Abstract
Crosstalk between histone modifications plays an important role in transcriptional regulation. In a recent study, an elegant chemical synthesis approach has been used to study the intricate relationship between two modifications associated with active transcription.
The eukaryotic genome is packaged with core histones and other chromosomal proteins to form chromatin. One unique feature of the core histones is their enrichment in covalent modifications, which include acetylation, phosphorylation, ubiquitylation and methylation. Interestingly, these modifications are intricately related. For example, genetic studies have revealed that histone methylation on lysine residues 4 and 79 of H3 (H3K4 and H3K79, respectively) is dependent on histone H2B ubiquitylation1–4. However, whether the effect of H2B ubiquitylation on H3 methylation is direct and how this effect is mediated remains unresolved. In a recent paper published in Nature5, McGinty et al. used a reconstituted nucleosome system that harbors a pure population of ubiquitylated H2B (uH2B) generated by chemical synthesis to demonstrate that uH2B directly contributes to H3K79 methylation.
Studies in the past several years have revealed differential patterns of histone modifications in regions of active versus inactive gene transcription. Actively transcribed genes are enriched in H3K4, H3K79 methylation and H2B ubiquitylation, whereas inactive genes are enriched in H3K9, H3K27 methylation and H2A ubiquitylation6,7. Notably, the active marks appear to have a sequential relationship. For example, genetic studies in budding yeast indicated that H2B ubiquitylation is upstream of H3K4 and H3K79 methylation, as deletion of the genes responsible for H2B ubiquitylation or mutation on the site of H2B ubiquitylation blocked H3K4 and H3K79 methylation1–4. The demonstrated interplay between H3 methylation and H2B ubiquitylation brought to light an important question: is the effect of H2B ubiquitylation on H3 methylation direct, and, if it is, how does H2B ubiquitylation contribute to H3 methylation?
Synthetic chemistry–based tools have previously been used to study histone modifications. For example, chemical ligation has been used to study the structural effect of H4K16 acetylation (H4K16ac)8. By incorporating the semisynthetic H4K16ac histone into nucleosomes, H4K16 acetylation has been shown to inhibit chromatin compaction, thus explaining how H4K16 acetylation facilitates transcription. Recently, a simple yet elegant method has been used to generate methyllysine analogs for histone methylation studies9. In this approach, lysines were replaced by cysteines, and specific alkylating agents were used to create N-methylated aminoethylcysteine9. To unambiguously address the relationship between H2B ubiquitylation and H3 methylation, it is necessary to use homogeneously ubiquitylated H2B. To avoid the previous challenges associated with enzyme-catalyzed in vitro H2B ubiquitylation or purification of endogenously ubiquitylated H2B, McGinty et al. again turned to chemical tools, using expressed protein ligation (EPL) technology to generate uH2B and thus ensuring chemical homogeneity5.
In their approach, two peptides—one containing an N-terminal cysteine and the other a C-terminal thioester—are ligated together (Fig. 1a). Due to its large size, the addition of ubiquitin to histones poses a serious challenge. The authors thus used a two-step chemical ligation strategy. First, they synthesized a peptide of the C-terminal part of H2B (residues 117–125) containing an N-terminal cysteine substitution for an alanine. This peptide contained a photoremovable group protecting the cysteine and a ligation auxiliary attached to the ε-NH2 group of Lys120 (H2B-C, Fig. 1a). In the first ligation step, this synthetic peptide was ligated to ubiquitin with a C-terminal α-thioester that was generated by thiolysis of an intein fusion protein. Upon removal of the ligation auxiliary and the cysteine protecting group by UV irradiation, the intermediate product was then ligated to recombinant N-terminal H2B–α-thioester (residues 1–116, H2B-N) in a second EPL reaction. As a final step, the authors carried out Raney nickel-mediated desulfurization to convert the cysteine at position 117 back to alanine, thus generating a traceless and native uH2B substrate (Fig. 1a).
Figure 1.
