Abstract
Cytomegalovirus (CMV) is an important pathogen in lung transplant recipients. Early detection of CMV end-organ disease should help with treatment management. We determined the CMV viral load by hybrid capture in bronchoalveolar lavage (BAL) fluid samples from patients who had undergone lung transplantation. For 39 of these samples (from 25 patients), corresponding transbronchial biopsy samples were available for CMV immunohistochemistry (IHC). The CMV IHC results were interpreted and categorized as positive or negative, and the positive results were subcategorized as typical if cells with both significant nuclear enlargement or Cowdry A-type inclusions and positive staining were present or as atypical if definitive nuclear staining was seen but significant nuclear enlargement was not. Diagnostic CMV viral inclusions were reported in the anatomic diagnosis, based on hematoxylin-eosin staining alone, for three (8%) of the biopsy samples. CMV was detected by IHC in 13 (33%) samples (5 typical, 8 atypical). The median CMV viral load in BAL samples was 0 copies/ml for BAL samples from patients with IHC-negative biopsy samples; 47,678 copies/ml for BAL samples from patients with biopsy samples with positive, atypical staining; and 1,548,827 copies/ml for BAL samples from patients with biopsy samples with positive, typical staining (P < 0.001). Compared to routine pathology of biopsy samples, the use of IHC increased the diagnostic yield of CMV. Also, the CMV viral load in BAL fluid samples increased along with immunoreactivity from negative to positive, atypical staining to positive, typical staining. The CMV viral load determined with the end-organ sample, the BAL fluid sample, was higher than the corresponding viral load determined with blood. Both IHC and determination of the CMV viral load in BAL samples may be useful for the detection of individuals at risk for the development of fulminant invasive CMV disease.
Cytomegalovirus (CMV) pneumonitis is associated with high rates of morbidity and mortality rate in solid-organ, blood, and bone marrow transplant recipients (5, 7, 10, 19, 20). CMV pneumonitis is quite common in lung transplant recipients in particular, as it develops in more than half of all lung transplant recipients over the first 3 years after transplantation (4, 14, 21). The diagnosis of CMV pneumonitis is based on the presence of signs and/or symptoms of lower respiratory tract infection and diffuse infiltrates on a chest radiograph, in combination with a CMV-positive culture of bronchoalveolar lavage (BAL) fluid, CMV-positive immunohistochemistry (IHC), positivity by in situ hybridization for CMV, or the presence of typical viral inclusions in routine hematoxylin and eosin (H&E)-stained sections (11). A diagnosis of CMV infection may be made in the absence of a positive culture, when definitive CMV viral inclusions are seen upon histopathologic evaluation of lung biopsy samples, sometimes aided by the use of an IHC stain or in situ hybridization for CMV (2, 22, 23). In the past, we noted definitive CMV-positive IHC staining in the nuclei of atypical cells in lung biopsy samples that lacked significant nuclear enlargement, Cowdry A-type inclusions, and the cytoplasmic changes that may sometimes be noted in cells infected with CMV.
Unfortunately, isolation of CMV from BAL fluid by culture methods does not discriminate between potentially lethal CMV pneumonitis and shedding of the virus. Furthermore, histopathologic detection of CMV in lung tissue, possibly because of sampling, has a low sensitivity and can lead to underdiagnosis and/or a delayed diagnosis of CMV pneumonitis (3, 9, 23). Hence, the limitations of these two methods for the diagnosis of CMV pneumonitis may lead to excessive treatment or a delay in the initiation of antiviral therapy.
The recent availability of new molecular diagnostic methods affords a more rapid means of CMV detection and even CMV quantification (i.e., the CMV viral load). Moreover, determination of the CMV viral loads in blood (6, 13, 16-18) and urine (8) has shown its utility in designating patients who at risk for disease (12). The clinical utility of the CMV viral load in BAL fluid has not been determined, however (1, 15; R. F. Chemaly, B. Yen-Lieberman, A. Reiley, S. M. Gordon, G. W. Procop, N. Shrestha, C. M. Isada, R. Schilz, M. DeCamp, and R. K. Avery, Proc. Annu. Meet. Am. Transplant Congr., abstr. 92, 2002). Therefore, in the present study, we investigated the utility of the CMV Hybrid Capture assay (Digene Corporation, Gaithersburg, Md.) for the quantification of CMV in BAL samples from lung transplant recipients and the concomitant blood CMV viral loads (if available), the routine detection of CMV in H&E-stained biopsy samples obtained concomitantly, and the results of IHC for CMV in the same samples.
