Abstract
The study aimed at investigating Gram-positive and Gram-negative bacteria in moldy and non-moldy homes, as defined by the home’s Environmental Relative Moldiness Index (ERMI) value. The ERMI values were determined from floor dust samples in 2010 and 2011 and homes were classified into low (<5) and high (>5) ERMI groups based on the average ERMI values as well as 2011 ERMI values. Dust and air samples were collected from the homes in 2011 and all samples were analyzed for Gram-positive and Gram-negative bacteria using QPCR assays, endotoxin by the LAL assay, and N-acetyl-muramic acid using HPLC. In addition, air samples were analyzed for culturable bacteria. When average ERMI values were considered, the concentration and load of Gram-positive bacteria determined with QPCR in house dust, but not air, were significantly greater in high ERMI homes than in low ERMI homes. Furthermore, the concentration of endotoxin, but not muramic acid, in the dust was significantly greater in high ERMI than in low ERMI homes. In contrast, when ERMI values of 2011 were considered, Gram-negative bacteria determined with QPCR in air, endotoxin in air, and muramic acid in dust were significantly greater in high ERMI homes. The results suggest that both short-term and long-term mold contamination in homes could be linked with the bacterial concentrations in house dust, however, only the current mold status was associated with bacterial concentrations in air. Although correlations were found between endotoxin and Gram-negative bacteria as well as between muramic acid and Gram-positive bacteria in the entire data set, diverging associations were observed between the different measures of bacteria and the home moldiness. It is likely that concentrations of cells obtained by QPCR and concentrations of cell wall components are not equivalent and represent too broad categories to understand the bacterial composition and sources of the home microbiota.
Keywords: Indoor air quality, Bacteria, Mold, ERMI, Endotoxin, Muramic acid
INTRODUCTION
The association between bacterial contamination and respiratory health has lagged behind studies of mold contamination and health (WHO, 2009). Human exposures to bacteria and their components in water-damaged homes have been reported to result in atopic and non-atopic inflammatory diseases (Douwes et al., 2003). Quantifying bacterial populations and their components in non-water-damaged and water-damaged moldy homes is needed to better understand building-related illnesses. Gram-positive and Gram-negative bacteria, including such genera as Acinetobacter, Bacillus, Flavobacterium, Nocardia, Thermomonospora and Streptomyces, have been isolated from moisture-damaged building materials and dust (Suihko et al., 2009; Rintala et al., 2008; Torvinen et al., 2006; Peltola et al., 2001; Andersson et al., 1997).
The major cell wall component of Gram-negative bacteria is lipopolysaccharide/endotoxin (hereafter, endotoxin), whereas N-acetyl-muramic acid (hereafter, muramic acid) is the major cell wall component of Gram-positive bacteria. For pathogenic bacteria, these components are important virulence factors (Feezor et al., 2003) but can also cause inflammatory responses that have been associated with both increased and decreased risk of the development or exacerbation of allergy and asthma (Heederik and von Mutius, 2012). Therefore, in addition to mold exposures, bacterial growth may affect occupant health in water-damaged homes but the quantification of bacteria has not been standardized as it has been for molds.
The quantification of mold contamination was standardized by using a DNA-based analytical method and the Environmental Relative Moldiness Index (ERMI) scale (Vesper et al., 2011). Developed by United States Environmental Protection Agency (US EPA) researchers in collaboration with the Department of Housing and Urban Development (HUD), the ERMI scale relies on measuring the concentrations of 26 mold species indicating water-damage (Group 1) and 10 species that were found in randomly selected non-water-damaged homes during the nationwide HUD 2006 American Healthy Homes Survey (Group 2) (Vesper et al., 2007). The ERMI scale was divided into quartiles and an ERMI value greater than 5 is in the upper quartile, indicating the highest mold contamination for the U.S. homes. Exposure of infants to high ERMI homes has been associated with the development of asthma (Reponen et al., 2012; 2011).
In this study, we investigated Gram-positive and Gram-negative bacteria, as well as their cell wall constituents, muramic acid and endotoxin, in dust and air samples collected from low and high ERMI homes. Both groups of bacteria were quantified using quantitative PCR (QPCR) and traditional cultivation methods. We also determined correlations between the measures of bacteria and some of the home’s environmental conditions, including temperature, relative humidity (RH), age of the home, and the number of occupants.
