Abstract
Despite the existence of formal models to explain how chromosomal rearrangements can be fixed in a population in the presence of gene flow, few empirical data are available regarding the mechanisms by which genome shuffling contributes to speciation, especially in mammals. In order to shed light on this intriguing evolutionary process, here we present a detailed empirical study that shows how Robertsonian (Rb) fusions alter the chromosomal distribution of recombination events during the formation of the germline in a Rb system of the western house mouse (Mus musculus domesticus). Our results indicate that both the total number of meiotic crossovers and the chromosomal distribution of recombination events are reduced in mice with Rb fusions and that this can be related to alterations in epigenetic signatures for heterochromatinization. Furthermore, we detected novel house mouse Prdm9 allelic variants in the Rb system. Remarkably, mean recombination rates were positively correlated with a decrease in the number of ZnF domains in the Prdm9 gene. The suggestion that recombination can be modulated by both chromosomal reorganizations and genetic determinants that control the formation of double-stranded breaks during meiosis opens new avenues for understanding the role of recombination in chromosomal speciation.
Keywords: house mouse, Robertsonian fusion, speciation, heterochromatinization, recombination, Prdm9
1. Introduction
The role of chromosomal reorganizations in speciation has been a long-standing question in biology. Understanding the genetic and mechanistic basis of these processes will provide insights into how biodiversity originates and is maintained. Compelling evidence supports that chromosomal rearrangements may reduce gene flow and therefore potentially contribute to speciation by the suppression of recombination [1–3]. Under this ‘suppressed recombination’ model, chromosome reorganizations in heterokaryotypes have a minimal influence on fitness, but rather affect recombination thus contributing to a reduction of gene flow within these genomic regions and the consequent accumulation of genetic incompatibilities [3]. Data supporting recombination suppression by inversions have been reported in different model organisms (see [4] and references therein), whereas studies regarding the effects of fusions have been restricted to the western house mouse, Mus musculus domesticus [5–14] and the common shrew, Sorex araneus [15,16]. The general view is that chromosomal reorganizations disturb the chromosomal distribution of recombinational events altering, in the long term, the final outcome of crossovers (COs). If this phenomenon was perpetuated within the population for long periods of time, gene flow within the reorganized regions would be reduced, thus eventually contributing to the establishment of genetic incompatibilities and subsequent progress towards complete reproductive isolation [3]. Despite recent progress in the field, the mechanisms and the genetic basis underlying the process remain elusive. In this regard, recent attention has focused on the Prdm9 gene, the only known speciation-associated gene described for mammals. The protein encoded by this gene, the PR domain zinc finger 9 (PRDM9) [17], is a meiotic-specific histone (H3) methyltransferase with a C-terminal tandem repeat zinc finger (ZnF) domain that accumulates at recombination sites through its recognition of a species-specific and highly mutagenic repetitive DNA motif [17–19]. In humans, the high nucleotide variation detected at this locus suggests that the protein (together with the repetitive DNA motif that it recognizes) is highly mutable enabling binding to new motifs as soon as they are formed [20,21]. Studies in laboratory strains of mice, moreover, have revealed that the PRDM9 protein could be directly involved in the recruitment of the recombination initiation machinery during meiosis [22]. It has also been reported that variations in the Prdm9 sequence affect the positioning of meiotic double-stranded breaks (DSBs), in the same way that the number and sequence of ZnF could determine the strength and specificity of DNA binding [19,22].
Despite the existence of formal models to explain how chromosomal rearrangements could be fixed in two parapatrically distributed populations under divergent selection, and by which mechanisms these may contribute to speciation [23], few empirical data are available, especially in mammals (see [4] and references therein). Therefore, the appreciation of how meiotic recombination is controlled during meiosis and how chromosomal reorganization affects the process is of critical importance for our understanding of speciation mechanisms. In this context, natural populations represent an excellent scenario for determining the genetic and mechanistic factors underlying the origin of meiotic disturbances triggered by chromosomal reorganizations. The standard karyotype of M. musculus domesticus consists of all-acrocentric chromosomes (2n = 40). However, a wide range of diploid numbers (from 22 to 40) have been described in natural populations resulting from the occurrence of Robertsonian (Rb) fusions and/or whole arm reciprocal translocations [24,25]. According to Piálek et al. [25], a Rb system is defined as a series of house mouse populations from a restricted geographical area that share a set of metacentrics with an apparently common evolutionary origin. In this context, a total of 97 Rb systems distributed across Europe, and the Mediterranean basin has been formally recognized [25]. Among them, the Barcelona Rb system is found in the provinces of Barcelona, Tarragona and Lleida (Spain), extending over approximately 5000 km2 (figure 1a). Diploid numbers (2n) range from 27 to 39 chromosomes, with the lowest 2n values observed at sites located approximately 30 km west of the city of Barcelona ([26] and references therein). This Rb system is characterized by a high level of chromosomal polymorphism, with seven different Rb chromosomes (Rb (3.8), (4.14), (5.15), (6.10), (7.17), (9.11) and (12.13)) showing non-geographically coincident (staggered) clines ([26] and references therein). Owing to the absence of a Rb race, as defined by Hausser et al. [27], this system has been considered to be a Rb polymorphism zone rather than be a typical hybrid zone ([26] and references therein). Although the chromosomal composition and structure of the Barcelona Rb system are known [26,28], the meiotic dynamics and genetic recombination of the mice that comprise it remain elusive. Here, we have studied the recombination features of this Rb system with two specific aims: (i) to analyse the effect of Rb fusions on both recombination rate and its chromosomal distribution, and (ii) to determine the genetic and mechanistic factors that are shaping the process.
Figure 1.