MATERIALS AND METHODS
From August 2000 to October 2001, 42 consecutive BAL samples and 39 transbronchial tissue biopsy samples were collected prospectively from 27 lung transplant recipients. The indications for bronchoscopy in these 27 patients varied from routine surveillance to respiratory signs and symptoms with or without abnormal radiologic findings. Only the 39 BAL samples with matching biopsy samples were analyzed in this study, and they were obtained from 25 different patients. The tissue biopsy samples were examined after H&E staining by an attending pathologist, and the results were reported in the routine manner. IHC staining of all of the biopsy samples for CMV was performed by batch testing in a blinded manner, and the quantitative CMV viral load in the cell pellet taken from the BAL fluid was obtained by hybrid capture. The same technologist (A.R.) performed the quantitative CMV testing with the BAL samples, and the same pathologist (G.W.P.) read the IHC staining results. Both of them were blinded to the clinical status of the patients and the test results for the patients.
Quantitative CMV Hybrid Capture assay.
The quantitative CMV Hybrid Capture (DNA) assay was performed with 3.5 ml of whole blood and 0.5 to 1.0 ml of BAL fluid. Briefly, red blood cells were lysed with the assay manufacturer's lysis solution. The remaining cells were transferred to hybridization tubes, pelleted, and stored at −20°C until batch testing. After denaturation of the cell pellet, a CMV RNA probe was added and allowed to hybridize with the single-stranded DNA target sequence at 70°C for 2 h to form an RNA-DNA hybrid. The contents of the hybridization tubes were then transferred to corresponding “capture tubes” coated with anti-RNA-anti-DNA hybrid antibodies. Immobilized hybrids were then reacted with alkaline phosphatase-conjugated monoclonal antibodies to the RNA-DNA hybrids and detected with a chemiluminescent substrate. The tubes were subsequently read with a luminometer. CMV DNA was quantitated in picograms per milliliter by comparing the relative light units for the specimens with those on a calibration curve of CMV DNA standards. The results were then expressed as the number of CMV DNA copies per milliliter. The test was performed according to the instructions of the manufacturer (Digene Corporation) for whole blood.
Histology and IHC of lung tissues.
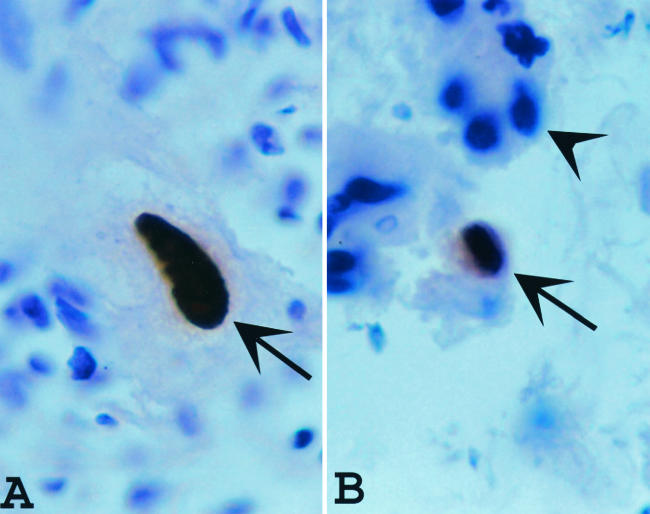
Whether CMV was present in or absent from the routine H&E-stained lung biopsy samples, as determined by the attending pathologist, was determined in a review of the surgical pathology reports. For IHC, the histologic sections were deparaffinized and stained with the CMV-specific IHC stain, which was a mixture of two mouse monoclonal antibodies. One of these antibodies reacts with a 76-kDa nonstructural protein from CMV, whereas the other reacts with delayed early DNA-binding protein p52 of CMV (both antibodies were from DAKO Corporation, Carpinteria, Calif.). This was followed by the addition of a biotinylated secondary antibody that targets the primary antibody (Basic DAB Detection kit; Ventana Medical Systems, Inc., Tucson, Ariz.). The complex was then visualized by using a precipitating enzyme-generated product. Positive and negative controls were used and were appropriately stained. The positive results were further categorized as typical if the CMV IHC-positive cells had nuclear enlargement and typical Cowdry A-type intranuclear inclusions (Fig. 1), atypical if definitive nuclear CMV-specific IHC staining was seen but significant nuclear enlargement was not (Fig. 1), and negative if CMV-positive IHC staining was not seen.
