“Plants Are Smarter Than We Thought” was the headline news recently in a leading journal (Science News, March 6, 2014; http://news.sciencemag.org/signal-noise/2014/03/plants-are-smarter-we-thought), highlighting an article published elsewhere (Meyer et al., 2014), which presented evidence that plants are able to make smart decisions in response to predation and environment. Plants are well known to have evolved a fascinating adaptability to environment likely because of their sessile nature. Among a long list of complex and unique processes that plants have evolved include the oxygen evolving process of photosynthesis (Ort and Yocum, 1996; Demmig-Adams et al., 2006), which is the life force of animal/mammalian kingdoms, carried out by the semi-autonomous organelle, the chloroplast (Wise and Hoober, 2007); totipotency such that any cell from any plant part can divide, differentiate and yield a fully functional plant (Chupeau et al., 2013); the ability of and restoring structural (Meyer et al., 2014) and metabolic memory (Mattoo et al., 2007; Mattoo and Handa, 2008); the differentiated chromoplasts (from chloroplasts) that store important nutrients for animal and human health (Egea et al., 2010); long distance signaling up and down the whole plant (Ruiz-Medrano et al., 2001; Köhler and Mueller-Roeber, 2004); and recognition and communication via the emission of select class of volatiles (Holopainen and Blande, 2012; Das et al., 2013). Although the phenomenology is well described, the molecular and biochemical mechanisms involved in these processes are better known of some than other of these complex processes. The application of chemistry (and physics) principles has considerably added to the progress made in our understanding of plant life thus far.
Life on earth became possible some 3.5 billion years ago because “chemistry begat biology” (Aberlin, 2014). However, little is known or understood of what led to the transition from chemistry to biology (Quoted from Jack Szostak, Harvard Medical School, February 2012, as appeared in The Scientist 03-2014). Nonetheless, the past two centuries witnessed a close merger between chemistry (and physics) and biology, producing a distinct platform for biochemistry (Neuberg, 1903 in en.wikipedia.org/wiki/Carl_Neubergı) to bear on our understanding of the functions of a living cell and its complex nature. Because of its very nature, this discipline unearthed common and distinct alphabets and trends of biochemical processes that led to a concept of “unity in diversity,” exemplifying common principles that underlie the uniformity of life in diverse kingdoms. Kluyver's studies on microorganisms led to the discovery that ecological microbiology has a biochemical basis (Kluyver, 1924 in Florkin, 1924). The discovery that all the diverse organisms harbor same macromolecules and genetic code led to the knowledge that all organisms, from microorganisms to human beings, are built from similar molecular components with some variations (Berg et al., 2002).
The past century was a witness to advances in diverse disciplines including genetics, and micro- and macro-elements of biological chemistry. Thus, major discoveries were made on biochemical pathways (by Krebs, encompassing 1932, 1937, and 1957), cofactors (for instance, coenzyme A, by Fritz A. Lippman), enzymes (by Wilhelm Kuhne, as early as 1878), proteins (by Sumner), nucleic acids—DNA (by Watson and Crick) and RNA world (http://en.wikipedia.org/wiki/History_of_RNA_biology), cell membrane function and signaling pathways (http://en.wikipedia.org/wiki/History_of_biochemistry). These advances and discoveries brought together pieces of the puzzle(s) and laid the foundation for modern day molecular biology, biotechnology and epigenetic regulation. Progress in the identification and quantification of low abundant molecules led to Metabolome, which delves into the plasticity and/or homeostasis of primary and secondary metabolites, while small RNAs (including snoRNAs and miRNAs) brought to the fore the regulation by non-coding RNAs. Now and again, chemistry is having a bearing on the tremendous progress made in life sciences and our understanding of the biological processes.
The application of the innovative recombinant DNA technology enabled transfer of genes across kingdoms, creating the modern day biotechnological intervention to revolutionize research, and changing for good the paradigms in agricultural and medical research (Cohen et al., 1973). Transgenic research is the buzzword for all sorts of remedial measures, be that to prevent/solve diseases, produce recombinant products, or grow more food. Because of the totipotency of the plant cells, plant biologists were able to transform and develop plants engineered with novel, heterologous and endogenous genes. This has enabled unambiguous confirmation of gene function in planta and testing the novel genotypes for global changes in macromolecules and micromolecules alike. The detection, unambiguous identification, quantification and fast analysis of minutest amounts of cellular micromolecules including plant hormones has materialized through high resolution superior gas chromatography, ultra high performance liquid chromatography coupled to triple-quadrupole mass spectrometer, and Nuclear Magnetic Resonance (NMR) Spectroscopy, and this has become instrumental in understanding the chemical footprints during different phases of growth and development of plants.
The human genome sequencing was completed ahead of expected time, and this success catalyzed moves to sequence other genomes including crop plants, all expedited by the progress made in Information Technology - mining the data and putting pieces of the puzzle together in shortest time possible. Progress in the Genomics (http://www.sciencemag.org/site/feature/misc/webfeat/plantgenomes/feature.html) field ushered in the Epigenomics (Schmitz and Zhang, 2011) science, and together they have thrown the whole biological kingdom wide open to new research, furthering our understanding of the fundamental basis of life, development and regulation. The length and breadth of data that accumulate each day is humongous and has led to the active collaboration in the interface of bioinformatics and biology. This merging of two disciplines has enhanced the timely solutions and brought to bear on the way science in this century is being conducted worldwide.
More importantly, these advances in technology and biology have prepared the groundwork for the benefit of the humankind by providing reagents and roadmaps to solve the issues world is faced with at this time. This is most applicable and needed for human welfare, to have agricultural sustainability and feed the world, because there clearly is an increasing population growth trend throughout the globe except in Europe. World human population is expected to reach close to 9.6 billion by 2050 (http://esa.un.org/wpp/). The production capacity to grow more food to meet the demands of the burgeoning population gets complicated in view of the limits in the arable land that is less and less available, declining trends in crop yields, less sustainability of resources such as water, and losses due to abiotic and biotic stresses. Added to the challenge in producing more (nutritious) food is the need to fight malnutrition and reduce extensive chemical use in agriculture for a cleaner environment. Breeding strategies employing marker-assisted selection for high yielding varieties as well as for identifying germplasm resistant to abiotic and biotic stresses are already in vogue. Another approach is to introduce agronomically important genes and those that can help crops withstand environmental extremes into major and minor crops using genetic engineering technology.
Making biotechnology more sustainable
Biological revolution—genetic engineering and biotechnology—has a promise to enhance crop resilience and make a breeder's dream come true: produce more in a shorter time, reduce our reliance on agricultural chemicals such as pesticides and fungicides, and add to environment-friendly sustainable agriculture (Mattoo, 2013). Some of the desirable traits that have been successfully introduced into crops by genetic engineering include insect resistance, disease resistance, herbicide tolerance, chilling tolerance, delayed fruit ripening, prolonged shelf-life, texture and processing attributes. Irrespective of this promise, the rapid pace in the development of novel engineered crops and the considerable interest generated among growers and consumers, the application of this technology and/or acceptance of genetically engineered foods world-wide has been hampered by continued debate on the safety of such produce. These issues include ethical concerns, potential toxicity, selection markers, undesired gene flow, development of resistance against herbicides and pesticides. Significant research efforts have gone into satisfying the consumers' concerns. Risk assessment studies have focused on determining the substantial equivalence of genetically engineered food and traditionally-bred wild type crops. In such studies, optimized unambiguous methodologies are required to search for differences between the engineered and non-engineered food. Many studies in the literature have not revealed any unusual compound in the genetically engineered crops, suggesting that they are basically substantially equivalent to non-engineered foods (Baker et al., 2006; Mattoo et al., 2006; Sobolev et al., 2010; Farre et al., 2011).
