Abstract
Coccidioides DNA was amplified from serum by a PCR using coccidioid-specific primers. A 239-bp product was visualized when 10 fg of exogenous coccidioidal DNA was subjected to amplification. This product was demonstrated in some human and mouse sera prior to the detection of coccidioidal antibodies.
Coccidioidomycosis is caused by inhalation of the diphasic, multimorphic fungus Coccidioides immitis. Diagnosis can be established by the recovery and identification of C. immitis or by the detection of coccidioidal antibodies. The detection of antibodies may be accomplished 1 to 4 weeks after symptoms develop (16). Coccidioidal immunoglobulin M (IgM) (precipitin) antibodies are usually not detectable before 7 days after the onset of illness, and IgG (complement fixation) antibodies are not detectable until later (14, 16). The diagnosis may be further delayed in patients who are unable to generate a detectable humoral response, e.g., immunocompromised individuals (14). Methods have been sought that are more sensitive than antibody detection. Galgiani et al. (6) detected coccidioid-specific antigens in the sera of patients suspected of having new infections. Clark and McAllister (3) used RNA amplification to detect C. immitis in clinical specimens that contained whole organisms, but they were not successful when testing cerebrospinal fluid (CSF) and they did not test serum. We sought a nonserologic approach to earlier diagnoses, and here we describe an effort to detect coccidioidal DNA in serum by PCR using C. immitis (or Coccidioides posadasii [4])-specific primers.
MATERIALS AND METHODS
PCR.
The primers used for this study were designed by Greene et al. (7) based on the internal transcribed spacer (ITS) sequences of C. immitis ribosomal DNA (accession no. X94142) (2) and were prepared by Invitrogen (Carlsbad, Calif.). These Coccidioides-specific primers, designated ITS C1A and ITS C2, amplify a 239-bp sequence but do not amplify DNAs from closely related, non-Coccidioides species (7). PCR mixtures (50 μl) contained 5 μl of GeneAmp 10× PCR Gold buffer (Applied Biosciences, Foster City, Calif.), 3 μl of 1.5 mM MgCl2, 4 μl of a 10 mM dNTP mix (a 2.5 mM concentration of each), 1.25 U of AmpliTaq Gold (a hot start enzyme), 10 pmol of each primer, and 30 μl of sample. Each reaction mixture was subjected to the following conditions: 1 cycle at 94°C for 10 min; 35 cycles at 94°C for 1 min, 53°C for 1 min, and 72°C for 1 min; and 1 cycle at 72°C for 10 min. After amplification, a 10-μl sample of each reaction was electrophoresed through a 1.5% agarose gel at 90 V for 60 min. Gels were stained by incubation for 15 min in ethidium bromide (0.5 μg/ml in water) or for 60 min in Vistra Green (Amersham Pharmacia Biotech, Piscataway, N.J.) diluted 1:10,000 in TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0). They were then analyzed and photographed in a Nucleotech Geltech imager (Nucleotech, San Carlos, Calif.).
Sensitivity assay.
C. immitis strain Silveira (ATCC 28868) genomic DNA, isolated from endosporulating spherules 36 h after inoculation (19), was quantitated spectrophotometrically, diluted in water, and added to the PCR master mix. Reaction mixtures contained between 100 pg and 1 fg of DNA, and after amplification, a 10-μl sample was electrophoresed and stained.
Mouse sera.
Sera were collected from Swiss Webster mice experimentally infected intranasally with 500 C. immitis strain Silveira arthroconidia. Uninfected mice received saline. On days 3, 7, 11, 16, and 21 after infection, the sera from three mice per group were collected, pooled, and then stored at −80°C until further use. Coccidioidal antibodies in the sera were detected by double immunodiffusion (9, 10, 14). Histologic examinations revealed C. immitis spherules in the lungs from day 7 on.
DNA isolation.
A QIAmp DNA blood mini kit (Qiagen, Valencia, Calif.) was used to isolate DNAs from human and mouse specimens (200-μl samples) and to remove inhibitory components. The kit was used according to the manufacturer's protocol except that the DNA was eluted with a 50-μl volume to effect concentration of the DNA. Thirty microliters of eluate was used for PCR amplification.
