. S. Park, T. C. Park, S. E. Namkoong, and J. S. Park. 2003. HPV oligonucleotide microarray-based detection of HPV genotypes on cervical neoplastic lesions. Gynecol. Oncol. 89:210-217. [DOI] [PubMed] [Google Scholar]
- 16.Li, J., S. Chen, and D. H. Evans. 2001. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J. Clin. Microbiol. 39:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, K. S., K. M. Xu, J. S. Peiris, L. L. Poon, K. Y. Yuen, K. Shortridge, R. G. Webster, and Y. Guan. 2003. Characterization of H9 subtype influenza viruses from the ducks of southern China: a candidate for the next influenza pandemic in humans? J. Virol. 77:6988-6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, Y. P., M. Shaw, V. Gregory, K. Cameron, W. Lim, A. Klimov, K. Subbarao, Y. Guan, S. Krauss, K. Shortridge, R. Webster, N. Cox, and A. Hay. 2000. Avian to human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA 97:9654-9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poddar, S. K. 2002. Influenza virus types and subtypes detection by single step single tube multiplex reverse transcription-polymerase chain reaction (RT-PCR) and agarose gel electrophoresis. J. Virol. Methods 99:63-70. [DOI] [PubMed] [Google Scholar]
- 20.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M.-H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Günther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 21.Sengupta, S., K. Onodera, A. Lai, and U. Melcher. 2003. Molecular detection and identification of influenza viruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 41:4542-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Special Report. 1993. Genosensors: the next step in biosensor technology. Genesis Rep. 2:6. [Google Scholar]
- 23.Steel, A., M. Torres, J. Hartwell, Y. Yu, N. Ting, G. Hoke, and H. Yang. 2000. The Flow-Thru Chip: a three-dimensional biochip platform, p. 97-117. In M. Schena (ed.), Microarray biochip technology. BioTechniques Books, Natick, Mass.
- 24.Stockton, J., J. S. Ellis, M. Saville, J. P. Clewley, and M. C. Zambon. 1998. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J. Clin. Microbiol. 36:2990-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemhill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 26.Vahey, M., M. E. Nau, S. Barrick, J. D. Cooley, R. Sawyer, A. A. Sleeker, P. Vickerman, S. Bloor, B. Larder, N. L. Michael, and S. A. Wegner. 1999. Performance of the Affymetrix GeneChip HIV PRT 440 platform for antiretroviral drug resistance genotyping of human immunodeficiency virus type 1 clades and viral isolates with length polymorphisms. J. Clin. Microbiol. 37:2533-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valassina, M., A. M. Cuppone, M. G. Cusi, and P. E. Valensin. 1997. Rapid detection of different RVA respiratory virus species by multiplex RT-PCR: application to clinical specimens. Clin. Diagn. Virol. 8:227-232. [DOI] [PubMed] [Google Scholar]
- 28.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson, J. W., P. Bean, T. Robins, F. Graziano, and D. H. Persing. 2000. Comparative evaluation of three human immunodeficiency virus genotyping systems: the HIV-GenotypR method, the HIV PRT GeneChip assay, and the HIV-1 RT line probe assay. J. Clin. Microbiol. 38:3022-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright, K. E., G. A. R. Wilson, D. Novosad, C. Dimock, D. Tan, and J. M. Weber. 1995. Typing and subtyping influenza viruses in clinical samples by PCR. J. Clin. Microbiol. 33:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]