Plant-breeding programs normally include assessing and accounting the influence of genetic background (G), ecosystem environment (E), and G × E interactions singly and/or together on their impact on the growth and development of crops/plants, particularly for producing suitable genotypes for multiple environments (El-Soda et al., 2014). Evaluating these parameters also helps understand plant fitness trade-offs and evolutionary ecology El-Soda et al. (2014). Because the genetically engineered plants harbor a modified gene(s) and the positioning of the introduced gene is mostly a random event, it seems important that such plants be tested for agronomic and biological performance in the fields under multiple ecosystem services to assess which production system is conducive for impact on different parameters of the engineered plant(s) grown side by side the non-engineered wild type control(s). Studies of this kind/nature reported in the literature are miniscule.
It is imperative that controlled field studies of genetically engineered and other crops are carried out in an unambiguous manner alongside the wild type to ascertain which production system may provide the best medium. Such studies are expected to provide new ways to leverage growth enhancement, crop resistance to stresses, and improve the nutrient content of the edible produce in an eco-friendly environment. For instance, a major agriculturally utilized genetic event is the introduction of the Bacillus thuringenesis (Bt) protein gene in diverse crops, particularly cotton and maize, to make these crops resistant to insect pests (Marvier et al., 2007; James, 2013). The Bt crops have generated good revenue both for the farmer and the industry. However, like the non-engineered crops that suffer losses because of the adaptability of insects and microbial pathogens, it is expected that Bt crops and other such novel genetic materials will also be manipulated by the pathogens in the long run. Thus, in the field evaluation, populations of western rootworm were identified that had developed resistance to multiple Bt-maize toxins (Gassmann et al., 2014). In the above-mentioned context, i.e., developing a friendlier ecosystem suitable for growing each crop with a unique genetic event(s), it is of critical importance to understand their long-term performance in practical terms as well as a novel resource to discern various players in biological adaptability. Thus, studies that were geared to test the effect of natural predators in a defined ecosystem on the performance of Bt crops demonstrated that, indeed, natural enemies of insects help delay the development of insect resistance to Bt crops (Liu et al., 2014). Just as in conventional breeding strategies, so with novel biotech crops any unusual observation(s) in field performance behavior will have to be scientifically tackled and rectified.
Developing robust crop plants to resist abiotic stressors
Plant growth in nature is compromised on a daily basis because plants expend energy to adjust and adapt to changing environment, which becomes more precarious under additional stresses due to drought and extreme temperatures, for instance, excessive summer temperatures of the tropics, or in cooler climatic situations. Thus, water availability, temperature, soil properties and ecosystem can dictate the growth response and yield of a crop plant. Moreover, each plant adapts to environment based on the genetic make up, accordingly impacting growth, development and yield (Porter and Semenov, 2005).
Molecular responses to unfavorable environment include a medley of genes and signal transduction pathways that are tightly regulated and empower plants to combat the stress conditions. Although much of this regulation is at transcriptional, post-transcriptional, and post-translational levels, the intricacy is of essence at the transcriptional level involving chromatin modification and remodeling, cis-regulatory elements located upstream and downstream the coding region of the gene, and trans regulatory transcription factors (Luo et al., 2012). Also, other important players that are directly or indirectly associated with imparting tolerance to abiotic stresses include protective proteins (including dehydrins, heat shock proteins—HSPs, Late Embryogenesis Abundant proteins—LEA Vierling, 1991; Wang et al., 2004; Kazuko and Shinozaki, 2006; Lipiec et al., 2013; Mu et al., 2013), osmolytes (proline/trehalose/sugars Fougere et al., 1991; Petrusa and Winicov, 1997; Wingler, 2002; Avonce et al., 2006; Ito et al., 2006; Ge et al., 2008; Zhang et al., 2010; Hayat et al., 2012; Yanhui et al., 2012), glycine betaine (Sakamoto and Murata, 2002; Quan et al., 2004; Wang et al., 2010; Chen and Murata, 2011), signaling molecules (polyamines Roy and Wu, 2002; Navakouidis et al., 2003; Capell et al., 2004; Kasukabe et al., 2006; Alcázar et al., 2010; Wi et al., 2006; Liu et al., 2007; Kusano et al., 2008; Wen et al., 2008; Cheng et al., 2009; Kalamaki et al., 2009; Gill and Tuteja, 2010; Shukla and Mattoo, 2010; inositol Xiong et al., 2001; Sengupta et al., 2008); and hormones (abscisic acid—ABA Davies and Zhang, 1991; Saradhi et al., 2000; ethylene—C2H4 Hinz et al., 2010; Quan et al., 2010; Xiong et al., 2013; and methyl jasmonate—meJA Bartels and Sunkar, 2005; Vincour and Altman, 2005; Wu et al., 2008; Jan et al., 2013), several of which have been validated for mitigating abiotic stresses.
Genome sequencing of model and crop plants before and after exposure to a given stress has identified candidate genes whose role(s) in response to different abiotic stresses can then be tested/validated by expression and down-regulation in homologous as well as in heterologous systems. Thus, stress responsive genes including specific transcription factors have been identified by comparative transcriptomics. Enormous activity regarding validated data on a few crops for the involvement of transcription factors (b-ZIP, ERF/AP2 family, DOF, HD-ZIP, MYB, NAC, WRKY, and Zn-finger) (Riechmann and Ratcliffe, 2000; Dubouzet et al., 2003; Hu et al., 2006; Ito et al., 2006; Mittler, 2006; Nakashima et al., 2007; Weiste et al., 2007; Wu et al., 2008; Xiang et al., 2008; Zou et al., 2008; Gao et al., 2009; Lu et al., 2009; Oh et al., 2009; Jeong et al., 2010; Su et al., 2010; Takasaki et al., 2010; Zhang et al., 2010; Zhao et al., 2010; Wan et al., 2011; Liu et al., 2012; Yang et al., 2012; Jan et al., 2013), and other genes (CDPKs, HAP/CAAT, HSPs-LEA family, MAPKKK) (Vierling, 1991; Saijo et al., 2000; Wang et al., 2004; Chandra Babu et al., 2004; Shou et al., 2004; Kazuko and Shinozaki, 2006; Xu et al., 1996; Nelson et al., 2007; Xiao et al., 2007; Ning et al., 2010; Duan and Cai, 2012; Lipiec et al., 2013; Mu et al., 2013) have shown the true promise of these candidates as stress modulators. Members from each transcription factor family show protective phenotypes against multiple stresses such as cold, drought and excess salt (summarized in Figure 1; Shukla and Mattoo, 2013; Mattoo et al., 2014). Similarly, engineering targeted metabolic pathways enable multi-throng efforts to produce and sustain agricultural commodities for the benefit of the farmer and the consumer (Reugera et al., 2012).
Figure 1.