Genetic recombination diversity in the Barcelona chromosomal polymorphism zone. (a) Map showing the sampling distribution. Diploid numbers are shown for each locality (see the electronic supplementary material, table S1). Locations with standard and Rb individuals are indicated in red and black, respectively. (b) Example of immunolocalization of meiotic recombination events in mouse spermatocytes at pachynema from Rb5 (2n = 37). MLH1 foci are depicted in green, centromeres in blue and the synaptomenal complexes in red. Asterisks indicate trivalent structures. (c) Fluorescence in situ hybridization on a metaphase spread from Rb3 (2n = 38) with the chromosomes involved in the Rb fusion painted in green (chromosome 4) and red (chromosome 14). (d) Distribution of the mean numbers of MLH1 foci per cell observed in the specimens analysed (n = 34): lab_strain, standard mice from laboratory strain (n = 310 cells); standard, wild standard mice (n = 349 cells); Rb, 2n = 39–37, Rb mice with diploid numbers between 2n = 37–39 (n = 270 cells); and Rb, 2n = 32–28, Rb mice with diploid numbers between 2n = 28–32 (n = 333 cells). Asterisks indicate statistical significance (Mann–Whitney U-test or Kruskal–Wallis test; *p-value ≤ 0.05, **p-value ≤ 0.001). (e) Percentage of chromosomal arms showing 0, 1 or 2 MLH1 foci. Three groups are differentiated: Ac_St, all acrocentric chromosomes belonging to standard wild mice (n = 1197); Ac_Rb, all acrocentric chromosomes from Rb mice (n = 2211); and Met, all chromosomal arms involved in Rb fusions (n = 2056). Asterisk indicates statistical significance (Kruskal–Wallis test, **p-value ≤ 0.001).
2. Results
(a). Recombination events are reduced in Robertsonian mice
We studied the effect of Rb fusions on meiotic recombination by analysing the variation in the number of autosomal MLH1 foci (marker of COs) detected at pachynema in males (figure 1a–c and the electronic supplementary material, table S1). The average of MLH1 foci per cell observed ranged from 20.16 (±1.18) to 21.65 (±2.43) in wild mice with standard (St) karyotype (2n = 40), and from 18.13 (±1.78) to 20.57 (±1.90) in Rb mice (figure 1d and the electronic supplementary material, table S1). Wild St mice presented, on average, significantly less COs (20.88 ± 1.83) per cell than did mice derived from laboratory strains (21.32 ± 2.12; Mann–Whitney U-test, p ≤ 0.05; figure 1d and the electronic supplementary material, table S1). More importantly, mice with Rb fusions showed significantly smaller mean number of COs than wild St mice (Kruskal–Wallis test, p ≤ 0.001; figure 1d and the electronic supplementary material, table S1). Among Rb mice, we could distinguish two different groups (Mann–Whitney U-test, p ≤ 0.001): mice with high diploid numbers (2n = 39–37, 19.93 ± 1.77 COs per cell) and mice with low diploid numbers (2n = 32–28, 19.39 ± 1.68 COs per cell). These results indicate that Rb mice have a significant decrease in the overall number of recombination events per bivalent as a result of the presence of fused chromosomes.
In order to disentangle the effect of different factors on the decreased number of COs observed in Rb mice, we evaluated correlations between diploid number and the degree of heterozygosity in terms of rearrangements (heterokaryotypes). Owing to the absence of a fixed metacentric race and the high level of chromosomal polymorphisms that characterizes this Rb system, we pooled Rb compositions in three categories: one, two and three Rb fusions in heterozygote state. When considering both St and Rb animals, we found a positive correlation between the mean number of COs and the diploid number (Spearman correlation test ρ = 0.757, p ≤ 0.001; electronic supplementary material, figure S1); however, when applying the same correlation only among the Rb specimens, this was not significant (Spearman correlation tests ρ = 0.236, p = 0.315). Additionally, the comparison of the mean number of COs in Rb mice revealed that recombination was not affected by the degree of heterozygosity (Kruskal–Wallis test, p = 0.345). These results suggest that additional factors (other than diploid number reduction and the degree of heterozygosity) play a role in the final outcome of COs in this Rb system.
Considering the low mean values of COs observed in Rb mice, we investigated whether a similar pattern was reflected by the proteins implicated in the repair of DSBs in the early stages of prophase I (early pachynema). Meiotic recombination is initiated by DSBs generated by the protein Spo11 [29]. The replication protein A (RPA protein) associates with single-stranded DNA following DSBs formation and subsequently accumulates at the DSB sites [30]. Therefore, by analysing the number of RPA foci in early pachynema, we can determine the progression of DSBs as early recombinational nodules. If the final outcome of CO events observed in Rb mice is directly related to the initial DSBs formed in early stages of prophase I, then we would expect to find a reduced number of RPA foci when compared with St mice. We found that the mean number of RPA foci per cell at early pachynema was similar in wild St (67.49 ± 18.55) and Rb mice (64.83 ± 17.40; Mann–Whitney test, p = 0.536). However, wild St and Rb mice differed significantly with respect to laboratory mice (83.32 ± 19.92; Kruskal–Wallis test, p ≤ 0.001; electronic supplementary material, figure S2), suggesting that the difference between wild and laboratory mice is probably related with a reduction in the initial number of DSBs at early meiosis (exemplified here as RPA foci, representative of early nodules).
(b). Robertsonian fusions alter the chromosomal distribution of recombination events
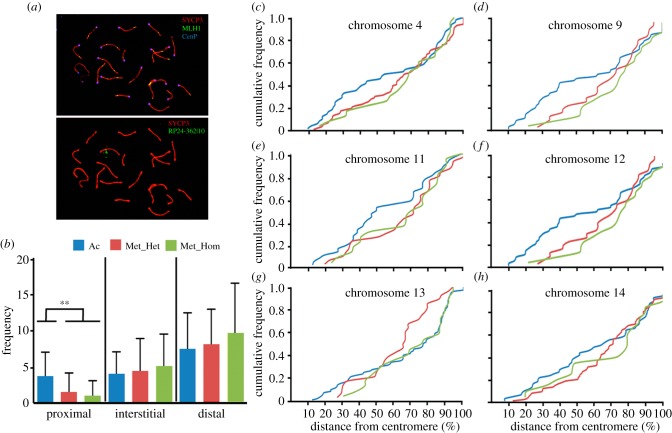
Moved by this striking pattern, we tested whether the overall decrease in recombination events observed was as a result of a reduction of COs in reorganized chromosomes. We analysed the percentage of chromosomal arms showing 0, 1 or 2 MLH1 foci considering three different groups based on the state of the chromosome (acrocentric, Ac, or metacentric, Met) and the type of specimen (St or Rb): (i) chromosomal arms involved in Rb fusions in Rb mice (Met), (ii) chromosomal arms not involved in Rb fusions belonging to wild St mice (Ac_St), and (iii) chromosomal arms not involved in Rb fusions in Rb mice (Ac_Rb; figure 1e and the electronic supplementary material, table S2). Our results show that when chromosomes are involved in Rb fusions (Met) the frequency of chromosomal arms with two COs decreases significantly when compared with acrocentric chromosomes (Kruskal–Wallis test, p ≤ 0.001; figure 1e and the electronic supplementary material, table S2). More importantly, the frequency of chromosomal arms with the absence of COs is not altered in the three groups (Kruskal–Wallis test, p = 0.071; figure 1e and the electronic supplementary material, table S2).