FIG. 1.
(A) Typical positive immunohistochemistry (IHC) staining for CMV (arrow) in a cytomegalic cell; (B) the type of cell designated atypical in this study (arrow); it was positive for CMV by IHC but not cytomegalic. The cells that stain positive for the presence of CMV (arrows) are a distinctive brown, whereas the cells that are negative for CMV by IHC (arrowhead) are blue, secondary to counterstaining with H&E. Magnification ×1,000.
Statistical analysis.
We compared the proportion of samples in which CMV was detected by H&E staining with the proportion of samples in which CMV was detected by IHC staining using McNemar's test. The Kruskal-Wallis test was used to compare the viral loads in specimens found to be positive by IHC staining; if significant differences were detected overall, pairwise tests were done by Dunn's multiple-comparison procedure. We also tested the relationship between increasing viral load and the ordered outcome of (i) negative, (ii) positive, atypical, or (iii) positive, typical staining using cumulative logit logistic regression analysis. To compare blood and BAL fluid viral levels, we used the Wilcoxon signed-rank test with the difference between the level in blood and that in BAL fluid. We treated the samples as independent, because, given the amount of data for each group, we could not properly use any methods that adjusted for within-patient correlations. All of the data were recorded as the medians, minimums, and maximums. Statistical analysis was performed with SAS (version 8) software (SAS Institute, Cary, N.C.).
RESULTS
Thirty-nine transbronchial tissue biopsy samples were available for CMV IHC evaluation. CMV viral inclusions were reported in three (8%) of the samples upon routine surgical pathology examination. However, CMV was detected by IHC in 13 (33%) biopsy samples (5 typical, 8 atypical) (P = 0.002). In patients in whom no CMV was detected by routine H&E staining, the viral load in BAL fluid was significantly higher than the viral load in blood (P = 0.014) (Table 1). Not surprisingly, the CMV viral loads in both blood and BAL fluid in patients in whom CMV was detected by routine H&E staining were significantly higher than those in patients who lacked routine histologic evidence of CMV disease (P = 0.012 and P = 0.005, respectively).
TABLE 1.
Comparison of viral loads in blood and BAL fluid and results of routine histology
| H & E staining result for CMV | BAL fluid
|
Blood
|
||||
|---|---|---|---|---|---|---|
| CMV viral load (no. of copies/ml)
|
No. of samples | CMV viral load (no. of copies/ml)
|
No. of samples | |||
| Median | Minimum-maximum | Median | Minimum-maximum | |||
| Positive | 2,164,180 | 1,548,827-2,450,702 | 3 | 189,013 | 11,499-1,491,020 | 3 |
| Negative | 74 | 0-595,109 | 36 | 0 | 0-915,800a | 29 |
| P value | 0.005b | 0.012b | ||||
P = 0.014 by the Wilcoxon signed-rank test for the difference in the load between blood and BAL fluid. For negative samples, the load in BAL fluid was higher than that in blood.
Wilcoxon rank-sum test.
The median BAL fluid CMV viral load was 0 copies/ml for BAL samples from patients whose biopsy samples were negative for CMV by IHC; 47,678 copies/ml for BAL samples from patients whose biopsy samples had positive, atypical staining; and 1,548,827 copies/ml for BAL samples from patients whose biopsy samples had positive, typical staining (Table 2). These groups had significantly different levels of viral expression (P < 0.001), with the levels for the negative samples being significantly different from those for the samples in both the positive, atypical and the positive, typical categories. The differences between the typical and atypical categories, however, were not found to be significantly significant.
TABLE 2.
Comparison of BAL fluid and blood CMV viral loads with CMV IHC staining result
| IHC staining result for CMVa | BAL fluid
|
Blood
|
||||
|---|---|---|---|---|---|---|
| CMV viral load (no. of copies/ml)
|
No. of samples | CMV viral load (no. of copies/ml)
|
No. of samples | |||
| Median | Minimum-maximum | Median | Minimum-maximum | |||
| Negative | 0 | 0-193,442 | 26 | 0 | 0-40,587 | 20 |
| Atypical | 47,678 | 2,791-312,175 | 8 | 1,176 | 0-14,019 | 7 |
| Typical | 1,548,827 | 555,505-2,450,702 | 5 | 189,013 | 0-1,491,020 | 5 |
| P value | 0.001b | 0.010c | 0.028d | 0.15c | ||
Comparison of the differences in CMV viral loads between blood and BAL fluid were significantly different for the negative category (P = 0.21) and atypical category (P = 0.016), as stratified according to IHC characteristics (Wilcoxon signed-rank test). For the atypical category, load in BAL fluid was higher than that in blood.