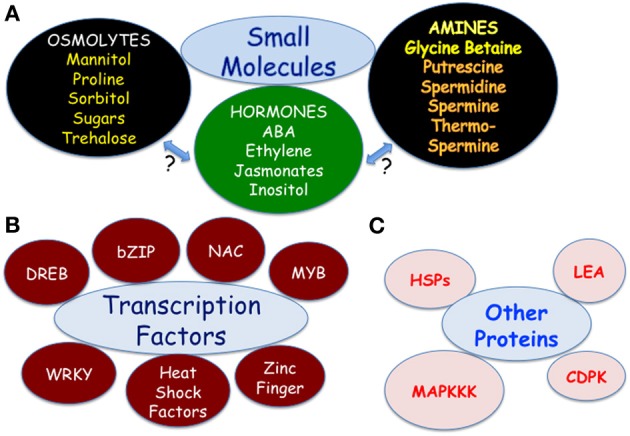
Players that empower crop plants to withstand abiotic stresses—drought, temperature extremes and high saline soils. (A) Small molecules such as osmolytes, biogenic amines and hormones. (B) Transcriptional factors. (C) Heat shock proteins (chaperone proteins) and protein kinases.
Such successful efforts on engineering crop plants for resistance to different abiotic stresses have started paying dividends since industry has generated some drought resistant germplasm for the farmers. This progress in translating basic research into viable products offers a roadmap for intensifying efforts to develop resistant germplasm for all the major and minor crops and test them under varied climatic conditions and different ecosystems worldwide.
Nutrient-enhanced produce and human health
It is more and more recognized that phytonutrient-rich diet containing high dose of antioxidants/vitamins, present in fruit and vegetables, potentially prevents polygenic diseases such as epithelial cancers, diabetes, atherosclerosis, hypertension, cardiovascular diseases and osteoporosis (Mattoo et al., 2010; Shukla and Mattoo, 2010; Fatima et al., 2013). This is the reason that antioxidant/vitamin supplements are available over the counter in pharmacies and grocery stores worldwide. The well-known health-promoting phytonutrients that have been proposed to alleviate disease symptoms and reduce incidence of diseases include carotenoids (β-carotene, lutein, lycopene), polyphenolics/flavonoids, vitamins C and E, isothiocyanates and glucosinolates. That disease and nutrition are intertwined in humans is becoming relevant in this science/technology, post-genomic era and the beneficial effects seem linked to interactions among different antioxidants present in food, although little is known about the nature of these interactions (Shukla and Mattoo, 2010; Fatima et al., 2013). Thus, the need for a dense nutrient intake through eating a variety of foods including grains, fruits and vegetables has been emphasized in the dietary guidelines for Americans (http://www.dietaryguidelines.gov).
The increasing interest in the bioactive molecules present in grains, fruits and vegetables has catalyzed research interest in developing definitive basic information on their content, with more than 40 such molecules having been identified and deemed essential for a healthy life (Failla, 2012). The quantity required of each nutrient to decrease “disease risk” is a critical factor while bioavailability of nutrients in a diet determines how much of the good nutrient's potential is realized (Shukla and Mattoo, 2010; Fatima et al., 2013). Although grains, vegetables, and fruits are sources of antioxidants and vitamin nutrients, the levels in general are low, likely due to tight genetic and developmental controls of their metabolic pathways during plant growth and development (Paine et al., 2005). Further, biosynthetic pathways for phytonutrients and their regulation are incomplete in many instances, and “germplasm” with higher accumulation of phytonutrients is not easily available. Modern tools of metabolomics have already overcome these limitations by identifying, purifying and quantifying hundreds of biochemicals (Mattoo et al., 2006; Saito and Matsuda, 2010). Mutagenesis and TILLING (Targeting Induced Local Lesions in Genomes) approaches are being used to use selection against genes that negatively regulate biosynthesis or accumulation of phytonutrients (Zhang et al., 2009; Handa et al., 2011).
Molecular genetics is now providing tools to identify and characterize genes regulating the biosynthesis of phytonutrients in plants. Thus, genetic/metabolic engineering of the rate limiting steps in the biosynthesis of a compound has facilitated increased levels of phytonutrients in plant tissues/organs (Shukla and Mattoo, 2010; Fitzpatrick et al., 2012; Fatima et al., 2013; Handa et al., 2014). Animal and human trials can help determine which nutrient(s) needs to be enhanced through molecular strategies designed to increase their contents in grains, vegetables, and fruits. Interestingly, the human health paradigm has been revised to include preventive, dietary intervention to ameliorate diseases and physiological disorders. More scientific research and validation are required before phytonutrients become a “mantra” for healthy living (Handa et al., 2014).
Regulatable promoters fused to heterologous genes allowed higher levels of carotenoids to accumulate in a fruit-specific manner in tomato (Rosati et al., 2000; Dharmapuri et al., 2002; Fraser et al., 2002; Mehta et al., 2002), and similarly in other instances where constitutive promoters were employed (Römer et al., 2000; D'Ambrosio et al., 2004). Use of regulatable promoters fused to the E. coli DXS gene (Enfissi et al., 2005) or suppression RNAi to downregulate the photomorphogenesis regulatory protein gene DET1 (Davuluri et al., 2005) also led to high levels of carotenoids in tomato fruit. Also, metabolic engineering of polyamine biosynthesis in tomato by introducing fruit-specific expression of yeast S-adenosylmethionine (SAM) decarboxylase gene led to 200–300% increase in lycopene content (Mehta et al., 2002) while constitutive expression of yeast spermidine synthase increased carotenoid content by 40% (Nambessan et al., 2010). TOMATO AGAMOUS-LIKE 1 (TAGL1), a MADS-box transcription factor, expression resulted in higher accumulation of lycopene and naringenin chalcone (Itkin et al., 2009). Another study used RNAi-mediated fruit-specific suppression of 9-cis-epoxycarotenoid dioxygenase 3 (NCED3) to suppress ABA synthesis to stimulate accumulation of upstream compounds such as β-carotene and lycopene in transgenic tomato fruits (Sun et al., 2012). In this regard, a mutation in zep1 caused ABA-deficiency in tomato plants with concomitant accumulation of 30% more carotenoids in mature red tomato fruit (Galpaz et al., 2008).
A few other selective examples of engineering phytonutrient content are:
GDP-L-galactose phosphorylase (VTC2) expression increased vitamin C level in tomato, strawberry, and potato by 2-6 fold (Bulley et al., 2012); mammalian GTP cyclohydrolase I caused 140-fold increase in pteridine and 2-fold increase in folate, and by combining with aminodeoxychorismate synthase (PABA biosynthesis), folate levels were increased 19-fold (Diaz de la Garza et al., 2007); expression of α-tocopherol methyltransferase and γ-tocopherol methyltransferase, respectively in soy oil and lettuce, increased vitamin E several-fold (see Fatima et al., 2013); simultaneous expression of genes for β-carotene, ascorbate, and folate biosynthetic pathways increased β-carotene (169-fold), ascorbate (6-fold), and folate (2-fold) in corn (Naqvi et al., 2009); expression of Rosea1 and Delila or flavonoid-related R2R3-MYB increased flavonoid content in tomato pericarp (Butelli et al., 2008); RNAi suppression of the DE-ETIOLATED1 (DET1) gene (a photomorphogenesis regulatory gene) caused several-fold increase in carotenoid, tocopherol, phenylpropanoids and flavonoids (Enfissi et al., 2010); metabolic engineering of diverse genes that encode enzymes for secondary metabolites all increased content of polyphenolic flavonoids in tomato fruit (Muir et al., 2001; Niggeweg et al., 2004; Giliberto et al., 2005; Schijlen et al., 2006), α-tocopherol (vitamin E) in potato tubers (Crowell et al., 2008), and some produced novel flavonoids in tomato fruit (Schijlen et al., 2006).