In order to analyse the redistribution of COs in Rb chromosomes in greater detail, we studied the chromosomal-specific distribution of MLH1 foci by applying sequential immunostaining and fluorescence in situ hybridization (FISH) with chromosome-specific bacterial artificial chromosomes (BAC) clones (electronic supplementary material, table S3 and figure 2a). This allowed for the identification of each of the chromosomes implicated in the Rb fusions in different mice, enabling us to test whether the overall reduced number of COs detected in Rb mice was due to the chromosomes involved in Rb fusions. We focused our efforts on the chromosomes most frequently involved in Rb fusions described in the area [26] by analysing the relative positions of MLH1 foci along each synaptonemal complex (SC; figure 2a–h). These were chromosomes 4, 9, 11, 12, 13 and 14. First, we established chromosome-specific recombination maps in acrocentric forms; these data serve as controls (figure 2b–h and the electronic supplementary material, figure S3). The COs distribution in acrocentrics was in accordance with the pattern previously reported for mice [31]; that is, a bimodal distribution of COs in long- and medium-sized chromosomal arms (i.e. chromosomes 4, 9, 11 and 12) and a telomeric distribution in short chromosomes (i.e. chromosomes 13 and 14; figure 2b–h and the electronic supplementary material, figure S3). However, and more importantly, we observed a trend in Rb chromosomes, that is, the distribution of CO events was displaced towards the telomeric regions (figure 2b–h and the electronic supplementary material, figure S3). In fact, we observed a significantly reduction of COs in the proximal area of the SCs (from the centromere to 30% of SC length) in Rb chromosomes, either in homozygosis (0.99 ± 2.10) or in heterozygosis (1.51 ± 2.67) when compared with acrocentric (3.70 ± 3.33; Kruskal–Wallis test, p ≤ 0.001). These results clearly show that when chromosomal arms are implicated in the Rb fusions, there is a reduction in the number of COs, displacing the recombination event towards the telomeric regions of the chromosomes, mirroring previous observations in mice [5,12,13].
Figure 2.
Cumulative frequencies of MLH1 foci in chromosomes implicated in Rb fusions. (a) Sequential image of a mouse spermatocyte at pachynema depicting a triple immunostaining with SYCP3 (red), MLH1 (green) and centromeres (blue) (upper panel) and fluorescent in situ hybridization (FISH; bottom panel) with a BAC probe corresponding to mouse chromosome 11 (RP23–362I10) in green. (b) Representation of the MLH1 foci frequencies obtained after merging data for all chromosomes analysed (no. 4, no. 9, no. 11, no. 12, no. 13 and no. 14). MLH1 foci frequencies along the SCs are shown for three regions depending on the relative distance from the centromere: (i) proximal, from the centromere to 30% of the SC; (ii) interstitial, between 30% and 70% of SC; and (iii) distal, from 70% to telomeric region (Kruskal–Wallis test; **p-value ≤ 0.001). Three different groups were considered: Ac, chromosomes in acrocentric form; Met_Hom, chromosomes involved in Rb fusions in homozygosis; and Met_Het, chromosomes involved in Rb fusions in heterozygosis. (c–h) Cumulative frequency plots representing the CO distribution of specific mouse chromosomes. In all instances, blue lines indicate chromosomes in the acrocentric form (n = 38 cells analysed for no. 4, n = 25 for no. 9, n = 36 for no. 11, n = 31 for no. 12, n = 71 for no. 13 and n = 46 for no. 14). Green lines represent the distribution when chromosomes are involved in Rb fusions in homozygosis (n = 21 cells analysed for no. 4, n = 22 for no. 9 n = 30 for no. 11, n = 21 for no. 12, n = 20 for no. 13 and n = 36 for no. 14) and red lines when in heterozygosis (n = 86 cells analysed for no. 4, n = 22 for no. 9, n = 20 for no. 11, n = 28 for no. 12, n = 26 for no. 13, n = 73 for no. 14).
(c). Robertsonian fusions alter the chromosomal distribution of H3K9me3 and γH2AX
We detected that the number of early recombinational nodules (RPA foci) was not altered in Rb mice when compared with wild St mice (electronic supplementary material, figure S2a–c). Moreover, the chromosomal distribution of RPA foci was not significantly different between acrocentric and metacentric chromosomes (electronic supplementary material, figure S2d), indicating that disturbances in the final outcome of COs occur during the resolution of early recombination nodules into COs, and not during the formation of DSBs. Therefore, we tested whether additional mechanistic factors (i.e. synapsis alterations and heterochromatinization) were altering the chromosomal distribution of COs in reorganized chromosomes.