Isolation and amplification of DNA added to serum.
Human serum (200 μl) that was negative for coccidioidal antibodies and for coccidioidal DNA by PCR was seeded with coccidioidal genomic DNA (166 pg). This quantity should yield 100 pg per 30 μl of eluate, the volume added to the PCR mixture. The DNA was then isolated from the serum and amplified.
Amplification of DNA from human clinical specimens.
Anonymous human specimens, either sera or CSF, were obtained from the Coccidioidomycosis Serology Laboratory (Davis, Calif.) in accordance with the policies of the Institutional Review Board at the University of California, Davis (protocol 2002210077-2). Sera from humans with coccidioidomycosis with various antibody responses were obtained, and if present, the DNAs were isolated and amplified. Samples were electrophoresed and the gels were stained. The PCR product from one representative reaction was cloned into a TA cloning vector (Invitrogen) and the sequence of the insert was determined.
Blinded study.
Sera that were previously evaluated for the presence of coccidioidal antibodies by immunodiffusion and by complement fixation were obtained in a blinded fashion. The specimen collection date and serologic results were revealed after the DNA amplifications were completed. Successive samples from some patients were evaluated for coccidioidal antibodies and DNAs.
RESULTS
Sensitivity assay.
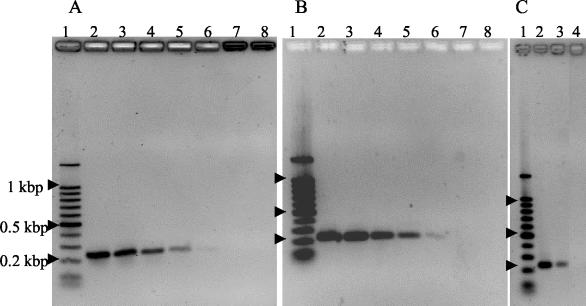
A 239-bp PCR product was readily visualized after staining with ethidium bromide when 1 pg, but not 100 fg, of coccidioidal genomic DNA was added to the reaction mixture (Fig. 1A). When a similar gel was stained with Vistra Green for comparison, the PCR product was easily visualized when 100 fg of DNA was added and there was a faint band when 10 fg was added (Fig. 1B). We calculated that 10 fg of genomic DNA would contain approximately 323 genome copies based on an estimated genome size of 28.2 Mb (13). While there was an increased sensitivity when the Vistra Green stain was used, some preliminary samples originally stained with ethidium bromide were unavailable for comparisons with Vistra Green-stained samples. Regardless of the staining method, the intensity of the stained bands was proportional to the amount of DNA added such that the relative quantity of DNA present prior to amplification could be estimated.
FIG. 1.
Demonstration of coccidioidal PCR products after agarose gel electrophoresis. A 100-bp DNA ladder was used to estimate the product sizes (lane 1). (A and B) Coccidioidal genomic DNAs were added to PCRs in the following quantities to determine the sensitivity of detetion. Lane 2, 100 pg; lane 3, 10 pg; lane 4, 1 pg; lane 5, 100 fg; lane 6, 10 fg; lane 7, 1 fg; lane 8, no DNA added. Gels were stained with ethidium bromide (A) or Vistra Green (B). (C) Recovery of coccidioidal DNA added to serum. Lane 2, coccidioidal DNA added to normal human serum after the isolation procedure and then amplified by PCR; lane 3, coccidioidal genomic DNA added to normal human serum prior to isolation and PCR amplification; lane 4, negative (no template) control reaction. The gel was stained with ethidium bromide.
Amplification of DNA added to serum.
Serum components that were inhibitory to amplification were removed by the QIAmp system. DNA that was deliberately added to a serum and then isolated was successfully amplified (Fig. 1C). However, the intensity of the product from this mixture, theoretically containing 100 pg of DNA/30 μl of eluate, was less than that of a QIAmp-treated serum that had 100 pg of DNA added after the isolation. This indicated that some DNA was lost during the isolation process.
Amplification of DNA from clinical specimens.