MATERIALS AND METHODS
Study Homes
The families recruited for this study (n=42) were a subset of the cohort participating in the Cincinnati Childhood Allergy and Air Pollution Study (CCAAPS) undertaken in Cincinnati, Ohio and Northern Kentucky in 2001 (Ryan et al., 2007; LeMasters et al., 2006). The ERMI values for these homes had been determined in 2010 (Reponen et al., 2012; 2011). Based on the ERMI scale, homes were selected for inclusion into a low ERMI (<5) group (n=21) or a high ERMI (>5) group (n=21). With the possibility that conditions had changed, dust was again collected in 2011 from the same homes and the ERMI analysis was repeated [all primer and probe sequences used in the ERMI QPCR assays are available online (US Environmental Protection Agency, 2012)]. Subsequently, the homes were reclassified into low (n=25) and high (n=17) ERMI groups based on the average ERMI for the two years and into low (n=31) and high (n=11) ERMI groups based solely on year 2011 ERMI values. The protocols for collection and analysis of household dust samples previously approved by the Institutional Review Board at the University of Cincinnati were followed during the home sampling.
On-site Home Visit and Sampling
On-site home visits were performed by two-person teams. Floor dust samples were obtained for the assessment of bacteria and ERMI in the child’s primary activity room (PAR), as described by Cho et al. (2006). Dust samples were collected with a vacuum cleaner (Filter Queen Majestic; HMI Industries Inc., Seven Hills, OH) at a flow rate of 800 l/min. A custom-made cone-shape HEPA filter trap (Midwest Filtration, Cincinnati, OH) with a collection efficiency exceeding 95% for particles larger than 0.3 μm was attached to the nozzle of the vacuum cleaner to collect the dust sample. For carpeted floor, dust samples were collected from an area of 2 m2 in the middle of the room at a vacuuming rate of 2 min/m2. For non-carpeted floor (hard wood, linoleum, tile, or plastic sheet floor), the entire room floor was vacuumed at a rate of 1 min/m2. Large dust particles were removed by sieving (355 μm mesh sieve), and the resulting dust was stored at −20°C before analyses. Temperature (°C) and RH (%) were recorded in each home by a portable thermo-hygrometer (Fisher Scientific, Pittsburgh, PA) and data on the number of occupants and age of the homes were collected through a questionnaire survey.
Air samples (5.47 ± 0.54 m3 air) were collected at 3.5 l/min over a 24-h period using a NIOSH-developed 2-stage cyclone sampler, which separates airborne particles into three size fractions: < 1.0 μm, 1.0–1.8 μm, and >1.8 μm (Lindsley et al., 2006). Dust and air samples were analyzed for muramic acid and endotoxin biochemically and for Gram-positive and Gram-negative bacteria using QPCR. Four air samples had insufficient amounts of extract available for QPCR assays.
DNA Extraction from Environmental Samples
An aliquot of 5.0 ± 0.1 mg of each dust sample was extracted by placing the sample in a ‘bead-beating’ tube with glass beads (Sigma glass beads G-1277; size: 212 – 300 μm; 0.3 g in each tube) and shaken for 1 min, as previously described (Haugland et al., 2004; 2002). The DNA was purified using the DNA-EZ extraction kit (GeneRite, Cherry Hill, NJ). The air samples were extracted as described in Singh et al. (2011). Each size fraction was separately analyzed and the results were combined to represent non-size-selective air concentrations for bacterial and bacterial cell components.
QPCR Analysis of Gram-Positive and Gram-Negative Bacterial Cells
QPCR assays of Gram-positive and Gram-negative bacteria were conducted separately. The primer and probe sequences for groups of bacteria are presented in Table 1. These groups were previously analyzed collectively by Kärkkäinen et al. (2010) using the same primers and probes; the amplicon sizes are also available in the quoted paper. Our attempt to perform the original combined assay was unsuccessful, perhaps due to the different sequence detector utilized. The QPCR assays targeted the 16s rRNA gene. Primers and probes were synthesized commercially (Applied Biosystems, Inc., Foster City, CA) and final concentrations were 1 μM for each primer and 80 nM for each probe.
Table 1.