We analysed the morphology of trivalents and sex chromosomes in order to detect pairing disturbances, because early studies in mice already noted the presence of chromosome unpairing in trivalents [32]. The γH2AX protein recognizes and localizes at DSBs, working as a marker for gene inactivation of asynapsed regions [33,34]. The sex body showed positive signal for γH2AX in all specimens analysed (electronic supplementary material, figure S4a,b) indicating a normal progression of the meiotic silencing of asynapsed chromatin that characterizes mammalian sex chromosomes [35]. However, we detected significantly higher numbers of cells with sex chromosomes totally asynapsed in Rb mice when compared with St animals (Fisher's test, p ≤ 0.05; electronic supplementary material, figure S4e–g). Regarding autosomes, when analysing the pairing dynamics of trivalents, we identified two different configurations: (i) closed trivalents and (ii) open trivalents (the electronic supplementary material, figure S4e). The frequency of spermatocytes detected with open (asynapsed) trivalents varied depending on the specimen analysed and these ranged from 4% to 21% of cells. As a general trend, open trivalents showed positive γH2AX labelling (electronic supplementary material, figure S4d,e). However, we also detected an abnormal pattern for γH2AX along chromosomes in specimens with Rb fusions, with γH2AX localizing along the SC in synapsed regions in both bivalents and trivalents in 48.6–52% of pachynema analysed (electronic supplementary material, figure S4f). This pattern was significantly less frequent in wild St mice (Fisher's test, p ≤ 0.001), and especially rare in the laboratory strain (only 10.9% of the cells analysed; Fisher's test p ≤ 0.001; electronic supplementary material, figure S4f). These results indicate that the persistence of regions with non-repaired DSBs through pachynema in animals with Rb fusions does not compromise the correct formation of the sex body, allowing the progression through prophase I and thus producing germ cells.
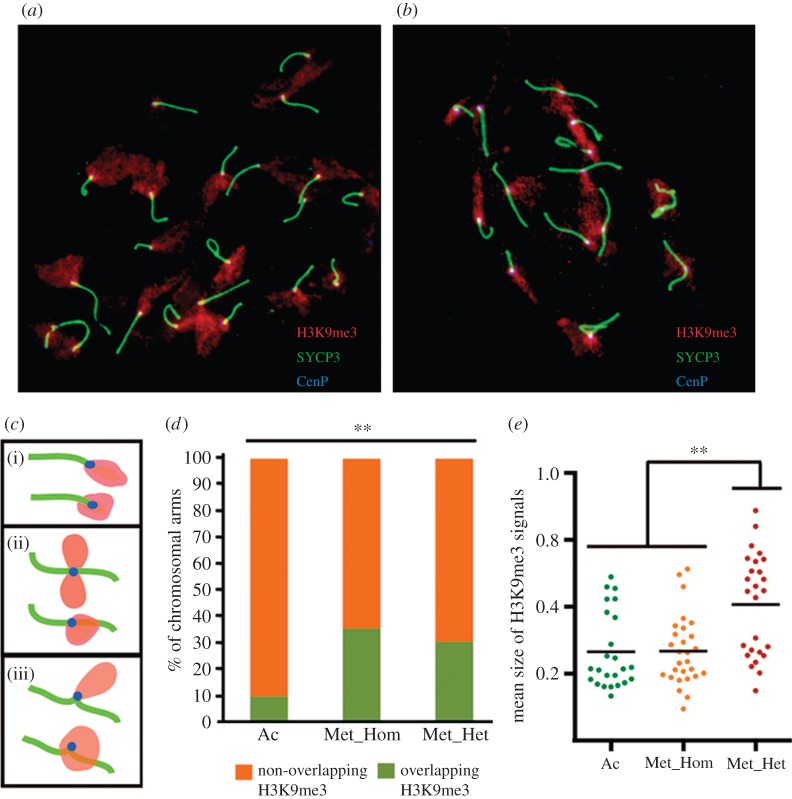
We also tested whether CO reduction in Rb mice was owing to a displacement towards telomeric regions triggered by an expansion centromeric interference effect as it has been previously postulated, although not experimentally tested [5]. In doing so, we analysed the distribution of the histone H3 lysine 9 tri-methylated (H3K9me3), an epigenetic signal for constitutive heterochromatin [36], along the SCs in several mice with different chromosomal configurations: 2n = 29 (11 Rb fusions; two specimens), 2n = 38 (two Rb fusions; one specimen), wild 2n = 40 (two specimens) and a laboratory mouse 2n = 40. In all mice analysed, H3K9me3 signals were located at autosomal centromeric regions in both acrocentric and metacentric chromosomes (figure 3a,b). However, the distribution pattern of H3K9me3 signals around the centromere differed between acrocentric and metacentric chromosomes (figure 3c). In fact, we identified two different configurations: (i) signals projected outside the SCs and only restricted to the centromeres, and (ii) H3K9me3 signals overlapping both the centromere and the pericentromeric area of the SCs. We detected that in the majority (90%) of the acrocentric chromosomes analysed, H3K9me3 signals did not overlap the SC. However, this proportion decreased significantly (Fisher's test, p-value ≤ 0.001) in chromosomes implicated in Rb fusions (70–60%; figure 3d). Moreover, the area (expressed in μm of SC length) occupied by the H3K9me3 signals from the centromere overlapping the SC in trivalents was, on average, significantly larger than in acrocentric chromosomes (Kruskal–Wallis test, p-value ≤ 0.001; figure 3e).
Figure 3.
Chromosomal distribution of H3K9me3 signals. (a,b) Examples of mouse spermatocytes from standard (a) and Rb (b) mice displaying the H3K9me3 distribution (red) with SC (green) and centromeres (blue). (c) Representation of the different H3K9me3 signal patterns observed in the sample: (i) acrocentric chromosomes, (ii) heterozygote metacentric, and (iii) homozygote metacentric. In all cases, H3K9me3 signals (red) were found in two different situations: overlapping and not-overlapping the SC. (d) Frequency of chromosomal arms with H3K9me3 signal overlapping (green) or non-overlapping (orange) the SC in acrocentric (Ac, n = 392) homozygote metacentric (Met_Hom, n = 172) and heterozygote metacentric chromosomal arms (Met_Het, n = 71). (e) Distribution of the mean size (expressed in μm of SC length) of the H3K9me3 signals overlapping the SC measured in the centromeric area (n = 78) in the three types of chromosomal arms described in (c,d). Asterisks indicate statistical significance (Fisher's test, **p-value ≤ 0.001; Kruskal–Wallis test, **p-value ≤ 0.001).