It is apparent that modern biotechnology in conjunction with Metabolomics is enabling tissue specific redesign of primary and secondary metabolic pathways so as to accumulate high levels of phytonutrients in different plant systems. Thus, transgenic crops are an addition to the genetic resource to further define genetic, biochemical, and physiological regulation of cellular metabolism pathways, including enhancing functional metabolites in produce and provide novel “specialty crops” to the public. In return, public awareness of benefits of consumer-driven products such as in human health will further add to new markets for specialty, highly nutritious crops.
Future perspective
Agricultural biologists have their work cut out for translating large database of fundamental nature from laboratory, growth chamber and greenhouses studies to the field for securing and producing (nutritious) food and making agriculture sustainable. All kinds of transgenic lines have been developed, including transgenic lines that have promise of withstanding environmental extremes (abiotic and biotic) and others that have a high dose of phytonutrients. How they fare in the field is the need of the day, effectiveness of this translation will require diligence and a thorough knowledge of the investigated trait in each crop (Ronald, 2011; Nelissen et al., 2014). Moreover, there appears to be a probability that ecological surprises could be more prevalent because of global climate change and interacting environment extremes (Lindenmayer et al., 2010). Also, the point to note is that nutrient levels in crops are influenced by genotype/cultivar, growth condition and developmental stage of the crop (Shukla and Mattoo, 2010; Lee and Scagel, 2014), therefore unambiguous analysis of edible crops grown under similar conditions in the field is needed to determine the robustness of a trait (Neelam et al., 2008; Mattoo and Teasdale, 2010). Convergence of agriculture with health and wealth is a distinct possibility (Dube et al., 2012), and would also be benefited by developing necessary toolkits to establish bon a fide natural products chemistry and translate it into alternative medicine.
Thus, with the available genetic toolkits together with advanced technologies, chemical genetics, and progression with alternative agricultural practices, future action plan is more or less laid out and roadmap defined for scientists and farmers to work together to meet the challenges the humankind faces in this new century. It is evident that there is a need to prioritize translational research as an important component of bench scientists' goals of research. Certainly, there is hope in the horizon for developing new types of crop plants that can yield more and be nutritious with less inputs, are resilient to harsher environment, and are disease tolerant.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I wish to thank Dr. Marie Soulière of Frontiers for her comments and suggestions for improving the original manuscript. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
References
- Aberlin M. B. (2014). Let there be life. The Scientist 13 [Google Scholar]
- Alcázar R., Altabella T., Marco F., Bortolotti C., Reymond M., Koncz C., et al. (2010). Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231, 1237–1249 10.1007/s00425-010-1130-0 [DOI] [PubMed] [Google Scholar]
- Avonce N., Mendoza-Vargas A., Morett E., Iturriaga G. (2006). Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 6:109 10.1186/1471-2148-6-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J. M., Hawkins N. D., Ward J. L., Lovegrove A., Napier J. A., Shewry P. R., Beale M. H. (2006). A metabolomic study of substantial equivalence of field-grown genetically modified wheat. Plant Biotechnol. J. 4, 381–392 10.1111/j.1467-7652.2006.00197.x [DOI] [PubMed] [Google Scholar]
- Bartels D., Sunkar R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58 10.1080/07352680590910410 [DOI] [Google Scholar]
- Berg J. M., Tymoczko J. L., Stryer L. (2002). Biochemistry, 5th Edn. New York, NY: W. H. Freeman [Google Scholar]
- Bulley S., Wright M., Rommens C., Yan H., Rassam M., Lin-Wang K., et al. (2012). Enhancing ascorbate in fruits and tubers through over-expression of the l-galactose pathway gene GDP-l-galactose phosphorylase. Plant Biotechnol. J. 10, 390–397 10.1111/j.1467-7652.2011.00668.x [DOI] [PubMed] [Google Scholar]
- Butelli E., Titta L., Giorgio M., Mock H. P., Matros A., Peterek S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308 10.1038/nbt.1506 [DOI] [PubMed] [Google Scholar]
- Capell T., Bassie L., Christou P. (2004). Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc. Natl. Acad. Sci. U.S.A. 101, 990–991 10.1073/pnas.0306974101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra Babu R., Zhang J., Blum A., Hod T. H. D., Wue R., Nguyen H. T. (2004). HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci. 166, 855–862 10.1016/j.plantsci.2003.11.023 [DOI] [Google Scholar]
- Chen T. H., Murata N. (2011). Glycinebetaine protects plants against abiotic stress: mechanisms and biotechnological applications. Plant Cell Environ. 34, 1–20 10.1111/j.1365-3040.2010.02232.x [DOI] [PubMed] [Google Scholar]
- Cheng L., Zou Y. J., Ding S. L., Zhang J. J., Yu X. L., Cao J. S., et al. (2009). Polyamine accumulation in transgenic tomato enhances the tolerance to high temperature stress. J. Integr. Plant Biol. 51, 489–499 10.1111/j.1744-7909.2009.00816.x [DOI] [PubMed] [Google Scholar]
- Chupeau M.-C., Granier F., Pichon O., Renou J.-P., Gaudin V., Chupeau Y. (2013). Characterization of the early events leading to totipotency in an Arabidopsis protoplast liquid culture by temporal transcript profiling. Plant Cell 25, 2444–2463 10.1105/tpc.113.109538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S., Chang A., Boyer H., Helling R. (1973). Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. U.S.A. 70, 3240–3244 10.1073/pnas.70.11.3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell E. F., McGrath J. M., Douches D. S. (2008). Accumulation of vitamin E in potato (Solanum tuberosum) tubers. Transgenic Res. 17, 205–217 10.1007/s11248-007-9091-1 [DOI] [PubMed] [Google Scholar]
- D'Ambrosio G., Griorio G., Marino I., Merendino A., Petrozza A., Salfi L., et al. (2004). Virtually complete conversion of lycopene into β-carotene in fruits of tomato plants transformed with the tomato lycopene β-cyclase (tlcy-b) cDNA. Plant Sci. 166, 207–214 10.1016/j.plantsci.2003.09.015 [DOI] [Google Scholar]
- Das A., Lee S.-H., Hyun T. K., Kim S.-W., Kim J.-Y. (2013). Plant volatiles as method of communication. Plant Biotechnol. Rep. 7, 9–26 10.1007/s11816-012-0236-1 [DOI] [Google Scholar]
- Davies W. J., Zhang J. (1991). Root signals and the regulation of growth and development of plants in drying soil. Annu. Rev. Plant Physiol. 42, 55–76 10.1146/annurev.pp.42.060191.000415 [DOI] [Google Scholar]