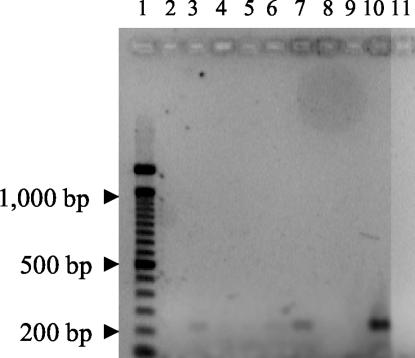
Of the nine patient samples that were initially evaluated, eight were seropositive and one was seronegative. Four patient sera (all positive for antibodies) yielded PCR products of the size expected. Figure 2 shows an ethidium bromide-stained agarose gel with eight of the amplified serum specimens. Positive 239-bp products are seen in lanes 2, 6, and 7. The intensely staining band in lane 10 is the 100-fg positive control. No product is apparent in lane 11, the negative (no template) control reaction. An additional positive product (not shown) was cloned and sequenced. This clone was found to contain the complete and correct 239-bp ITS sequence.
FIG. 2.
PCR amplification of coccidioidal DNAs from clinical specimens. Products were separated by electrophoresis through a 1.5% agarose gel and stained with ethidium bromide. Lanes 2, 3, 4, 6, 7, 8, and 9, sera that were positive for coccidioidal antibodies; lane 5, seronegative sample; lane 10, 100 fg of coccidioidal DNA as a positive control; lane 11, negative (no template) control.
Mouse study.
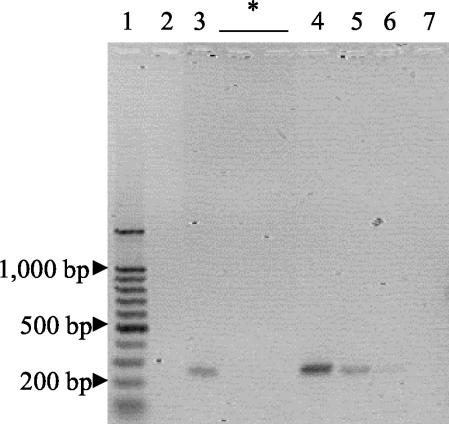
All sera collected from uninfected mice were negative by PCR. A single pooled specimen collected 3 days after infection with 500 arthroconidia was PCR positive (Fig. 3). All mouse sera were negative for coccidioidal antibodies by double immunodiffusion (data not shown). Spherules were observed in the lungs of mice 7 days after infection.
FIG. 3.
PCR amplification of coccidioidal DNAs from mouse sera. DNAs present in sera collected from mice after infection were amplified, separated by electrophoresis, and stained with Vistra Green. Amplified samples collected 3 days after infection are shown. Lane 2, uninfected mice; lane 3, mice infected with 500 arthroconidia. While samples collected after 3 days were also subjected to amplification and electrophoresis, no products were visible, and representative lanes are indicated by an asterisk. Lane 1, 100-bp DNA ladder; lane 4, 10-pg PCR control; lane 5, 1-pg PCR control; lane 6, 100-fg PCR control; lane 7, negative (no template) control.
Blinded human study.
A total of 90 human serum and 4 human CSF specimens were evaluated in a blinded fashion. After the completion of the PCR testing, the code was broken (Table 1). Fifty-eight serum specimens had no detectable coccidioidal antibodies, as determined by immunodiffusion and/or complement fixation. Twenty-three had a detectable antibody (11 were positive for IgM, 5 were positive for IgG, and 7 were positive for both IgM and IgG), and 9 were equivocal. All four CSF specimens were negative for coccidioidal antibodies. One of these specimens was from an individual whose serum later became positive for antibodies. Additional serologic information on subsequent samples was recorded for 17 patients who were initially seronegative or equivocal. Six of the 14 seronegative and 3 of the 4 equivocal patients became seropositive. Three sets of successive samples were included in the study; two were sera and one was CSF. The serum sets both included an initial specimen that had no detectable antibody and a subsequent sample that was seropositive. The CSF set was seronegative, and successive samples had no detectable antibodies.