(d). Prdm9 allelic background and recombination rates
To directly assay the genetic basis underlying the alterations in the number and distribution of COs observed in Rb mice, we screened our sample for new Prdm9 allelic variants that might account for such diversity. Prdm9 plays a key role in determining the patterning of recombination events [22]; therefore, our working hypothesis was to consider that the observed diversity in recombination rates (figure 1 and the electronic supplementary material, table S1) was related to the Prdm9 genetic background. With this in mind, we sequenced the Prmd9 exon 12 containing the Zn finger domain from Zn repeat +3 towards the C-terminal domain in 27 mice from our sample (electronic supplementary material, table S1). We detected that 20 specimens had alleles of the same length, whereas the remaining specimens had alleles of two different lengths (electronic supplementary material, table S1 and figure S5). Our genetic screening detected five Prdm9 allelic variants present in the study area, which differed both in nucleotide sequences and the number of Zn finger repeats from previous studies [17,37]. These were referred to as 10A, 10B, 11B, 12B and 12C (electronic supplementary material, figures S6 and S7). The alleles 12B and 12C shared the same number of ZnF domains as the alleles previously described in M. musculus domesticus [17,37], but differed in the DNA sequence of the ZnF domains (electronic supplementary material, figures S7 and S8). The allele 11B shared the same number of ZnF domains as previously described in Mus musculus castaneus [37], but, as with the above, differed in the DNA sequence of ZnF domains (electronic supplementary material, figure S7). More importantly, we detected two alleles with 10 ZnF repeats (allele 10A and 10B), which are specific for the Barcelona chromosomal polymorphism zone (electronic supplementary material, figures S6–S8). The analysis of the nucleotide and amino acid sequence revealed that the highest replacement rates for all alleles were detected at positions −1, +3 and +6, all regions that were previously described as being highly polymorphic (electronic supplementary material, figures S7 and S8). These positions correspond to the amino acids that recognize the DNA repeat sequence-specific for the Prdm9 protein in the mouse.
Allele frequency and distribution varied among specimens and among localities (electronic supplementary material, figure S6a,b). Allele 10A was the most frequently observed (77.77%) in our sample followed by 12B (14.81%), 12C (3.70%) and to a lesser extent 11B (1.85%) and 10B (1.85%; electronic supplementary material, figure S6a). The highest number of different alleles was observed within St wild mice that surrounded the localities containing Rb mice (electronic supplementary material, figure S6b and table S1). Remarkably, these Rb mice were genetically more homogeneous, being mostly homozygous for allele 10A (electronic supplementary material, figure S6b and table S1).
We additionally investigated whether Prdm9 genetic background influences recombination, as has been previously shown in humans [38]. As it has been suggested that low numbers of ZnF repeats are correlated with lower recombination rates [38], three different groups were considered: mice carrying two common 10 alleles (10/10), mice carrying one non-10 allele (10/N) and mice carrying two non-10 alleles (N/N). The comparison of CO frequency (mean number of MLH1 foci/cell) in specimens with 10/10, 10/N and N/N genotypes revealed that those carrying the 10 alleles in homozygosis state showed, on average, a significantly lower number of COs than N/10 and N/N mice (Kruskal–Wallis test, p-value ≤ 0.001; electronic supplementary material, figure S6c). Moreover, we applied different correlation analysis between Prdm9 allelic diversity, diploid number and mean number of MLH1 foci per cell. We detected a highly similar correlation value between these three factors: ρ = 0.56 between total number of PRDM9 ZnF domains and recombination rate (p-value < 0.05), ρ = 0.58 between total number of PRDM9 ZnF domains and diploid number (p-value < 0.05), and ρ = 0.75 between diploid number and recombination rate (p-value < 0.001), suggesting that the three (mechanistic factors, genetic factors and recombination rates) are somehow related.
3. Discussion
In an evolutionary context, our study represents a detailed empirical demonstration that Rb fusions affect meiotic recombination and that this can be related to alterations in epigenetic signatures for heterochromatinization. Although some analyses performed were based on a limited number of individuals, this is, to our knowledge, the first meiotic study on a Rb polymorphism zone. This has allowed us to detect important trends. First, we found that Rb males have significantly lower recombination rates that wild St mice, despite the variability observed in diploid numbers. Second, our results suggest that Rb fusions indeed have an effect on the chromosomal distribution of COs, altering their chromosomal distribution. The average number of recombination events is reduced in chromosomal arms involved in Rb fusions when compared with acrocentrics, reflecting a reduction in the percentage of chromosomal arms with two COs in Rb animals. These data add to preliminary observations that have reported a reduction in chiasmata number (in latter stages of meiosis; i.e. metaphase II) and MLH1 foci in different house mice populations with Rb fusions [5,6,11–13]. Our approach, however, provided a more detailed and chromosomal-specific analysis of CO redistribution. In fact, we observed that the distribution of COs along chromosomal arms that occur in acrocentrics is altered in Rb chromosomes; recombinational events closer to the centromeric region are frequently lost and COs tend to be more terminal in metacentrics. It is well known that meiotic recombination is repressed close to the centromere, a pattern that has been conserved in all eukaryote species. This is because COs occurring close to centromere interfere with normal chromatid segregation, inducing aneuploidy [39]. Although the molecular mechanisms underlying centromere interference are still largely unknown, pericentric heterochromatin formation (here exemplified by H3K9me3 signals) is considered crucial for the process [40]. In fact, the epigenetic status of the chromatin is, in general, important for recombination. Meiotic DSBs tend to occur in open and highly transcribed euchromatic regions [19], whereas DNA methylation suppresses CO formation. Recent studies in plants have suggested that DNA methylation can affect the distribution of recombination events in cis, this alteration being chromosomal-dependent [41]. This fact has important implications for our observations given that chromosomes involved in the Rb fusions locally affect the overall reduction of COs detected in Rb mice. We observed larger and more expanded H3K9me3 signals over the SC in trivalents, which are characterized by the presence of three centromeres non-completely aligned. These results, together with the presence of altered γH2AX pattern along the SC in synapsed regions in both bivalents and trivalents in Rb mice, suggest that DSBs are not properly repaired, especially in trivalent structures, affecting the final outcome of COs.
