Abstract
A TaqMan-based real-time PCR assay was established to quantify the periodontopathic bacteria Tannerella forsythensis and Fusobacterium spp. With this assay, the prevalence and proportion of these bacteria in clinical specimens were evaluated. Our preliminary results suggest a positive colocalization of T. forsythensis and Fusobacterium spp. in periodontal pockets.
Periodontitis is thought to arise from the complex microflora consisting of putative periodontopathic bacteria, such as Porphyromonas gingivalis, Treponema denticola, Tannerella forsythensis (formerly Bacteroides forsythus), and Fusobacterium spp. (1, 5). These bacterial species are isolated together from the affected periodontal sites (3, 4), and it has been suggested that bacterial cooperation might promote the infectious process of periodontitis. Consequently, it is important to know the composition of microorganisms in periodontal pockets for diagnosis and rational treatment. A TaqMan-based real-time PCR assay recently developed for the quantitative detection of periodontopathic species has actually contributed to the diagnosis and monitoring of periodontal disease (2, 8). One advantage of using a real-time PCR assay is the detection of both absolute and relative amounts of bacteria. This assay is also advantageous for detecting species such as T. forsythensis that are extremely difficult to grow in broth. In this study, we developed a real-time PCR assay to quantify T. forsythensis and Fusobacterium spp. and evaluated the prevalence and proportion of these bacteria by using clinical specimens to describe their colocalization in periodontal pockets.
The oligonucleotide primers and TaqMan probes used, designed from the 16S rRNA genes with the Primer Express 1.5 software (Applied Biosystems, Foster City, Calif.), are listed in Table 1. The specificities of the primers and probes were confirmed by conventional PCR and by dot blot analysis with chromosomal DNAs extracted from T. forsythensis ATCC 43037; F. nucleatum ATCC 10953, ATCC 25586, ATCC 49256, and ATCC 51191; Fusobacterium russii ATCC 25533; Fusobacterium periodonticum ATCC 33693; and 18 other oral bacteria (data not shown).
TABLE 1.
Oligonucleotide primers and probes used in this studya
| Primer or probe | Sequence | Amplicon size (bp) | Source or reference |
|---|---|---|---|
| Primers | |||
| Fs619F | 5′-CGCAGAAGGTGAAAGTCCTGTAT-3′ | 101 | ATCC 10953 |
| Fs719R | 5′-TGGTCCTCACTGATTCACACAGA-3′ | ||
| Tf18F | 5′-ATCCTGGCTCAGGATGAACG-3′ | 226 | 338 |
| Tf243R | 5′-TACGCATACCCATCCGCAA-3′ | ||
| Uni152F | 5′-CGCTAGTAATCGTGGATCAGAATG-3′ | 69 | 7 |
| Uni220R | 5′-TGTGACGGGCGGTGTGTA-3′ | ||
| Fluorescent probes | |||
| Fs663T | 5′-FAM-ACTTTGCTCCCAAGTAACATGGAACACGAG-TAMRA-3′ | ATCC 10953 | |
| Tf124T | 5′-FAM-ATGTAACCTGCCCGCAACAGAGGGATAAC-TAMRA-3′ | 338 | |
| Uni177T | 5′-FAM-CACGGTGAATACGTTCCCGGGC-TAMRA-3′ | 7 |
The target of each primer or probe was the 16S rRNA gene.