TABLE 1.
Comparison of coccidioidal serology results with PCR results
| Coccidioidal serology result or sample type | No. of isolates | No. with PCR result
|
||
|---|---|---|---|---|
| Negative | Positive | Equivocal | ||
| Serum | 90 | 80 | 6 | 4 |
| Seronegative | 58 | 54 | 3 | 1 |
| Subsequent sample seronegative | 7 | 7 | 0 | 0 |
| Subsequent sample seropositive | 6 | 4 | 2 | 0 |
| Seropositive | 23 | 20 | 1 | 2 |
| IgM only | 11 | 10 | 1 | 0 |
| IgG only | 5 | 5 | 0 | 0 |
| IgM and IgG | 7 | 5 | 0 | 2 |
| Equivocal | 9 | 6 | 2 | 1 |
| Subsequent sample seropositive | 3 | 1 | 2 | 0 |
| Subsequent sample equivocal | 1 | 1 | 0 | 0 |
| CSF | ||||
| Seronegative | 4 | 4 | 0 | 0 |
| Total | 94 | 84 | 6 | 4 |
Samples were designated PCR positive if an amplified product of the appropriate size was both visualized and photographed. In some instances (four specimens), a faint band of the appropriate size was visualized; however, the intensity was below the photographic sensitivity. These specimens were designated equivocal. As the sample volume was limited, it was not possible to repeat the isolation and amplification with these specimens.
Of the 94 specimens evaluated (from 88 patients), six patient samples contained coccidioidal DNAs that were amplified by PCR (Table 1). PCR-positive specimens were most often seronegative or equivocal (Table 2), although one PCR-positive specimen was IgM positive. This specimen was from a patient who had an initial diagnosis of eosinophilic pneumonia and died from extensive coccidioidal pulmonary disease. Four of the seronegative specimens (DN40, DN73, DN78, and DN91) were from individuals who later became seropositive. The time until the collection of additional specimens ranged from 5 to 36 days. One PCR-positive specimen (DN63) was seronegative, and there were no additional specimens from this patient. In two instances, the second specimen was also subjected to PCR. In both of these cases (DN73-DN74 and DN91-DN92), the second specimen was seropositive but PCR negative. All of the CSF samples studied were PCR negative.
TABLE 2.
Results of PCR amplification of select sera
| Specimen no. | Collection date (mo/day/yr) | Serology result | PCR result |
|---|---|---|---|
| DN40a | 8/20/02 | Equivocal | Positive |
| DN55b | 2/6/02 | Negative | Negative |
| DN56b | 2/19/02 | Negative | Negative |
| DN63 | 8/7/01 | Negative | Positive |
| DN66 | 3/19/02 | IgM positive | Positive |
| DN73c | 9/21/01 | Negative | Positive |
| DN74c | 10/16/01 | IgM and IgG positive | Negative |
| DN78d | 9/10/01 | Equivocal | Positive |
| DN91a | 1/31/01 | Negative | Positive |
| DN92e | 2/7/02 | IgM positive | Negative |
A later sample, collected 8/25/02 was positive for coccidioidal IgM.
DN55 and DN56 were CSF samples and were collected from the same individual.
DN73 and DN74 were collected from the same individual.
A later sample, collected 9/19/01, was positive for coccidioidal IgM.
DN91 and DN92 were collected from the same individual.
DISCUSSION
The diagnosis of coccidioidomycosis may be delayed by the dependence on the demonstration of an antibody response, which may take a week or longer after the development of symptoms, or by the failure of some patients to mount an adequate humoral response. Galgiani et al. found circulating coccidioidal antigens in 35 of 233 patients with symptoms that were compatible with coccidioidomycosis (6). Thirty-three of the patients had no detectable coccidioidal antibodies at that time. An alternative approach is the detection of fungal nucleic acids present in clinical specimens or swabs. These nucleic acids could represent those released from whole fungal cells after lysis or circulating free nucleic acids produced in the host. Indeed, many groups have explored nucleic acid detection for the diagnosis of coccidioidomycosis or other medically important fungi (1, 3, 5, 8, 11, 12, 15, 17, 18). All of these methods employed some means of amplifying the nucleic acids. Clark and McAllister (3) evaluated the detection of rRNA that was released after the lysis of C. immitis. They used transcription-mediated amplification of rRNAs released from fungal cells. While their method was successful for demonstrating C. immitis in tissues and other specimens that were culture positive, they were not able to demonstrate C. immitis in specimens that were culture negative (e.g., CSF). A more common approach is the amplification of DNA by PCR, and this was the method we chose to employ.