Kruskal-Wallis test. For the negative samples, the CMV viral load in BAL samples was significantly different from those in both the atypical and typical samples (Dunn's multiple-comparison procedure).
Ordered logistic regression analysis.
Kruskal-Wallis test. The results for the negative and positive, typical samples were significantly different (Dunn's multiple-comparison procedure).
Thirty-two concomitant blood samples drawn for CMV viral load determination were available for review. The median blood CMV viral load was 0 copies/ml for blood samples from patients whose biopsy samples were negative for CMV by IHC; 1,176 copies/ml for blood samples from patients whose biopsy samples had positive, atypical staining; and 189,013 copies/ml for blood samples from patients whose biopsy samples had positive, typical staining. Only the difference between the viral loads in blood from patients with CMV-negative and -positive biopsy samples with typical staining was statistically significant (P = 0.028) (Table 2).
The viral load in positive, atypical specimens was significantly higher in BAL fluid specimens than in blood specimens (P = 0.016) (Table 2). Although the trend for the CMV viral loads in BAL fluid to be higher than those in blood seems consistent, we could not find a significant difference by the Wilcoxon signed-rank test with paired data for less than six samples.
DISCUSSION
Viral culture by the shell vial assay or a traditional method may be too sensitive for the detection of CMV in BAL fluid, as viral shedding without demonstrable disease has been documented. Quantitation of CMV (i.e., determination of the viral load) may be more useful in differentiating between shedding and CMV pneumonitis.
Our data showed that the use of CMV IHC with transbronchial biopsy samples increased the diagnostic yield of CMV in lung transplant recipients compared with that from the use of H&E staining alone. In addition, IHC aided in the earlier detection of lesions that had CMV viral loads intermediate between those in biopsy samples from patients who were clearly negative for CMV pneumonitis and those in biopsy samples from patients who were clearly positive for CMV pneumonitis. Therefore, we concluded that the sensitivity of H&E staining alone for the detection of CMV-infected cells in lung tissue biopsy samples is low and that it should not be used as the sole method for the diagnosis of CMV pneumonitis. Tamm et al. (23) also demonstrated the superior role of IHC staining in the diagnosis of CMV pneumonitis compared with the role of routine histopathology for viral inclusions alone.
Riise et al. (15) also found that patients with CMV disease had a significantly higher number of CMV copies per milliliter of BAL fluid, but they did not demonstrate that the method sufficiently discriminated between patients with and without CMV pneumonitis to use this method as a diagnostic tool. The copy numbers reported by those investigators were obtained by a method (quantitative PCR) different from the one that we used in our study and did not demonstrate the large range of CMV viral loads that we obtained. In addition, the patients with CMV disease reported by Riise et al. (15) had a mean CMV viral load of 1,120 DNA copies/ml, whereas all of the patients in the present study characterized as having a definitive diagnosis of CMV pneumonitis due to the presence of typical CMV inclusions by IHC had CMV viral loads in BAL fluid of >500,000 DNA copies/ml. The difference between the results of our study and those reported by Riise et al. (15) may reflect differences in the quantification methodologies used.
We stratified the CMV viral load data in our study into three categories on the basis of the results of CMV-specific IHC staining. These categories were negative, positive with atypical characteristics of CMV (i.e., nuclear IHC staining without typical cytomegalic cells), and positive with cytomegalic changes typical of CMV. As the pathology changed along this spectrum, the CMV viral load increased, suggesting that increasing numbers of viral copies may be associated with categories such as unlikely CMV pneumonitis, possible or incipient CMV pneumonitis, and definite CMV pneumonitis. Of most interest to us was the intermediate category of patients with elevated CMV viral loads and evidence of CMV by IHC. This patient population has histologic and virologic evidence suggestive of incipient CMV pneumonitis and would potentially benefit from antiviral therapy. Further studies should be performed to confirm this hypothesis.
