Abstract
We present a case of invasive pneumococcal infection in a healthy 10-month-old infant from whom Streptococcus pneumoniae serotype 23F was isolated from the blood and serotype 23B was isolated from the cerebrospinal fluid. Both serotypes were penicillin nonsusceptible. Pulsed-field gel electrophoresis analysis demonstrated that the two serotypes had distinct DNA patterns, indicating that infection did not occur as a result of capsular transformation but as a result of a mixed infection with two distinct pneumococcal serotypes.
CASE REPORT
In July 2001, a previously healthy 10-month-old boy was admitted to the Tropical Diseases Hospital in GoiÂnia, Central Brazil, with a 2-day history of fever, irritability, and vomiting. There was no history of otitis media, pulmonary infection, or immunosuppressive illness. He had received two doses of an oral antimicrobial 12 h before admission, but his parents did not know its name. He had previously received three doses of Haemophilus influenzae type b conjugate vaccine, but he had not been vaccinated against Streptococcus pneumoniae. On admission, he was afebrile. He had meningeal signs and a bulging fontanelle, but the rest of the physical exam was unremarkable. Blood cultures were obtained and a lumbar puncture was performed immediately. The cerebrospinal fluid (CSF) was cloudy, with a white cell count of 6,750 cells/ml (90% polymorphonuclear cells), a protein level of 85 mg/dl, and a glucose level of 63 mg/dl. A Gram stain of the CSF was negative. A complete blood count showed 26,500 leukocytes/mm3 (14% bands, 56% neutrophils, 26% lymphocytes, and 4% monocytes). The hemoglobin was 9.4 g/dl, the hematocrit was 28.3%, and the platelet count was 274,000/mm3. Treatment was begun with ceftriaxone (100 mg/kg of body weight/day) and dexamethasone, and after a 10-day course of treatment, the patient was discharged in good health.
Cultures of the patient's blood and CSF grew S. pneumoniae. According to NCCLS standards (9), both isolates were nonsusceptible to penicillin (MIC = 0.125 μg/ml) and trimethoprim-sulfamethoxazole (MIC = 4.0 μg/ml) and susceptible to chloramphenicol (MIC = 1.0 μg/ml for blood and 2.0 μg/ml for CSF), ceftriaxone (MIC = 0.06 μg/ml), vancomycin (MIC = 0.5 μg/ml), and erythromycin (MIC = 0.03 μg/ml). Both isolates were serotyped by the Quellung reaction with sera produced by the Statens Seruminstitute, Copenhagen, Denmark. The tests were performed at the Adolfo Lutz Institute, the national reference center for S. pneumoniae located in São Paulo, Brazil. Serotype 23F was identified in the blood, and serotype 23B was identified in the CSF. These results were confirmed by the National Center for Streptococcus in Edmonton, Canada.
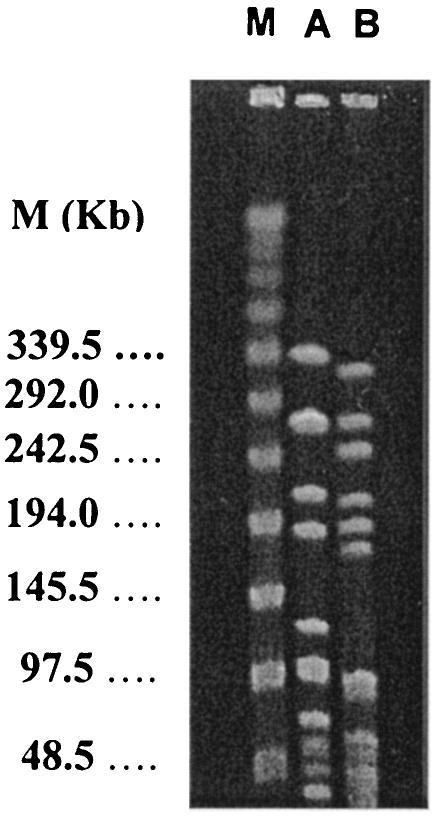
The genetic relatedness of the two pneumococcal isolates was investigated at the Adolfo Lutz Institute by pulsed-field gel electrophoresis (PFGE) of chromosomal DNA after SmaI digestion, as previously described (8). As shown in Fig. 1, the two isolates presented different DNA restriction patterns (lanes A and B) with seven band differences according to the criteria of Tenover et al. (13).