Our study included three phases: (i) the detection of coccidioidal DNA deliberately added to normal human serum, (ii) the detection of DNA in the sera of mice that had been deliberately infected with coccidioides, and (iii) the detection of coccidioidal DNA in the sera of humans with coccidioidomycosis (who were seropositive or became seropositive).
The PCR assay evaluated in the present study used C. immitis species-specific primers that amplified 239 bp of the ITS region of ribosomal DNA. These primers were selected since they were previously shown to be specific and since they amplified a multicopy region (7). We evaluated body fluids that would not likely contain fungal cells but would possibly contain circulating DNA that could be amplified. The sensitivity was enhanced by the use of Vistra Green (detection of as little as 10 fg of DNA) staining as opposed to ethidium bromide, which required at least 100 fg of DNA (Fig. 1A and B).
We considered it possible that coccidioidal DNAs might be present in patients' early-phase sera. Preliminary amplification studies indicated that DNA was present in four of nine seropositive human specimens. To test this experimentally, we evaluated the method by using sera of mice infected with C. immitis. A PCR-positive sample was collected 3 days after infection with 500 arthroconidia. All other samples were PCR negative as well as seronegative. This suggested that DNA may be transiently present, as was the circulating coccidioidal antigen (6). It is not yet known whether circulating DNA might reappear in the serum with the progression of coccidioidal disease and the proliferation of coccidioidal cells.
The coccidioidal DNA PCR was further evaluated by using a panel of sera and CSF that were previously evaluated for coccidioidal antibodies but that were provided for this study in a blinded fashion. Of the 94 specimens evaluated, 6 were PCR positive. Four of these six patients who were positive by PCR were seronegative, but subsequent specimens from these patients were seropositive, providing confirmation that they had developed coccidioidal infections. Thus, the PCR was able to demonstrate coccidioidal infections before antibodies became detectable by immunodiffusion or complement fixation. However, the PCR positivity appeared to be transient, as two patients whose initial specimens were seronegative and PCR positive later became seropositive and PCR negative. Only one specimen included in this portion of the study was both PCR positive and seropositive.
The present study indicates the feasibility but perhaps limited utility of DNA amplification from serum specimens for the diagnosis of coccidioidomycosis. However, as has been suggested to us, the testing of whole blood or the buffy coat may enhance the recovery of fungal DNA from the infected host. While the current protocol will yield a product if 10 fg of DNA or more is present in 200 μl of serum (approximately 323 genome copies), the sensitivity might be improved by using an objective, instead of visual, quantitative amplification system. Alternatively, the PCR could be coupled to a second assay, such as an initial amplification with universal fungal primers and then hybridization of the PCR product to labeled species-specific probes (5, 12, 15). Further studies are also required to address the effect of antibodies on the presence of circulating DNA. An animal model may provide insights into these questions.
Acknowledgments
This work was supported by a grant from the California HealthCare Foundation and by the Department of Health Services of the State of California and California State University, Bakersfield.
REFERENCES
- 1.Bialek, R., A. Ibricevic, C. Aepinus, L. K. Najvar, A. W. Fothergill, J. Knobloch, and J. R. Graybill. 2000. Detection of Paracoccidioides brasiliensis in tissue samples by nested PCR assay. J. Clin. Microbiol. 38:2940-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burt, A., D. A. Carter, G. L. Koenig, T. J. White, and J. W. Taylor. 1996. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc. Natl. Acad. Sci. USA 93:770-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark, K. A., and D. McAllister. 1996. Direct detection of Coccidioides immitis in clinical specimens using target amplification, p. 129-136. In H. E. Einstein and A. Cantanzaro (ed.), Coccidioidomycosis. Proceedings of the 5th International Conference. National Foundation for Infectious Diseases, Bethesda, Md.