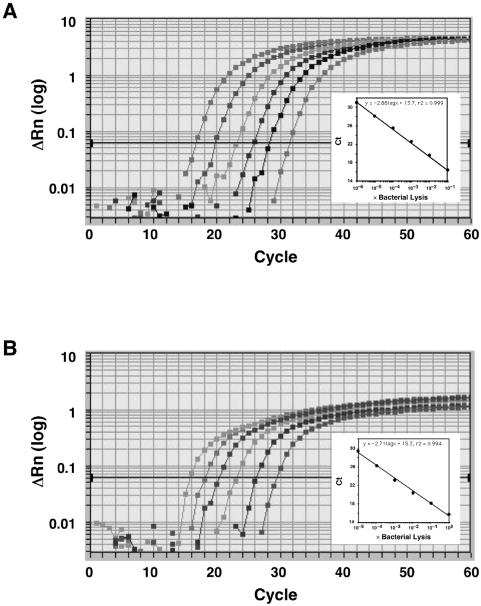
Amplification and detection by real-time PCR were performed with the ABI PRISM 7700 Sequence Detection System (Applied Biosystems). For each real-time PCR, 20-μl reaction mixtures containing 1 μl of lysed cells, 1× qPCR Master Mix (Eurogentec S.A., Seraing, Belgium), each sense and antisense primer at 200 nM, and the TaqMan probe at 250 nM were placed in each well of a 96-well plate. The reaction conditions were set at 50°C for 2 min and 95°C for 10 min, followed by 60 cycles of 95°C for 15 s and 58°C for 1 min. The critical threshold cycle (Ct) is defined as the cycle at which the fluorescence becomes detectable above the background and is inversely proportional to the logarithm of the initial number of template molecules. Standard curves for each organism were plotted for each primer-probe set with the Ct values obtained by amplifying successive 10-fold dilutions of a known concentration of DNA extracted from bacterial cells containing 1.2 × 106 CFU (230 ng/μl) for F. nucleatum ATCC 10953 and 4.2 × 105 CFU (370 ng/μl) for T. forsythensis ATCC 43037 (Fig. 1). On the basis of this approach, correlations between Ct and CFU counts were observed. Detection and quantification were linear over a range of 1.2 × 100 to 1.2 × 105 cells per reaction mixture for F. nucleatum (y = −2.88 log x + 31.3, r2 = 0.999) and over a range of 4.2 × 100 to 4.2 × 105 cells per reaction mixture for T. forsythensis (y = −2.70 log x + 30.6, r2 = 0.995). The relative amounts of these bacteria in oral specimens were calculated by the comparative Ct (ΔΔCt) method, with a simple modification (7).
FIG. 1.
Amplification plots of genomic DNA from lysed cells. Serial dilutions of genomic DNA from F. nucleatum ATCC 10953 (A) or T. forsythensis ATCC 43037 (B) are shown. The log-transformed relative fluorescence [ΔRn (log)] was monitored as the increase in reporter dye intensity relative to that of the passive internal reference dye. The threshold fluorescence, or the level at which the threshold cycle was determined, is shown. The standard curves were generated from the amplification plots in the insets (correlation coefficients, 0.999 for F. nucleatum and 0.994 for T. forsythensis). Ct is the cycle number at which the threshold fluorescence was reached.
Subgingival plaque samples were collected from 10 Japanese adults (mean age, 52.5 ± 16.9 years) referred to Kyushu University Dental Hospital for treatment of periodontitis. None of the subjects had taken antibiotics or undergone scaling or root planing within the 6 months before the study. All of the subjects were positive for Fusobacterium spp. and T. forsythensis by conventional PCR with saliva as the template. The probing pocket depth of each tooth was measured at six sites of each tooth (distal, mid, and mesial for both the buccal and lingual surfaces of each tooth) with a periodontal probe. Following World Health Organization criteria (6), six segments were evaluated for each mouth. From these measurements, the deepest periodontal pockets in each segment were selected for microbial sampling from each patient, resulting in 59 samples being collected. The mean periodontal pocket depth of the samples was 4.20 ± 1.67 mm. The template DNA was extracted as described previously (8). Statistical analyses were performed with SPSS version 11.0 (SPSS Japan Inc., Tokyo, Japan).
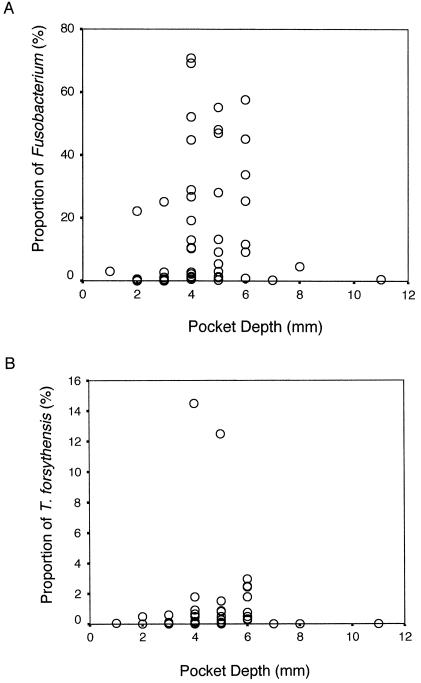
Figure 2 shows the proportions of Fusobacterium spp. and T. forsythensis in all of the samples according to pocket depth, respectively. The Spearman rank correlation between the proportions of each organism and the probing depth of each sample were calculated. Both bacteria significantly correlated with deeper pockets (r = 0.48 for T. forsythensis and r = 0.50 for Fusobacterium spp., P < 0.001). The proportions of both bacteria for each pocket were significantly correlated with each other (r = 0.75, P < 0.001), and this is also significant when Pearson's correlation is used (r = 0.292, P = 0.025). Successively, a between-subjects analysis was performed to avoid cross-contamination of sites. The Spearman rank correlations between the mean proportion of the organism and the mean pocket depth for each subject were 0.63 (P < 0.05) for T. forsythensis and 0.72 (P < 0.05) for Fusobacterium spp. The correlation between the mean proportions of these two bacteria was very strong (r = 0.88, P < 0.001), and this was also significant when Pearson's correlation (r = 0.678, P = 0.031) was used. The significant relationship between both bacteria and pocket depth observed in this study suggests that both species grow well in the environment of a deeper periodontal pocket.