- Davuluri G. R., van Tuinen A., Fraser P. D., Manfredonia A., Newman R., Burgess D., et al. (2005). Fruit-specific RNAi-mediated suppression of DET1 enhances tomato nutritional quality. Nat. Biotechnol. 7, 825–826 10.1038/nbt1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B., Adams W. W., 3rd, Mattoo A. K. (eds.). (2006). Photoprotection, Photoinhibition, Gene Regulation, and Environment. Advances in Photosynthesis and Respiration, Vol. 21. Dordrecht: Springer [Google Scholar]
- Dharmapuri S., Rosati C., Pallara P., Aquilani R., Bouvier F., Camara B., et al. (2002). Metabolic engineering of xanthophyll content in tomato fruits. FEBS Lett. 519, 30–34 10.1016/S0014-5793(02)02699-6 [DOI] [PubMed] [Google Scholar]
- Diaz de la Garza R. I., Gregory J. F., 3rd, Hanson A. D. (2007). Folate biofortification of tomato fruit. Proc. Natl. Acad. Sci. U.S.A. 104, 4218–4222 10.1073/pnas.0700409104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J., Cai W. (2012). OsLEA3-2, an abiotic sress induced gene of rice plays a key role in salt and drought tolerance. PLoS ONE 7:e45117 10.1371/journal.pone.0045117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube L., Pingali P., Webb P. (2012). Paths of convergence for agriculture, health, and wealth. Proc. Natl. Acad. Sci. U.S.A. 109, 12294–12301 10.1073/pnas.0912951109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet J. G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E. G., Miura S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought, high salt and cold responsive gene expression. Plant J. 33, 751–763 10.1046/j.1365-313X.2003.01661.x [DOI] [PubMed] [Google Scholar]
- Egea I., Barson C., Bian W., Purgatto E., Latche A., Chervin C., et al. (2010). Chromoplast differentiation: current status and perspectives. Plant Cell Physiol. 51, 1601–1611 10.1093/pcp/pcq136 [DOI] [PubMed] [Google Scholar]
- El-Soda M., Malosetti M., Zwaan B. J., Koornneef M., Aarts M. G. M. (2014). Genotype × environment interaction QTL mapping in plants: lessons from Arabidopsis. Trends Plant Sci. [Epub ahead of print]. 10.1016/j.tplants.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Enfissi E. M. A., Fraser P. D., Lois L. M., Boronat A., Schuch W., Bramley P. M. (2005). Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids. Plant Biotechnol. J. 2, 17–27 10.1111/j.1467-7652.2004.00091.x [DOI] [PubMed] [Google Scholar]
- Enfissi E. M., Barneche F., Ahmed I., Lichtle C., Gerrish C., McQuinn R. P., et al. (2010). Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22, 1190–1215 10.1105/tpc.110.073866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failla M. (2012). Interdisciplinary team-work for providing global citizens with a safe, adequate and healthy diet, Proceedings 7th International Postharvest Symposium (Malaysia: ), 6. [Google Scholar]
- Farre G., Twyman R., Zhu C., Capell T., Christou P. (2011). Nutritionally enhanced crops and food security: scientific achievements versus political expediency. Curr. Opin. Biotechnol. 22, 245–251 10.1016/j.copbio.2010.11.002 [DOI] [PubMed] [Google Scholar]
- Fatima T., Handa A. K., Mattoo A. K. (2013). Functional foods: genetics, metabolome, and engineering phytonutrient levels, in Natural Products, Part V, eds Ramawat K. G., Merillon J. -M. (Berlin; Heidelberg: Springer-Verlag; ), 1715–1749 [Google Scholar]
- Fitzpatrick T. B., Basset G. J., Borel P., Carrari F., DellaPenna D., Fraser P. D., et al. (2012). Vitamin deficiencies in humans: can plant science help? Plant Cell 24, 395–414 10.1105/tpc.111.093120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florkin M. (1960). Unity and diversity in biochemistry. Pergamon. (Kluyver, A. J. (1924). Eenheid en verscheidenheid in de stofwisseling der microben'. Cbemiscb Week- blud, 21, 266. Unity and diversity in the metabolism of micro-organisms.1924 ALbert J. Kluyver Translation from Albert Jan Kluyver, His Life and Work, 1959, North- Holland Publishing Company, Amsterdam). [Google Scholar]
- Fougere F., Le-Rudulier D., Streeter J. G. (1991). Effects of salt stress on amino acid, organic acid, and carbohydrate composition of roots, bacteroids, and cytosol of alfalfa (Medicago sativa L.). Plant Physiol. 96, 1228–1236 10.1104/pp.96.4.1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P. D., Romer S., Shipton C. A., Mills P. B., Kiano J. W., Misawa N., et al. (2002). Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc. Natl. Acad. Sci. U.S.A. 99, 1092–1097 10.1073/pnas.241374598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N., Wang Q., Menda N., Zamir D., Hirschberg J. (2008). Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 53, 717–730 10.1111/j.1365-313X.2007.03362.x [DOI] [PubMed] [Google Scholar]
- Gao S. Q., Chen M., Xia L.-Q., Xiu H. J., Xu Z. S., Li L.-C., et al. (2009). A cotton (Gossypium hirusutum) DRE-binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Rep. 28, 301–311 10.1007/s00299-008-0623-9 [DOI] [PubMed] [Google Scholar]
- Gassmann A. J., Petzold-Maxwell J. L., Clifton E. H., Dunbar M. W., Hoffmann A. M., Ingber D. A., et al. (2014). Field-evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. Proc. Natl. Acad. Sci. U.S.A. 111, 5141–5146 10.1073/pnas.1317179111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L. F., Chao D. Y., Shi M., Zhu M. Z., Gao J. P., Lin H. X. (2008). Overexpression of the trehlose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228, 191–201 10.1007/s00425-008-0729-x [DOI] [PubMed] [Google Scholar]
- Giliberto L., Perrotta G., Pallara P., Weller J. L., Fraser P. D., Bramley P. M., et al. (2005). Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol. 137, 199–208 10.1104/pp.104.051987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. S., Tuteja N. (2010). Polyamines and abiotic stress tolerance in plants. Plant Signal. Behav. 5, 26–33 10.4161/psb.5.1.10291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa A. K., Anwar R., Mattoo A. K. (2014). Biotechnology of fruit quality, in Fruit Ripening: Physiology, Signalling and Genomics, eds Nath P., Bouzayen M., Mattoo A. K., Pech J.-C. (Oxfordshire, UK: CAB International Publishers; ). [Google Scholar]
- Handa A. K., Tiznado-Hernández M. E., Mattoo A. K. (2011). Fruit development and ripening: a molecular perspective, in Plant Biotechnology and Agriculture: Prospects for the 21st Century, eds Altman A., Hasegawa P. M. (New York, NY: Oxford Academic Press; ), 405–424 [Google Scholar]
- Hayat S., Hayat Q., Alyemeni M. N., Wani A. S., Pichtel J., Ahmad A. (2012). Role of proline under changing environments: a review. Plant Signal. Behav. 1456–1466 10.4161/psb.21949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz M., Wilson I. W., Yang J., Buerstenbinder K., Llewellyn D., Dennis E. S., et al. (2010). Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 153, 757–772 10.1104/pp.110.155077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holopainen J. K., Blande J. D. (2012). Molecular plant volatile communication. Adv. Exp. Med. Biol. 739, 17–31 10.1007/978-1-4614-1704-0_2 [DOI] [PubMed] [Google Scholar]