FIG. 1.
PFGE patterns of two S. pneumoniae strains recovered from the same child. Lanes A and B, DNA restriction patterns revealed by PFGE performed with isolates from CSF (serotype 23B) and blood (serotype 23F), respectively; lane M, molecular size markers in kilobases.
Previous studies in the laboratory and in vivo have demonstrated the transfer of a large number of capsular polysaccharide genes between different pneumococcal serotypes (11). In our study, molecular typing provided crucial information for determining whether the two isolates belonged to the same or different clonal groups. PFGE showed that they differed in their DNA restriction patterns. These findings indicated that the two strains most likely belonged to different genetic lineages and that our patient had developed a mixed dual infection caused by two distinct pneumococcal serotypes. The two serotypes most likely had not arisen as a result of capsular transformation.
The frequency of dual infections with S. pneumoniae strains of different phenotypes and genotypes is unknown. The occurrence of dual infections might be underestimated since blood cultures are not routinely obtained in cases of suspected bacterial meningitis in Brazil. Moreover, CSF isolates are not usually submitted for genotyping in the majority of health center laboratories, and serotype results obtained in local laboratories are not normally sent to reference centers for confirmation. In this case it is conceivable to expect that both isolates were present in both the CSF and the blood, but they might have been present in insufficient numbers to be detected by culturing. Also, the current method of serotyping may underestimate the occurrence of dual infections. Coinfection with different pneumococcal strains has been reported for immunodepressed individuals. Jordens et al. (7) detected pneumococcal coinfection in a human immunodeficiency virus-positive patient with two different strains recovered from lung aspirate (serotype 1) and blood (serotype 7C). A mixed meningeal infection with genetically distinct pneumococcal strains (serotypes 7 and 9V) was recently reported for an alcoholic, diabetic patient (3). To our knowledge, this is the first reported case of bacteremia and meningitis due to different S. pneumoniae serotypes in an immunocompetent host.
The nasopharynx is often colonized by more than one pneumococcal serotype (10), raising the possibility that mixed invasive pneumococcal infections may not be uncommon events. Serotype 23F is an isolate frequently recovered from the nasopharynx. Although any pneumococcal serotype may invade a sterile site, some serotypes seem to prefer particular locations. For instance, serotypes 1, 3, 5, 6A or 6B, 9V, 14, 19F, and 23F have been associated with invasive infections (2, 4, 6). In our patient, finding serotype 23B in the CSF was unusual, since it is rarely found among S. pneumoniae strains causing invasive infections. In the study by Hausdorff and colleagues, serotype 23B was seldom found among serotypes that cause meningitis in Latin America (6). Similarly, in the analysis of 3,066 S. pneumoniae isolates from CSF derived from the Brazilian pneumococcal surveillance system, serotype 23B was identified in only 1% of isolates (2). In contrast, serotype 23F ranks as the second most commonly found serotype in CSF or blood in studies conducted in industrialized countries (1, 5) and is the sixth most common serotype in pneumococcal meningitis in children less than 5 years old in Brazil (2, 12). The serotype 23B isolate from the CSF of our patient was not only a nonvaccine serotype, it was also not susceptible to penicillin. Serotype 23B is not included in any formulation of the 7- to 11-valent pneumococcal conjugate vaccines.
Our observations highlight the importance of molecular typing as an additional tool to optimize surveillance systems for invasive bacterial diseases. Continued surveillance will be necessary to determine the extent to which mixed pneumococcal infections occur and their implications for routine case management.