We also detected that the formation of the sex body at pachynema is not compromised in the Rb males by either the presence of asynapsed regions or asynapsed sex chromosomes. But more importantly, the number of chromosomal arms with no CO is not significantly altered when comparing acrocentric and metacentric chromosomes in Rb animals. COs are highly regulated in mammals to ensure the proper disjunction of homologous chromosomes during meiosis [42] and mammalian species normally present (on average) one CO per chromosomal arm [43,44]. Our observations have important implications for CO homeostasis [42], indicating that cells carrying Rb fusions modulate the final CO outcome without losing the obligatory one chiasmata per arm necessary to allow even chromosomal segregation, thus not compromising the viability of germ cells. Therefore, and despite the presence of unrepaired DNA regions in both open trivalents and synapsed autosomes at pachynema in Rb mice, the sex body is probably established and cells escape the pachytene checkpoint [34]. Given the widespread distribution of Rb animals in the Barcelona chromosomal polymorphic zone, the low recombination rates observed and the presence of asynapsed regions in the trivalents probably would have a mild negative effect on fertility, mirroring previous observations [34]. This interpretation is in line with analyses performed on mice from the Barcelona Rb system [26,45,46], which suggest that Rb fusions have a reduced effect on mice fertility, thus permitting the de novo occurrence of chromosomal rearrangements within this zone.
Together with the CO mechanistic disturbances observed, the Prdm9 allelic diversity has been revealed as an additional factor modulating genetic recombination. The PRDM9 protein is directly involved in the recruitment of the recombination initiation machinery during meiosis [22]. Previous studies have described the presence of four different haplotypes in mice with differences in the number of Zn fingers: 9-Zn fingers, 11-Zn fingers, 12-Zn fingers, 13-Zn fingers and 14-Zn fingers [17,19,37], and our data add new Prdm9 allelic variants to the picture, confirming the high Prdm9 sequence variability in nature. More importantly, our results suggest that the number and sequence of Zn fingers might influence meiotic recombination outcome, most probably by regulating strength and specificity of DNA binding, thus in agreement with what has been reported in humans and mice [17,19,38,47]. Mice carrying the 10 allele showed, on average, a significantly lower number of COs than mice with longer Zn finger repeats. In fact, the allele 10A was the most frequently observed in the Barcelona chromosomal polymorphism zone (16 of 18 were homozygote for 10A). Despite this, we still could distinguish two different groups among Rb mice in terms of MLH1 mean values, irrespective of their Prmd9 allelic composition: mice with high (2n = 37–39) and low diploid numbers (2n = 28–32). The fact that both groups did not differ in genetic background (i.e. homozygote for 10A) suggests that additional factors (rather than genetic ones) could explain such differences. Further analysis of the population structure of the Barcelona Rb system would reveal novel Prdm9 allelic variants, as well as possible demographic or stochastic effects underling its allelic distribution and the genetic modulation of recombination. In fact, our analysis of RPA foci (representative of early nodules) indicated that both wild St and Rb mice (carrying the 10 allele) presented a similar pattern in terms of overall numbers of RPA foci per cell detected, but different with respect to the laboratory mouse strain, suggesting that mice from the Barcelona chromosomal polymorphic zone have a unique genetic background that is affecting recombination.
Which are the evolutionary implications of our results? Here, we demonstrate that the average number of recombination events is indeed reduced in chromosomal arms involved in the Rb fusions when compared with acrocentric chromosomes, especially in proximal chromosomal regions, and that this can be related to alterations in epigenetic signatures for heterochromatinization. In mammals, recent studies have detected reduced recombination rates and gene flow within reorganized regions [8,14,16]. This has been the case of inversions, where recombination is specially reduced in heterokaryotypes [1–4]. By contrast, the effect of Rb fusions based on the state of the fusion (hetero- versus homozygous) has been less explored [5,6,8,11,12]. The fact that we observed a reduction in recombination in both heterokaryotypes and homokaryotypes suggest that fusions might evolve differently than inversions do. Although at this stage it would be premature to discuss the evolutionary forces behind this pattern, both the absence of a fixed metacentric race and the high level of chromosomal polymorphisms that characterizes the Barcelona Rb system highlights its importance as an informative model. Further analysis of the genetic structure of this system would help us to elucidate whether suppressed recombination triggered by Rb fusions is indeed leaving a signature of genetic divergence.
4. Conclusion
Here, we show that mice with Rb fusions present a substantially reduced number of the total recombinational events as a result of a redistribution of COs in those chromosomal arms involved in Rb fusions. Moreover, the detection of novel Prdm9 allelic variants in the Barcelona Rb polymorphic zone has permitted the examination of recombination variability observed within the population. Overall, our results suggest that changes in both number and distribution of recombination events are probably modulated by heterochromatinization disturbances produced by Rb fusions and influenced by the Prdm9 genetic background.
5. Material and methods
A total of 31 wild male mice (2n = 40, 11 mice; 2n = 39, 2 mice; 2n = 38, 3 mice; 2n = 37, 3 mice; 2n = 32, 2 mice; 2n = 31, 3 mice; 2n = 30, 4 mice; 2n = 29, 2 mice; 2n = 28, 1 mouse) were live-trapped in commensal habitats from 10 different localities representative of the Barcelona Rb system (figure 1 and the electronic supplementary material, table S1). See the electronic supplementary material for details on sample processing, immunofluorescence, FISH, image processing and data analysis, and Prdm9 genotyping.
Acknowledgements
The authors acknowledge Josep Medarde for his valuable help in the fieldwork and J. Davidian-Britton, T. J. Robinson and M. García-Caldés for insightful comments on earlier versions of the manuscript and to M. Fritzler for the sclerodactyly and telangiectasia (CREST) serum.