- Hu H., Dai M., Yao J., Xiao B., Zhang Q., Xiong L. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103, 12987–12992 10.1073/pnas.0604882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M., Seybold H., Breitel D., Rogachev I., Meir S., Aharoni A. (2009). The TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60, 1081–1095 10.1111/j.1365-313X.2009.04064.x [DOI] [PubMed] [Google Scholar]
- Ito Y., Maruyama K., Taji T., Kobayashi M., Seki M., Shinozaki K., et al. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153 10.1093/pcp/pci230 [DOI] [PubMed] [Google Scholar]
- James C. (2013). Global status of commercialized biotech/GM crops: 2013, in International Service for the Acquisition of Agri-biotech Applications. Brief No. 46 (Ithaca, NY: ISAAA; ). [Google Scholar]
- Jan A., Maruyama K., Todaka D., Kidokorom S., Abo M., Yoshimura E., et al. (2013). OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 161, 1202–1216 10.1104/pp.112.205385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J. S., Kim Y. S., Baek K. H., Jung H., Ha S.-H., Choi Y. D., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197 10.1104/pp.110.154773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamaki M. S., Merkouropoulos G., Kanellis A. K. (2009). Can ornithine accumulation modulate abiotic stress tolerance in Arabidopsis? Plant Signal. Behav. 4, 1099–1101 10.4161/psb.4.11.9873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasukabe Y., He L., Watakabe Y., Otani M., Shimada T., Tachibana S. (2006). Improvement of environmental stress tolerance of sweet potato by introduction of genes for spermidine synthase. Plant Biotechnol. 23, 75–83 10.5511/plantbiotechnology.23.75 [DOI] [Google Scholar]
- Kazuko Y. S., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- Köhler B., Mueller-Roeber B. (2004). Remote control – cell and organ communication within plants. New Phytol. 161, 321–324 10.1111/j.1469-8137.2004.00988.x [DOI] [PubMed] [Google Scholar]
- Kusano T., Berberich T., Tateda C., Takahashi Y. (2008). Polyamines: essential factors for growth and survival. Planta 228, 367–381 10.1007/s00425-008-0772-7 [DOI] [PubMed] [Google Scholar]
- Lee J., Scagel C. F. (2014). Chicoric acid: chemistry, distribution, and production. Front. Chem. 1:40 10.3389/fchem.2013.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenmayer D. B., Likens G. E., Krebs C. J., Hobbs R. J. (2010) Improved probability of detection of ecological “surprises.” Proc. Natl. Acad. Sci. U.S.A. 107, 21957–21962 10.1073/pnas.1015696107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipiec J., Doussan C., Nosalewicz A., Kondracka K. (2013). Effect of drought and heat stresses on plant growth and yield: a review. Int. Agrophys. 27, 463–477 10.2478/intag-2013-001721415892 [DOI] [Google Scholar]
- Liu C., Wu Y., Wang X. (2012). bZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice. Planta 235, 1157–1169 10.1007/s00425-011-1564-z [DOI] [PubMed] [Google Scholar]
- Liu J. H., Kitashiba H., Wang J., Ban Y., Moriguchi T. (2007). Polyamines and their ability to provide environmental stress tolerance to plants. Plant Biotechnol. 24, 117–122 10.5511/plantbiotechnology.24.117 [DOI] [Google Scholar]
- Liu X., Chen M., Collins H. L., Onstad D. W., Roush R. T., et al. (2014). Natural enemies delay insect resistance to Bt crops. PLoS ONE 9:e90366 10.1371/journal.pone.0090366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Gao C., Zheng X., Han B. (2009). Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229, 605–615 10.1007/s00425-008-0857-3 [DOI] [PubMed] [Google Scholar]
- Luo M., Liu X., Singh P., Cui Y., Zimmerli L., Wu K. (2012). Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 129–136 10.1016/j.bbagrm.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Marvier M., McCreedy C., Regetz J., Kareiva P. (2007). A meta-analysis of effects of Bt cotton and maize on nontarget invertebrates. Science 316, 1475–1477 10.1126/science.1139208 [DOI] [PubMed] [Google Scholar]
- Mattoo A. K. (2013). Agricultural biology in the 3rd millennium: Nutritional food security and specialty crops through sustainable agriculture and biotechnology, in Diamond Jubilee Commemoration Volume: Research and Development from the Past Decade, eds Amla D. V., Nath P., Uprati D. K., Singh N., Nair K. M. (Lucknow: Army Printing Press; ), 9–14 [Google Scholar]
- Mattoo A. K., Chung S.-H., Goyal R. K., Fatima T., Srivastava A., Solomos T., et al. (2007). Over-accumulation of higher polyamines in ripening transgenic tomato fruit revives metabolic memory, upregulates anabolism-related genes, and positively impacts nutritional quality. J. AOAC Intl. 90, 1456–1464 [PubMed] [Google Scholar]
- Mattoo A. K., Handa A. K. (2008). Higher polyamines resuscitate metabolic memory in fruit. Plant Sci. 174, 386–393 10.1016/j.plantsci.2008.01.011 [DOI] [Google Scholar]
- Mattoo A. K., Shukla V., Fatima T., Handa A. K., Yachha S. K. (2010). Genetic engineering to enhance crop-based phytonutrients (nutraceuticals) to alleviate diet-related diseases, in Bio-Farms for Nutraceuticals: Functional Food and Safety Control by Biosensors, eds Giardi M. T., Rea G., Berra B. (New York, NY: Landes Bioscience and Springer; ), 122–143 [DOI] [PubMed] [Google Scholar]
- Mattoo A. K., Sobolev A. P., Neelam A., Goyal R. K., Handa A. K., Segre A. L. (2006). NMR spectroscopy-based metabolite profiling of transgenic tomato fruit engineered to accumulate spermidine and spermine reveals enhanced anabolic and nitrogen-carbon interactions. Plant Physiol. 142, 1759–1770 10.1104/pp.106.084400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo A. K., Teasdale J. R. (2010). Ecological and genetic systems underlying sustainable horticulture. Hort. Revs. 37, 331–362 [Google Scholar]
- Mattoo A. K., Upadhyay R. K., Rudrabhatla S. (2014). Abiotic stress in crops: candidate genes, osmolytes, polyamines and biotechnological intervention, in Elucidation of Abiotic Stress Signaling in Plants: a Functional Genomic Perspective, ed Pandey G. K. (New York, NY: Springer; ). (in press). [Google Scholar]
- Mehta R. A., Cassol T., Li N., Ali N., Handa A. K., Mattoo A. K. (2002). Engineered polyamine accumulation in tomato enhances phytonutrient content, juice quality and vine life. Nat. Biotechnol. 20, 613–618 10.1038/nbt0602-613 [DOI] [PubMed] [Google Scholar]