In summary, two diagnostic methods, quantitative CMV viral load testing of the BAL fluid and CMV IHC, may prove useful in identifying patients with an intermediate pathological state suggestive of incipient CMV pneumonitis. Quantitative testing of BAL fluid for viral load may be performed by the CMV Hybrid Capture assay, using a simple modification of the procedure used for CMV viral load testing in the blood. The results that we obtained by this method correlated better with end-organ (lung) disease than the blood viral load levels did. The viral loads in both the blood and the BAL fluid of patients whose biopsy samples demonstrated typical, positive CMV-specific IHC staining were significantly different from those in the blood and BAL fluid of patients whose biopsy samples were CMV IHC negative. However, the only significant differences between the viral loads for the patients with positive, atypical CMV-specific IHC staining and those for the patients with negative CMV-specific IHC staining was for the BAL specimens, not the blood specimens. This may reflect a heightened sensitivity of the viral load in BAL fluid compared with that in blood due to direct end-organ sampling. Alternatively, one could suggest that IHC for CMV should routinely be performed with transbronchial biopsy samples from patients who have received a lung transplant. If CMV-specific IHC staining for a patient is atypical, as described here, it would suggest an elevated pulmonary burden of CMV compared to that in CMV IHC-negative patients and, possibly, incipient CMV pneumonitis. No data are available to support the possibility that individuals with the atypical CMV IHC pattern or the intermediate CMV viral loads in BAL fluid will eventually develop CMV pneumonitis. However, the presence of an elevated CMV viral load level in patients with atypical CMV IHC findings compared with those who are negative for CMV by IHC demonstrates elevated CMV replication and is suggestive as a marker for individuals at risk for progressive disease. This hypothesis, however, remains to be proven.
REFERENCES
- 1.Cagle, P. T., G. Buffone, V. A. Holland, T. Samo, G. J. Demmler, G. P. Noon, and E. C. Lawrence. 1992. Semiquantitative measurement of cytomegalovirus DNA in lung and heart-lung transplant patients by in vitro DNA amplification. Chest 101:93-96. [DOI] [PubMed] [Google Scholar]
- 2.Cathomas, G., P. Morris, K. Pekle, I. Cunningham, and D. Emanuel. 1993. Rapid diagnosis of cytomegalovirus pneumonia in marrow transplant recipients by bronchoalveolar lavage using the polymerase chain reaction, virus culture, and the direct immunostaining of alveolar cells. Blood 81:1909-1914. [PubMed] [Google Scholar]
- 3.Crawford, S. W., R. A. Bowden, R. C. Hackman, C. A. Gleaves, J. D. Meyers, and J. G. Clark. 1988. Rapid detection of cytomegalovirus pulmonary infection by bronchoalveolar lavage and centrifugation culture. Ann. Intern. Med. 108:180-185. [DOI] [PubMed] [Google Scholar]
- 4.Dauber, J. H., I. L. Paradis, and J. S. Dummer. 1990. Infectious complications in pulmonary allograft recipients. Clin. Chest Med. 11:291-308. [PubMed] [Google Scholar]
- 5.Duncan, A. J., J. S. Dummer, I. L. Paradis, J. H. Dauber, S. A. Yousem, M. A. Zenati, R. L. Kormos, and B. P. Griffith. 1991. Cytomegalovirus infection and survival in lung transplant recipients. J. Heart Lung Transplant. 10:638-644. [PubMed] [Google Scholar]
- 6.Emery, V. C., C. A. Sabin, A. V. Cope, D. Gor, A. F. Hassan-Walker, and P. D. Griffiths. 2000. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet 355:2032-2036. [DOI] [PubMed] [Google Scholar]
- 7.Ettinger, N. A., T. C. Bailey, E. P. Trulock, G. A. Storch, D. Anderson, S. Raab, E. L. Spitznagel, C. Dresler, and J. D. Cooper. 1993. Cytomegalovirus infection and pneumonitis: impact after isolated lung transplantation. Am. Rev. Respir. Dis. 147:1017-1023. [DOI] [PubMed] [Google Scholar]