Permission to capture was granted by the Departament de Medi Ambient of the Generaltitat de Catalunya (Spain). Animals were handled in compliance with the guidelines and ethical approval by the Comissió d'Ètica en l'Experimentación Animal y Humana (CEEAH) of the Universitat Autònoma de Barcelona and by the Departament d'Agriculta, Romaderia, Pesca, Alimentació i Medi Natural (Direcció General de Medi Natural i Biodiversitat) of the Generalitat de Catalunya.
Funding statement
Financial support from Ministerio de Economia y Competitividad is gratefully acknowledged (CGL2010-15243 to J.V. and CGL-2010-20170 to A.R.H.). M.O.B. is supported by Instituto de Salud Carlos III (CP07/ 0258 and PI08/1185). L.C. is the beneficiary of an FPI pre-doctoral fellowship (BES-2011-047722), whereas A.A.S. is sponsored by a pre-doctoral fellowship from the Conselleria d'Educació, Cultura i Universitats (Govern Illes Balears) and Fons Social Europeu.
References
- 1.Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16, 351–358. ( 10.1016/S0169-5347(01)02187-5) [DOI] [PubMed] [Google Scholar]
- 2.Faria R, Navarro A. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol. Evol. 25, 660–669. ( 10.1016/j.tree.2010.07.008) [DOI] [PubMed] [Google Scholar]
- 3.Navarro A, Barton NH. 2003. Chromosomal speciation and molecular divergence-accelerated evolution in rearranged chromosomes. Science 300, 321–324. ( 10.1126/science.1080600) [DOI] [PubMed] [Google Scholar]
- 4.Farré M, Micheletti D, Ruiz-Herrera A. 2013. Recombination rates and genomic shuffling in human and chimpanzee: a new twist in the chromosomal speciation theory. Mol. Biol. Evol. 30, 853–864. ( 10.1093/molbev/mss272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumas D, Britton-Davidian J. 2002. Chromosomal rearrangements and evolution of recombination: comparison of chiasma distribution patterns in standard and Robertsonian populations of the house mouse. Genetics 162, 1355–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castiglia R, Capanna E. 2002. Chiasma repatterning across a chromosomal hybrid zone between chromosomal races of Mus musculus domesticus. Genetica 114, 35–40. ( 10.1023/A:1014626330022) [DOI] [PubMed] [Google Scholar]
- 7.Ruiz-Herrera A, Farré M, Ponsà M, Robinson TJ. 2010. Selection against Robertsonian fusions involving housekeeping genes in the house mouse: integrating data from gene expression arrays and chromosome evolution. Chromosome Res. 18, 801–808. ( 10.1007/s10577-010-9153-8) [DOI] [PubMed] [Google Scholar]
- 8.Franchini P, Colangelo P, Solano E, Capanna E, Verheyen E, Castiglia R. 2010. Reduced gene flow at pericentromeric loci in a hybrid zone involving chromosomal races of the house mouse Mus musculus domesticus. Evolution 64, 2020–2032. ( 10.1111/j.1558-5646.2010.00964.x) [DOI] [PubMed] [Google Scholar]
- 9.Nunes AC, Catalan J, Lopez J, Ramalhinho MDG, Mathias MDL, Britton-Davidian J. 2011. Fertility assessment in hybrids between monobrachially homologous Rb races of the house mouse from the island of Madeira: implications for modes of chromosomal evolution. Heredity 106, 348–356. ( 10.1038/hdy.2010.74) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Förster DW, Mathias ML, Britton-Davidian J, Searle JB. 2013. Origin of the chromosomal radiation of Madeiran house mice: a microsatellite analysis of metacentric chromosomes. Heredity 110, 380–388. ( 10.1038/hdy.2012.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merico V, Giménez MD, Vasco C, Zuccotti M, Searle JB, Hauffe HC, Garagna S. 2013. Chromosomal speciation in mice: a cytogenetic analysis of recombination. Chromosome Res. 21, 523–533. ( 10.1007/s10577-013-9377-5) [DOI] [PubMed] [Google Scholar]
- 12.Bidau CJ, Giménez MD, Palmer CL, Searle JB. 2001. The effects of Robertsonian fusions on chiasma frequency and distribution in the house mouse (Mus musculus domesticus) from a hybrid zone in northern Scotland. Heredity 87, 305–313. ( 10.1046/j.1365-2540.2001.00877.x) [DOI] [PubMed] [Google Scholar]
- 13.Merico V, Pigozzi MI, Esposito A, Merani MS, Garagna S. 2003. Meiotic recombination and spermatogenic impairment in Mus musculus domesticus carrying multiple simple Robertsonian translocations. Cytogenet. Genome Res. 103, 321–329. ( 10.1159/000076820) [DOI] [PubMed] [Google Scholar]
- 14.Giménez MD, White TA, Hauffe HC, Panithanarak T, Searle JB. 2013. Understanding the basis of diminished gene flow between hybridizing chromosome races of the house mouse. Evolution 67, 1446–1462. ( 10.1111/evo.12054) [DOI] [PubMed] [Google Scholar]
- 15.Borodin PM, Karamysheva TV, Belonogova NM, Torgasheva AA, Rubtsov NB, Searle JB. 2008. Recombination map of the common shrew, Sorex araneus (Eulipotyphla, Mammalia). Genetics 178, 621–632. ( 10.1534/genetics.107.079665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yannic G, Basset P, Hausser J. 2009. Chromosomal rearrangements and gene flow over time in an inter-specific hybrid zone of the Sorex araneus group. Heredity 102, 616–625. ( 10.1038/hdy.2009.19) [DOI] [PubMed] [Google Scholar]
- 17.Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323, 373–375. ( 10.1126/science.1163601) [DOI] [PubMed] [Google Scholar]
- 18.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840. ( 10.1126/science.1183439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472, 375–378. ( 10.1038/nature09869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wegmann D, et al. 2011. Recombination rates in admixed individuals identified by ancestry-based inference. Nat. Genet. 43, 847–853. ( 10.1038/ng.894) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffreys AJ, Cotton VE, Neumann R, Lam KG. 2013. Recombination regulator PRDM9 influences the instability of its own coding sequence in humans. Proc. Natl Acad. Sci. USA 110, 600–605. ( 10.1073/pnas.1220813110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645. ( 10.1038/nature11089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirkpatrick M, Barton N. 2006. Chromosome inversions, local adaptation and speciation. Genetics 173, 419–434. ( 10.1534/genetics.105.047985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gazave E, Catalan J, Da Graça Ramalhinho M, Da Luz Mathias M, Claudia Nunes A, Dumas D, Britton-Davidian J, Auffray J-C. 2003. The non-random occurrence of Robertsonian fusion in the house mouse. Genet. Res. 81, 33–42. ( 10.1017/S001667230200602X) [DOI] [PubMed] [Google Scholar]