- Meyer K. M., Soldaat L. I., Auge H., Thulke H.-H. (2014). Adaptive and selective seed abortion reveals complex conditional decision making in plants. Am. Nat. 183, 376–383 10.1086/675063 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19 10.1016/j.tplants.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Mu C., Zhang S., Yu G., Chen N., Li X. (2013). Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLoS ONE 8:e82264 10.1371/journal.pone.0082264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir S. R., Collins G. J., Robinson S., Hughes S., Bovy A., Rich De Vos C. H., et al. (2001). Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Biotechnol. 19, 470–474 10.1038/88150 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Tran L. S., van Nguyen D., Fujita M., Maruyama K., Todaka D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress responsive gene expression in rice. Plant J. 51, 617–630 10.1111/j.1365-313X.2007.03168.x [DOI] [PubMed] [Google Scholar]
- Nambessan S., Datsenka T., Ferruzzi M. G., Malladi A., Mattoo A. K., Handa A. K. (2010). Overexpression of yeast spermidine synthase impacts ripening, senescence and decay symptoms in tomato. Plant J. 63, 836–847 10.1111/j.1365-313X.2010.04286.x [DOI] [PubMed] [Google Scholar]
- Naqvi S., Zhu C., Farre G., Ramessar K., Bassie L., Breitenbach J., et al. (2009). Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc. Natl. Acad. Sci. U.S.A. 106, 7762–7767 10.1073/pnas.0901412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navakouidis E., Lütz C., Langebartels C., Lütz-Meindl U., Kotzabasis K. (2003). Ozone impact on the photosynthetic apparatus and the protective role of polyamines. Biochem. Biophys. Acta 1621, 160–169 10.1016/S0304-4165(03)00056-4 [DOI] [PubMed] [Google Scholar]
- Neelam A., Cassol T., Mehta R. A., Abdul-Baki A. A., Sobolev A., Goyal R. K., et al. (2008). A field-grown transgenic tomato line expressing higher levels of polyamines reveals legume cover crop mulch-specific perturbations in fruit phenotype at the levels of metabolite profiles, gene expression and agronomic characteristics. J. Exp. Bot. 59, 2337–2346 10.1093/jxb/ern100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H., Moloney M., Inz D. (2014). Translational research: from pot to plot. Plant Biotechnol. J. 12, 277–285 10.1111/pbi.12176 [DOI] [PubMed] [Google Scholar]
- Nelson D. E., Repetti P. P., Adams T. R., Creelman R. A., Wu J., Warner D. C., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. U.S.A. 104, 16450–16455 10.1073/pnas.0707193104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggeweg R., Michael A. J., Martin C. (2004). Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 22, 746–754 10.1038/nbt966 [DOI] [PubMed] [Google Scholar]
- Ning J., Li X., Hicks L. M., Xiong L. (2010). A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 152, 876–890 10.1104/pp.109.149856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. J., Kim Y. S., Kwon C. W., Park H. K., Jeong J. S., Kim J. K. (2009). Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 250, 1368–1379 10.1104/pp.109.137554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort D. R., Yocum C. F. (eds). (1996). Oxygenic Photosynthesis: The Light Reactions. Advances in Photosynthesis, Vol. 4. Dordrecht: Kluwer Academic Publishers [Google Scholar]
- Paine J. A., Shipton C. A., Chaggar S., Howells R. M., Kennedy M. J., Vernon G., et al. (2005). Improving the nutritional value of golden rice through increased pro-vitamin A content. Nat. Biotechnol. 23, 482–487 10.1038/nbt1082 [DOI] [PubMed] [Google Scholar]
- Petrusa L. M., Winicov I. (1997). Proline status in salt tolerant and salt sensitive alfalfa cell lines and plants in response to NaCl. Plant Physiol. Biochem. 35, 303–310 [Google Scholar]
- Porter J. R., Semenov M. A. (2005). Crop responses to climatic variation. Philos. Trans. R. Soc. B Biol. Sci. 360, 2021–2035 10.1098/rstb.2005.1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R., Hu S., Zhang Z., Zhang H., Zhang Z., Huang R. (2010). Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol. J. 8, 476–488 10.1111/j.1467-7652.2009.00492.x [DOI] [PubMed] [Google Scholar]
- Quan R., Shang M., Zhang H., Zhao Y., Zhang J. (2004). Engineering of enhanced glycinebetaine synthesis improves drought tolerance in maize. Plant Biotechnol. J. 2, 477–486 10.1111/j.1467-7652.2004.00093.x [DOI] [PubMed] [Google Scholar]
- Reugera M., Peleg Z., Blumwald M. (2012). Targeting metabolic pathways for genetic engineering abiotic stress-tolerance in crops. Biochim. Biophys. Acta 1819, 186–194 10.1016/j.bbagrm.2011.08.005 [DOI] [PubMed] [Google Scholar]
- Riechmann J. L., Ratcliffe O. J. (2000). A genomic perspective on plant transcription factors. Curr. Opin. Plant. Biol. 3, 423–434 10.1016/S1369-5266(00)00107-2 [DOI] [PubMed] [Google Scholar]
- Römer S., Fraser P. D., Kiano J. W., Shipton C. A., Misawa N., Schuch W., et al. (2000). Eevation of the provitamin A content of transgenic tomato plants. Nat. Biotechnol. 18, 666–669 10.1038/76523 [DOI] [PubMed] [Google Scholar]
- Ronald P. (2011). Plant genetics, sustainable agriculture and global food security. Genetics 188, 11–20 10.1534/genetics.111.128553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati C., Aquilani R., Dharmapuri S., Pallara P., Marusic C., Tavazza R., et al. (2000). Metabolic engineering of beta carotene and lycopene content in tomato fruit. Plant J. 24, 413–419 10.1046/j.1365-313x.2000.00880.x [DOI] [PubMed] [Google Scholar]
- Roy M., Wu R. (2002). Overexpression of S-adenosylmethionine decarboxylase gene in rice increases polyamine level and enhances sodium chloride-stress tolerance. Plant Sci. 163, 987–992 10.1016/S0168-9452(02)00272-8 [DOI] [Google Scholar]
- Ruiz-Medrano R., Xoconostle-Cazares B., Lucas W. J. (2001). The phloem as a conduit for inter-organ communication. Curr. Opin. Plant Biol. 4, 202–209 10.1016/S1369-5266(00)00162-X [DOI] [PubMed] [Google Scholar]
- Saijo Y., Hata S., Kyozuka J., Shimamoto K., Izui K. (2000). Over-expression of a single Ca2+-dependednt protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 23, 319–327 10.1046/j.1365-313x.2000.00787.x [DOI] [PubMed] [Google Scholar]
- Saito K., Matsuda F. (2010). Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 61, 463–489 10.1146/annurev.arplant.043008.092035 [DOI] [PubMed] [Google Scholar]
- Sakamoto A., Murata N. (2002) The role of glycine betaine in the protection of plants from stress: clues from transgenic plants. Plant Cell Environ. 25, 163–171 10.1046/j.0016-8025.2001.00790.x [DOI] [PubMed] [Google Scholar]
- Saradhi P. P., Suzuki I., Katoh A., Sakamoto A., Sharmilla P., Shi D. J., et al. (2000). Protection against the photo-induced inactivation of the photosystem II complex by abscisic acid. Plant Cell Environ. 23, 711–718 10.1046/j.1365-3040.2000.00579.x [DOI] [Google Scholar]
- Schijlen E., Ric de Vos C. H., Jonker H., van den Broeck H., Molthoff J., van Tunen A., et al. (2006). Pathway engineering for healthy phytochemicals leading to the production of novel flavonoids in tomato fruit. Plant Biotechnol. J. 4, 433–444 10.1111/j.1467-7652.2006.00192.x [DOI] [PubMed] [Google Scholar]