FIG. 2.
Proportions of Fusobacterium spp. (A) and T. forsythensis (B) in samples from various pocket depths (n = 59).
The frequencies with which Fusobacterium spp. and T. forsythensis were detected in samples from periodontal pockets of different depths were analyzed (Table 2). Both bacteria were detected more frequently in samples from periodontal pockets ≥4 mm deep (P < 0.001, χ2 test). T. forsythensis was not detected in subgingival plaque, where Fusobacterium spp. were absent (P < 0.001, Fisher's exact probability test). In fact, in samples from shallow pockets ≤3 mm deep, T. forsythensis was not detected at any Fusobacterium-negative site (P = 0.03). This suggests a possible colocalization of T. forsythensis and Fusobacterium spp. without considering pocket depth as a confounder. Our preliminary tests show the ability of the real-time PCR assay to contribute to not only quantification of the target organisms but also to clarification of the correlation among oral bacteria in periodontal pockets.
TABLE 2.
Detection of T. forsythensis and Fusobacterium spp. within the same subgingival plaque samples
| Presence of T. forsythensis | Presence of Fusobacterium spp.
|
|||
|---|---|---|---|---|
| All samples (n = 59)a
|
Probing depth of ≤3mm (n = 21)b
|
|||
| + | − | + | − | |
| + | 34 | 0 | 6 | 0 |
| − | 17 | 8 | 7 | 8 |
P < 0.001 by Fisher's exact probability test.
P = 0.03 by Fisher's exact probability test.
Acknowledgments
We thank Kazuyuki Ishihara, Department of Microbiology, Tokyo Dental College, for providing bacterial strains.
This work was supported in part by a research grant from the Nakatomi Foundation (A.Y.) and by research fellowships from the Japan Society for the Promotion of Science for Young Scientists (N.S.).
REFERENCES
- 1.Haffajee, A. D., and S. S. Socransky. 1994. Microbial etiological agents of destructive periodontal diseases. Periodontology 2000 5:78-111. [DOI] [PubMed] [Google Scholar]
- 2.Maeda, H., C. Fujimoto, Y. Haruki, T. Maeda, S. Kokeguchi, M. Petelin, H. Arai, I. Tanimoto, F. Nishimura, and S. Takashiba. 2003. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol. Med. Microbiol. 39:81-86. [DOI] [PubMed] [Google Scholar]
- 3.Simonson, L. G., K. T. McMahon, D. W. Childers, and H. E. Morton. 1992. Bacterial synergy of Treponema denticola and Porphyromonas gingivalis in a multinational population. Oral Microbiol. Immunol. 7:111-112. [DOI] [PubMed] [Google Scholar]
- 4.Socransky, S. S., A. D. Haffajee, J. L. Dzink, and J. D. Hillman. 1988. Associations between microbial species in subgingival plaque samples. Oral Microbiol. Immunol. 3:1-7. [DOI] [PubMed] [Google Scholar]
- 5.Takamatsu, N., K. Yano, T. He, M. Umeda, and I. Ishikawa. 1999. Effect of initial periodontal therapy on the frequency of detecting Bacteroides forsythus, Porphyromonas gingivalis, and Actinobacillus actinomycetemcomitans. J. Periodontol. 70:574-580. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization. 1998. Oral health surveys: basic methods, 4th ed. World Health Organization, Geneva, Switzerland.
- 7.Yoshida, A., N. Suzuki, Y. Nakano, M. Kawada, T. Oho, and T. Koga. 2003. Development of a 5′ nuclease-based real-time PCR assay for quantitative detection of cariogenic dental pathogens Streptococcus mutans and Streptococcus sobrinus. J. Clin. Microbiol. 41:4438-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida, A., N. Suzuki, Y. Nakano, T. Oho, M. Kawada, and T. Koga. 2003. Development of a 5′ fluorogenic nuclease-based real-time PCR assay for quantitative detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J. Clin. Microbiol. 41:863-866. [DOI] [PMC free article] [PubMed] [Google Scholar]