Acknowledgments
This investigation was financially supported by the Division of Vaccines and Immunization from the Pan American Health Organization/WHO, the Bill and Melinda Gates Children's Vaccine Program, the Brazilian Council for Research and Development/CNPq (research grant numbers 470792/01-9, 520399/00-5, and 520580/00-1), and the Secretariat of Health of GoiÂnia Municipality.
We are indebted to David Fedson for critically reviewing the manuscript. We also thank Albert Ko for his comments on the earlier phase of this report.
REFERENCES
- 1.Alpern, E. R., E. A. Alessandrini, K. L. McGowan, L. M. Bell, and K. N. Shaw. 2001. Serotype prevalence of occult pneumococcal bacteremia. Pediatrics 108(2):E23. [Online.] http://www.pediatrics.org/cgi/content/full/108/2/e23. [DOI] [PubMed]
- 2.Brandileone, M. C., A. L. de Andrade, J. L. Di Fabio, M. L. Guerra, and R. Austrian. 2003. Appropriateness of a pneumococcal conjugate vaccine in Brazil: potential impact of age and clinical diagnosis, with emphasis on meningitis. J. Infect. Dis. 187:1206-1212. [DOI] [PubMed] [Google Scholar]
- 3.Chaves, F., C. Campelo, F. Sanz, and J. R. Otero. 2003. Meningitis due to mixed infection with penicillin-resistant and penicillin-susceptible strains of Streptococcus pneumoniae. J. Clin. Microbiol. 41:512-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Fabio, J. L., E. Castaneda, C. I. Agudelo, F. De La Hoz, M. Hortal, T. Camou, G. Echaniz-Aviles, M. Noemi, C. Barajas, I. Heitmann, J. C. Hormazabal, M. C. Brandileone, V. S. Dias Vieira, M. Regueira, R. Ruvinski, A. Corso, M. Lovgren, J. A. Talbot, C. De Quadros, et al. 2001. Evolution of Streptococcus pneumoniae serotypes and penicillin susceptibility in Latin America, Sireva-Vigia Group, 1993 to 1999. Pediatr. Infect. Dis. J. 20:959-967. [DOI] [PubMed] [Google Scholar]
- 5.Doit, C., C. Loukil, P. Geslin, and E. Bingen. 2002. Phenotypic and genetic diversity of invasive pneumococcal isolates recovered from French children. J. Clin. Microbiol. 40:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hausdorff, W. P., J. Bryant, C. Kloek, P. R. Paradiso, and G. R. Siber. 2000. The contribution of specific pneumococcal serogroups to different disease manifestations: implications for conjugate vaccine formulation and use, part II. Clin. Infect. Dis. 30:122-140. [DOI] [PubMed] [Google Scholar]
- 7.Jordens, J. Z., J. Paul, J. Bates, C. Beaumont, J. Kimari, and C. Gilks. 1995. Characterization of Streptococcus pneumoniae from human immunodeficiency virus-seropositive patients with acute and recurrent pneumonia. J. Infect. Dis. 172:983-987. [DOI] [PubMed] [Google Scholar]
- 8.Lefevre, J. C., G. Faucon, A. M. Sicard, and A. M. Gasc. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 31:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Committee for Clinical Laboratory Standards. 2001. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 10.O'Brien, K. L., H. Nohynek, and World Health Organization Pneumococcal Vaccine Trials Carriage Working Group. 2003. Report from a WHO working group: standard method for detecting upper respiratory carriage of Streptococcus pneumoniae. Pediatr. Infect. Dis. J. 22:133-140. [DOI] [PubMed] [Google Scholar]
- 11.Ottolenghi-Nightingale, E. 1972. Competence of pneumococcal isolates and bacterial transformations in man. Infect. Immun. 6:785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reis, J. N., S. M. Cordeiro, S. J. Coppola, K. Salgado, M. G. S. Carvalho, L. M. Teixeira, T. A. Thompson, R. R. Facklam, M. G. Reis, and A. I. Ko. 2002. Population-based survey of antimicrobial susceptibility and serotype distribution of Streptococcus pneumoniae from meningitis patients in Salvador, Brazil. J. Clin. Microbiol. 40:275-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]