- Schmitz R. J., Zhang X. (2011). High-throughput approaches for plant epigenomic studies. Curr. Opin. Plant Biol. 14, 130–136 10.1016/j.pbi.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S., Patra B., Ray S., Majumder A. L. (2008). Inositol methyl transferase from a halophytic wild rice, Porteresia coarctata Roxb. (Tateoka): regulation of pinitol synthesis under abiotic stress. Plant Cell Environ. 31, 1442–1459 10.1111/j.1365-3040.2008.01850.x [DOI] [PubMed] [Google Scholar]
- Shou H., Bordallo P., Fan J. B., Bibikova J. M., Sheen J., Wang K. (2004). Expression of an active tobacco mitogen–activated protein kinase kinase kinase enhances freezing tolerance in transgenic maize. Proc. Natl. Acad. Sci. U.S.A. 101, 3298–3303 10.1073/pnas.0308095100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla V., Mattoo A. K. (2010). Potential for engineering horticultural crops with high antioxidant capacity. CAB Reviews 4, 1–22 10.1079/PAVSNNR20094066 [DOI] [Google Scholar]
- Shukla V., Mattoo A. K. (2013). Developing robust crop plants for sustaining growth and yield under adverse climatic changes, in Climate Change and Plant Abiotic Stress Tolerance, eds Tuteja N., Gill S. (Germany: Wiley-VCH Verlag; ), 27–56 [Google Scholar]
- Sobolev A. P., Capitani D., Giannino D., Nicolodi C., Testone G., Santoro F., et al. (2010). NMR-metabolic methodology in the study of GM foods. Nutrients 2, 1–15 10.3390/nu2010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C.-F., Wang Y.-C., Hsieh T.-H., Lu C.-H., Tseng T.-H., Yu S.-M. (2010). A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 153, 145–158 10.1104/pp.110.153015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Yuan B., Zhang M., Wang L., Cui M., Wang Q., et al. (2012). Fruit-specific RNAi-mediated suppression of SlNCED1 increases both lycopene and β-carotene contents in tomato fruit. J. Exp. Bot. 63, 3097–3108 10.1093/jxb/ers026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki H., Maruyama K., Kidokoro S., Ito Y., Fujita Y., Shinozaki K., et al. (2010). The abiotic stress responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genom. 84, 173–183 10.1007/s00438-010-0557-0 [DOI] [PubMed] [Google Scholar]
- Vierling E. (1991). The roles of heat shock proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 579–620 10.1146/annurev.pp.42.060191.003051 [DOI] [Google Scholar]
- Vincour B., Altman A. (2005). Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr. Opin. Biotechnol. 16, 123–132 10.1016/j.copbio.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Wan L., Zhang J., Zhang H., Zhang Z., Quan R., Zhou S., et al. (2011). Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE 6:e25216 10.1371/journal.pone.0025216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. P., Zhang X. Y., Li F., Luo Y., Wang W. (2010). Over accumulation of glycinebetaine enhances tolerance to drought and heat stress in wheat leaves in the protection of photosynthesis. Photosynthetica 48, 117–126 10.1007/s11099-010-0016-5 [DOI] [Google Scholar]
- Wang W., Vincour B., Shoseyov O., Altman A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252 10.1016/j.tplants.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Weiste C., Iven T., Fischer U., Oñate-Sánchez L., Dröge-Laser W. (2007). In planta ORFeome analysis by large-scale over-expression of GATEWAY-compatible cDNA clones: screening of ERF transcription factors involved in abiotic stress defense. Plant J. 52, 382–390 10.1111/j.1365-313X.2007.03229.x [DOI] [PubMed] [Google Scholar]
- Wen X. P., Pang X. M., Matsuda N., Kita M., Inoue M., Hao Y. J., et al. (2008). Overexpression of the apple spermidine synthase gene in pear confers multiple abiotic stress tolerance by altering polyamine titers. Transgenic Res. 17, 251–263 10.1007/s11248-007-9098-7 [DOI] [PubMed] [Google Scholar]
- Wi S. J., Kim W. T., Park K. Y. (2006). Over expression of carnation S-adenosylmethionine decarboxylase gene generates a broad-spectrum tolerance to abiotic stresses in transgenic tobacco plants. Plant Cell Rep. 25, 1111–1121 10.1007/s00299-006-0160-3 [DOI] [PubMed] [Google Scholar]
- Wingler A. (2002). The function of trehalose biosynthesis in plants. Phytochemistry 60, 437–440 10.1016/S0031-9422(02)00137-1 [DOI] [PubMed] [Google Scholar]
- Wise R. R., Hoober J. K. (2007). The Structure and Function of Plastids. Advances in Photosynthesis and Respiration, Vol. 23. New York, NY: Springer [Google Scholar]
- Wu X., Shiroto Y., Kishitani S., Ito Y., Toriyama K. (2008). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30 10.1007/s00299-008-0614-x [DOI] [PubMed] [Google Scholar]
- Xiang Y., Tang N., Du H., Ye H., Xiong L. (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 148, 1938–1952 10.1104/pp.108.128199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B., Huang Y., Tang N., Xiong L. (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 115, 35–46 10.1007/s00122-007-0538-9 [DOI] [PubMed] [Google Scholar]
- Xiong A. S., Jiang H. H., Zhuang J., Peng R. H., Jin X. F., Zhu B., et al. (2013). Expression and function of a modified AP2/ERF transcription factor from Brassica napus enhances cold tolerance in transgenic Arabidopsis. Mol. Biotechnol. 53, 198–206 10.1007/s12033-012-9515-x [DOI] [PubMed] [Google Scholar]
- Xiong L., Lee B. L., Ishitani M., Lee H., Zhang C., Zhu J.-K. (2001). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15, 1971–1984 10.1101/gad.891901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Duan X., Wang B., Hong B., Ho T. H. D., Wu R. (1996). Expression of a late embryogenesis abundant protein gene HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 110, 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Dai X., Zhang W.-H. (2012). A R2R3-typre MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63, 2541–2556 10.1093/jxb/err431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C., Xiaoyuan Y., Kun H., Meihua L., Jigang L., Zhaofeng G., et al. (2012). A R2R3-typre MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63, 2541–2556 10.1093/jxb/err431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu W., Wan L., Li F., Dai L., Li D., et al. (2010). Functional analyses of ethylene response factor JERF3 with the aim of improving tolerance to drought and osmotic stress in transgenic rice. Transgenic Res. 19, 809–818 10.1007/s11248-009-9357-x [DOI] [PubMed] [Google Scholar]
- Zhang W., Lorence A., Gruszewski H. A., Chevone B. I., Nessler C. L. (2009). AMR1, an Arabidopsis gene that coordinately and negatively regulates the mannose/l-galactose ascorbic acid biosynthetic pathway. Plant Physiol. 150, 942–950 10.1104/pp.109.138453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Hu Y., Chong K., Wang T. (2010). ARAG1, an ABA-responsive DREB gene, plays a role in seed germination and drought tolerance of rice. Ann. Bot. 105, 401–409 10.1093/aob/mcp303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M., Guan Y., Ren H., Zhang F., Chen F. (2008). A bZIP transcription factor, OsABI5 is involved in rice fertility and stress tolerance. Plant Mol. Biol. 66, 675–683 10.1007/s11103-008-9298-4 [DOI] [PubMed] [Google Scholar]