- 25.Piálek J, Hauffe HC, Searle JB. 2005. Chromosomal variation in the house mouse. Biol. J. Linn. Soc. 84, 535–563. ( 10.1111/j.1095-8312.2005.00454.x) [DOI] [Google Scholar]
- 26.Medarde N, López-Fuster MJ, Muñoz-Muñoz F, Ventura J. 2012. Spatio-temporal variation in the structure of a chromosomal polymorphism zone in the house mouse. Heredity 109, 78–89. ( 10.1038/hdy.2012.16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hausser J, Fedyk S, Fredga K, Searle JB, Volobouev V, Wojcik JM, Zima J. 1994. Definition and nomenclature of the chromosome races of Sorex Araneus. Folia Zool. 43, 1–9. [Google Scholar]
- 28.Gündüz I, López-Fuster MJ, Ventura J, Searle JB. 2001. Clinal analysis of a chromosomal hybrid zone in the house mouse. Genet. Res. 77, 41–51. ( 10.1017/S0016672300004808) [DOI] [PubMed] [Google Scholar]
- 29.Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384. ( 10.1016/S0092-8674(00)81876-0) [DOI] [PubMed] [Google Scholar]
- 30.Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B. 2007. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J. Cell Sci. 120, 1017–1027. ( 10.1242/jcs.03394) [DOI] [PubMed] [Google Scholar]
- 31.Froenicke L, Anderson LK, Wienberg J, Ashley T. 2002. Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 71, 1353–1368. ( 10.1086/334714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace BM, Searle JB, Everett CA. 1992. Male meiosis and gametogenesis in wild house mice (Mus musculus domesticus) from a chromosomal hybrid zone; a comparison between ‘simple’ Robertsonian heterozygotes and homozygotes. Cytogenet. Cell Genet. 61, 211–220. ( 10.1159/000133410) [DOI] [PubMed] [Google Scholar]
- 33.Matveevsky SN, Pavlova SV, Acaeva MM, Kolomiets OL. 2012. Synaptonemal complex analysis of interracial hybrids between the Moscow and Neroosa chromosomal races of the common shrew Sorex araneus showing regular formation of a complex meiotic configuration (ring-of-four). Comp. Cytogenet. 6, 301–314. ( 10.3897/compcytogen.v6i3.3701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manterola M, et al. 2009. A high incidence of meiotic silencing of unsynapsed chromatin is not associated with substantial pachytene loss in heterozygous male mice carrying multiple simple Robertsonian translocations. PLoS Genet. 5, e1000625 ( 10.1371/journal.pgen.1000625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgoyne PS, Mahadevaiah SK, Turner JM. 2009. The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet. 10, 207–216. ( 10.1038/nrg2505) [DOI] [PubMed] [Google Scholar]
- 36.Hublitz P, Albert M, Peters AH. 2009. Mechanisms of transcriptional repression by histone lysine methylation. Int. J. Dev. Biol. 53, 335–354. ( 10.1387/ijdb.082717ph) [DOI] [PubMed] [Google Scholar]
- 37.Parvanov ED, Petkov PM, Paigen K. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327, 835 ( 10.1126/science.1181495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berg IL, Neumann R, Lam K-WG, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ. 2010. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 42, 859–863. ( 10.1038/ng.658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynn A, Ashley T, Hassold T. 2004. Variation in human meiotic recombination. Annu. Rev. Genomics Hum. Genet. 5, 317–349. ( 10.1146/annurev.genom.4.070802.110217) [DOI] [PubMed] [Google Scholar]
- 40.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542. ( 10.1126/science.1064027) [DOI] [PubMed] [Google Scholar]
- 41.Mirouze M, Lieberman-Lazarovich M, Aversano R, Bucher E, Nicolet J, Reinders J, Paszkowski J. 2012. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc. Natl Acad. Sci. USA 109, 5880–5885. ( 10.1073/pnas.1120841109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole F, Kauppi L, Lange J, Roig I, Wang R, Keeney S, Jasin M. 2012. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 14, 424–430. ( 10.1038/ncb2451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Cruz R, Pacheco S, Brieño MA, Steinberg ER, Mudry MD, Ruiz-Herrera A, Garcia-Caldés M. 2011. A comparative study of the recombination pattern in three species of Platyrrhini monkeys (primates). Chromosoma 120, 521–530. ( 10.1007/s00412-011-0329-6) [DOI] [PubMed] [Google Scholar]
- 44.Segura J, et al. 2013. Evolution of recombination in eutherian mammals: insights into mechanisms that affect recombination rates and crossover interference. Proc. R. Soc. B 280, 20131945 ( 10.1098/rspb.2013.1945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sans-Fuentes MA, García-Valero J, Ventura J, López-Fuster MJ. 2010. Spermatogenesis in house mouse in a Robertsonian polymorphism zone. Reproduction 140, 569–581. ( 10.1530/REP-10-0237) [DOI] [PubMed] [Google Scholar]
- 46.Medarde N, Muñoz-Muñoz F, López-Fuster MJ, Ventura J. 2013. Variational modularity at the cell level: insights from the sperm head of the house mouse. BMC Evol. Biol. 13, 179 ( 10.1186/1471-2148-13-179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berg IL, Neumann R, Sarbajna S, Odenthal-Hesse L, Butler NJ, Jeffreys AJ. 2011. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proc. Natl Acad. Sci. USA 108, 12 378–12 383. ( 10.1073/pnas.1109531108) [DOI] [PMC free article] [PubMed] [Google Scholar]