- 4.Fisher, M. C., G. L. Koenig, T. J. White, and J. W. Taylor. 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 94:73-84. [PubMed] [Google Scholar]
- 5.Fujita, S.-I., B. A. Lasker, T. J. Lott, E. Reiss, and C. J. Morrison. 1995. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J. Clin. Microbiol. 33:962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galgiani, J. N., G. M. Grace, and L. L. Lundergan. 1991. New serologic tests for early detection of coccidioidomycosis. J. Infect. Dis. 163:671-674. [DOI] [PubMed] [Google Scholar]
- 7.Greene, D. R., G. Koenig, M. C. Fisher, and J. W. Taylor. 2000. Soil isolation and molecular identification of Coccidioides immitis. Mycologia 92:406-410. [Google Scholar]
- 8.Hidalgo, J. A., G. J. Alangaden, D. Eliott, R. A. Akins, J. Puklin, G. Abrams, and J. A. Vazquez. 2000. Fungal endophthalmitis diagnosis by detection of Candida albicans DNA in intraocular fluid by use of species-specific polymerase chain reaction assay. J. Infect. Dis. 181:1198-1201. [DOI] [PubMed] [Google Scholar]
- 9.Huppert, M., and J. W. Bailey. 1965. The use of immunodiffusion tests in coccidioidomycosis. I. The accuracy and reproducibility of the immunodiffusion test which correlates with complement fixation. Am. J. Clin. Pathol. 44:364-368. [PubMed] [Google Scholar]
- 10.Huppert, M., and J. W. Bailey. 1965. The use of immunodiffusion tests in coccidioidomycosis. II. An immunodiffusion test as a substitute for the tube precipitin test. Am. J. Clin. Pathol. 44:369-373. [PubMed] [Google Scholar]
- 11.Lin, M.-T., H.-C. Lu, and W.-L. Chen. 2001. Improving efficacy of antifungal therapy by polymerase chain reaction-based strategy among febrile patients with neutropenia and cancer. Clin. Infect. Dis. 33:1621-1627. [DOI] [PubMed] [Google Scholar]
- 12.Martin, C., D. Roberts, M. Van Der Weide, R. Rossau, G. Jannes, T. Smith, and M. Maher. 2000. Development of a PCR-based line probe assay for identification of fungal pathogens. J. Clin. Microbiol. 38:3735-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan, S., and G. T. Cole. 1992. Electrophoretic karyotypes of clinical isolates of Coccidioides immitis. Infect. Immun. 60:4872-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pappagianis, D., and B. L. Zimmer. 1990. Serology of coccidioidomycosis. Clin. Microbiol. Rev. 3:247-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandhu, G. S., B. C. Kline, L. Stockman, and G. D. Roberts. 1995. Molecular probes for diagnosis of fungal infections. J. Clin. Microbiol. 33:2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith, C. E., M. T. Saito, and S. A. Simons. 1956. Pattern of the 39,500 serologic tests in coccidioidomycosis. JAMA 160:546-552. [DOI] [PubMed] [Google Scholar]
- 17.Turin, L., R. Riva, G. Galbiati, and R. Cainelli. 2000. Fast, simple and highly sensitive double-rounded polymerase chain reaction assay to detect medically relevant fungi in dermatological specimens. Eur. J. Clin. Investig. 30:511-518. [DOI] [PubMed] [Google Scholar]
- 18.Wahyuningsih, R., H.-J. Freisleben, H.-G. Sonntag, and P. Schnitzler. 2000. Simple and rapid detection of Candida albicans DNA in serum by PCR for diagnosis of invasive candidiasis. J. Clin. Microbiol. 38:3016-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmermann, C. R., C. J. Snedker, and D. Pappagianis. 1994. Characterization of Coccidioides immitis isolates by restriction length polymorphisms. J. Clin. Microbiol. 32:3040-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]