- 8.Fox, J. C., I. M. Kidd, P. D. Griffiths, P. Sweny, and V. C. Emery. 1995. Longitudinal analysis of cytomegalovirus load in renal transplant recipients using a quantitative polymerase chain reaction: correlation with disease. J. Gen. Virol. 76:309-319. [DOI] [PubMed] [Google Scholar]
- 9.Gleaves, C. A., E. C. Reed, R. C. Hackman, and J. D. Meyers. 1987. Rapid diagnosis of invasive cytomegalovirus infection by examination of tissue specimens in centrifugation culture. Am. J. Clin. Pathol. 88:354-358. [DOI] [PubMed] [Google Scholar]
- 10.Konoplev, S., R. E. Champlin, S. Giralt, N. T. Ueno, I. Khouri, I. Raad, K. Rolston, K. Jacobson, J. Tarrand, M. Luna, Q. Nguyen, and E. Whimbey. 2001. Cytomegalovirus pneumonia in adult autologous blood and marrow transplant recipients. Bone Marrow Transplant. 27:877-881. [DOI] [PubMed] [Google Scholar]
- 11.Ljungman, P., P. Griffiths, and C. Paya. 2002. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin. Infect. Dis. 34:1094-1097. [DOI] [PubMed] [Google Scholar]
- 12.Lore, K., G. Tyden, I. Lewensohn-Fuchs, J. Andersson, B. G. Ericzon, G. Lundgren, and A. Ehrnst. 1999. Determination of cytomegalovirus DNA load for monitoring of cytomegalovirus disease and antiviral treatment in solid organ transplant patients, comparing limiting-dilution PCR and hybrid capture assay with cytomegalovirus isolation. Clin. Microbiol. Infect. 5:78-87. [DOI] [PubMed] [Google Scholar]
- 13.Michaelides, A., L. Liolios, E. M. Glare, D. W. Spelman, M. J. Bailey, E. H. Walters, T. J. Williams, G. I. Snell, and T. C. Kotsimbos. 2001. Increased human cytomegalovirus (HCMV) DNA load in peripheral blood leukocytes after lung transplantation correlates with HCMV pneumonitis. Transplantation 72:141-147. [DOI] [PubMed] [Google Scholar]
- 14.Paradis, I. L., and P. Williams. 1993. Infection after lung transplantation. Semin. Respir. Infect. 8:207-215. [PubMed] [Google Scholar]
- 15.Riise, G. C., R. Andersson, T. Bergstrom, A. Lundmark, F. N. Nilsson, and S. Olofsson. 2000. Quantification of cytomegalovirus DNA in BAL fluid: a longitudinal study in lung transplant recipients. Chest 118:1653-1660. [DOI] [PubMed] [Google Scholar]
- 16.Rollag, H., S. Sagedal, K. I. Kristiansen, D. Kvale, E. Holter, M. Degre, and K. P. Nordal. 2002. Cytomegalovirus DNA concentration in plasma predicts development of cytomegalovirus disease in kidney transplant recipients. Clin. Microbiol. Infect. 8:431-434. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez, J. L., R. M. Kruger, S. Paranjothi, E. P. Trulock, J. P. Lynch, C. Hicks, W. D. Shannon, and G. A. Storch. 2001. Relationship of cytomegalovirus viral load in blood to pneumonitis in lung transplant recipients. Transplantation 72:733-735. [DOI] [PubMed] [Google Scholar]
- 18.Schafer, P., W. Tenschert, L. Cremaschi, M. Schroter, B. Zollner, and R. Laufs. 2001. Area under the viraemia curve versus absolute viral load: utility for predicting symptomatic cytomegalovirus infections in kidney transplant patients. J. Med. Virol. 65:85-89. [PubMed] [Google Scholar]
- 19.Smith, C. B. 1989. Cytomegalovirus pneumonia. Chest 95:182S-187S. [Google Scholar]
- 20.Smyth, R. L., J. P. Scott, L. K. Borysiewicz, L. D. Sharples, S. Stewart, T. G. Wreghitt, J. J. Gray, T. W. Higenbottam, and J. Wallwork. 1991. Cytomegalovirus infection and survival in heart-lung transplant recipients: risk factors, clinical associations and response to treatment. J. Infect. Dis. 164:1045-1050. [DOI] [PubMed] [Google Scholar]
- 21.Smyth, R. L., J. Sinclair, J. P. Scott, J. J. Gray, T. W. Higenbottam, T. G. Wreghitt, J. Wallwork, and L. K. Borysiewicz. 1991. Infection and reactivation with cytomegalovirus strains in lung transplant recipients. Transplantation 52:480-482. [DOI] [PubMed] [Google Scholar]
- 22.Solans, E. P., E. R. Garrity, M. McCabe, R. Martinez, and A. N. Hussain. 1995. Early diagnosis of CMV pneumonitis in lung transplant patients. Arch. Pathol. Lab. Med. 119:33-35. [PubMed] [Google Scholar]
- 23.Tamm, M., P. Traenkle, B. Grilli, M. Soler, C. T. Bolliger, P. Dalquen, and G. Cathomas. 2001. Pulmonary cytomegalovirus infection in immunocompromised patients. Chest 119:838-843. [DOI] [PubMed] [Google Scholar]