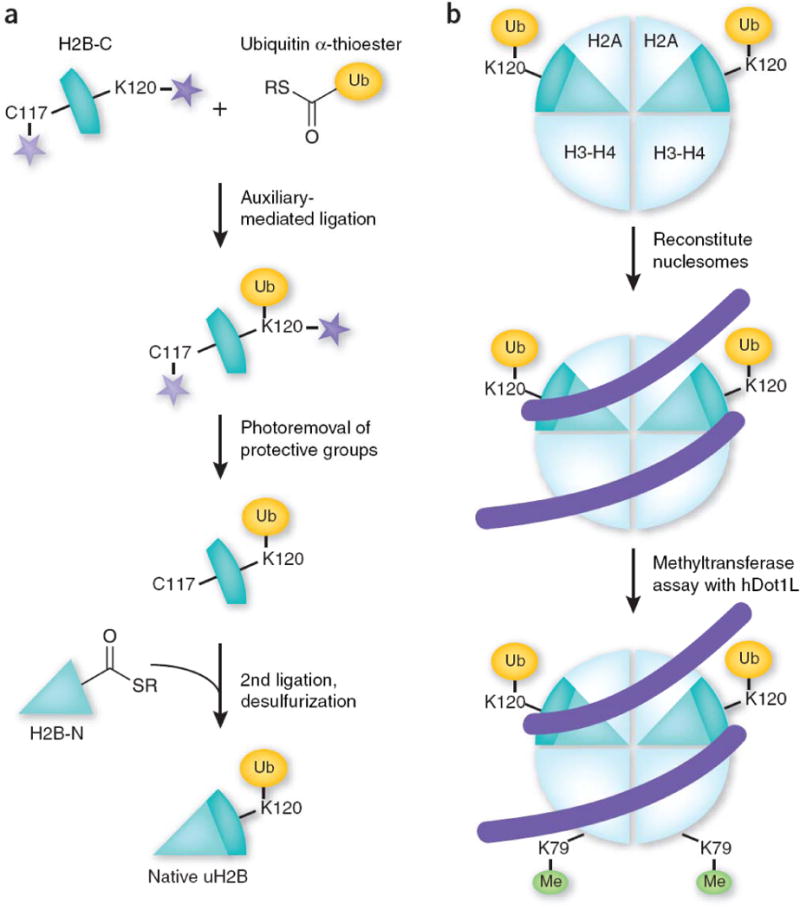
Making of synthetic uH2B for hDot1L-mediated methyltransferase assay. (a) Strategy for making native uH2B using a double native ligation approach. A C-terminal H2B peptide encompassing residues 117–125 and containing a cysteine substitution for Ala117 (H2B-C) is ligated to ubiquitin α-thioester. After photoremoval of the protective group and the ligation auxiliary (stars), the intermediate is ligated to the N-terminal part of H2B (residues 1–116) bearing a C-terminal α-thioester. Raney nickel desulfurization is then performed to convert the cysteine at position 117 back to alanine, which results in the formation of native uH2B. (b) uH2B molecules are incorporated into nucleosomes and subsequently subjected to a histone methyltransferase assay using the H3K79 methyltransferase hDot1L.
With the synthetic uH2B in hand, the authors asked how uH2B affects H3K79 methylation. Toward this end, uH2B was incorporated into nucleosomes and subjected to a hDot1L-mediated methyltransferase assay, which resulted in a significant enhancement in the enzymatic activity of hDot1L (Fig. 1b). This finding provided unequivocal evidence that uH2B directly contributes to H3K79 methylation. Furthermore, the authors investigated whether uH2B can stimulate methylation of H3K79 located in an adjacent nucleosome. To this end, dinucleosomes with only one nucleosome containing uH2B were generated by ligating two mononucleosomes with complementary DNA overhangs. Results indicated that hDot1L efficiently methylated only the uH2B-containing nucleosome, which demonstrates that uH2B specifically stimulates H3K79 methylation on the same nucleosome. The implication of this result is that H3K79 methylation might only occur in the nucleosomes that contain uH2B. However, in yeast the level of H2B ubiquitylation is ~10%10, whereas the level of H3K79 methylation is as high as 90%11. This apparent discrepancy might be explained by the fact that the ubiquitin moiety can be actively removed from uH2B by a deubiquitinase, whereas an enzyme capable of demethylating H3K79 may not exist12,13.
In an attempt to elucidate how uH2B contributes to hDot1L-mediated H3K79 methylation, the authors compared the affinity of hDot1L for mononucleosomes assembled with H2B or uH2B, which revealed no difference, thus leaving the question unresolved. Careful kinetic studies comparing the enzymatic activity of hDot1L toward nucleosomal substrates that contain either H2B or uH2B in combination with co-crystallization of hDot1L in complex with H2B- or uH2B-assembled nucleosomes should shed light on how uH2B contributes to hDot1L-mediated H3K79 methylation. With the advent of synthetic uH2B, we anticipate that similar studies will extend the understanding of how uH2B facilitates H3K4 methylation. Given that PRC1-mediated histone H2A ubiquitylation plays an important role in polycomb gene silencing14, an analogous EPL approach is likely to be adopted for the generation of uH2A as well, which will serve as an important reagent for understanding how H2A ubiquitylation impedes transcription.
The new chemical tools mentioned above open up the possibility of generating nucleosomal substrates carrying multiple modifications by combining the EPL strategy with the incorporation of methyllysine analogs. This will allow the intricate relationship of different histone modifications to be addressed at an unprecedented pace. We have every reason to believe that new chemical approaches will play more and more important roles in understanding the complicated crosstalk of various histone modifications that ultimately determine the transcriptional outcome of specific genes.
References
- 1.Briggs SD, et al. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 2.Dover J, et al. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 3.Ng HH, Xu RM, Zhang Y, Struhl K. J Biol Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 4.Sun ZW, Allis CD. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 5.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Nature. 2008 Apr 30; doi: 10.1038/nature06906. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 7.Minsky N, et al. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 8.Shogren-Knaak M, et al. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 9.Simon MD, et al. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robzyk K, Recht J, Osley MA. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 11.van Leeuwen F, Gafken PR, Gottschling DE. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 12.Henry KW, et al. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klose RJ, Zhang Y. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, et al. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]