Primer sequences for both Gram-positive and Gram-negative bacteria and probe sequences, with their respective reporter and quencher dyes, for each assay (Kärkkäinen et al., 2010). Assays for Gram-positive and Gram-negative bacteria were run in separate wells.
| Primers/probes | Sequence (5′-3′) |
|---|---|
| Forward primer | GGGTTAAGTCCCGCAACGA |
| Reverse primer | CATTGTAGCACGTGTGTAGCCC |
| Gram-positive probe | FAM – AAATCATCATGCCCCTTAT – MGBNFQ |
| Gram-negative probe | FAM – TGACGTCAAGTCATCATGGCCCTTACG – TAMRA |
Each QPCR reaction contained 12.5 μl of “Universal Master Mix” (Applied Biosystems, Inc.), 1 μl of a mixture of forward and reverse primers at 25 μM each, 2.5 μl of a 400 nM TaqMan probe (Applied Biosystems, Inc.), 2.5 μl of 2 mg/ml fraction V bovine serum albumin (Sigma Chemical, St. Louis, MO), 1.5 μl of DNA free water (Cepheid, Sunnyvale, CA), and 5 μl of DNA extract from the sample (DNA from 5.0 ± 0.1 mg of dust extracted in a final volume of 200 μl and then 5 μl of that was used here for analysis). Assays were performed using the Roche LightCycler® 480 System (Roche Applied Science, Indianapolis, IN) following manufacturer’s instructions.
The QPCR program for the Gram-positive assay consisted of an initial incubation step at 95°C for 15 min, followed by 45 cycles of denaturation at 95°C for 15 s and annealing/extension at 57°C for 45 s. The QPCR program for the Gram-negative assay consisted of an initial incubation step at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 10 s and annealing/extension at 60°C for 30 s. Forty-five cycles were chosen because this was the Roche instrument standard protocol for use of the 2nd derivative in calculating the cycle threshold.
Standard curves were generated from pure cultures of Bacillus subtilis (ATCC 6051, American Type Culture Collection, Manassas, VA) and Escherichia coli (ATCC 25922) for the Gram-positive and Gram-negative assays, respectively. Cell numbers were based on hemacytometer (Hausser Scientific, Horsham, PA) counts in the highest concentration in the standard curve. DNA extracted from the highest concentration of cells in the standard curve was used to generate a dilution series for the standard curve. Positive controls (extracts of B. subtilis or E. coli cells) and negative controls (Cepheid DNA free water) were run with each assay mix. The internal control was the addition of a known concentration of Geotrichum candidum cells to each sample before extraction (Haugland et al., 2004). The analysis was discarded and repeated if any abnormalities observed. Detection limits per PCR reaction, defined at a Ct value of 40, were 82 cells for B. subtilis and approximately one cell for E. coli.
Bacterial dust concentrations were expressed as average cell equivalents per milligram dust. Bacterial dust loading, expressed as cell equivalents per m2 floor area, was derived from concentration by multiplying concentration with total mass of dust vacuumed and dividing by m2 floor area vacuumed. Bacterial air concentrations were expressed as cell equivalents per m3 of air sampled.
Analysis of Endotoxin and Muramic Acid
Dust and air samples were analyzed for endotoxin using the Limulus Amebocyte Lysate assay (LAL; Pyrochrome LAL; Associates of Cape Cod Inc, Falmouth, MA), as described previously (Adhikari et al., 2010; 2009). An aliquot of 25 mg of sieved dust was used for each analysis. The samples were spiked with endotoxin standard of 0.50 EU/ml to assure that there was no inhibition or enhancement between the extract and the reagents. Endotoxin concentrations were expressed as endotoxin units per mg of dust (EU/mg). The lower detection limit (LDL) for endotoxin was 0.002 EU/mg in dust and 0.076 EU/m3 in air. The concentrations in all measured dust samples were above the LDL.
For analyzing muramic acid, vacuum dried dust (100 mg) and air sample extracts (1.0 ml) were hydrolyzed with 6 N HCl at 95°C for four hours. After cooling, centrifuging, and drying-off the HCl under vacuum, the amino acid/amino sugar analysis was performed by neutralization with triethylamine, modification of free amino groups with phenyl isothiocyanate (PITC), and high-performance liquid chromatography (HPLC) separation with detection by absorbance at 254 nm. The detection limits were 0.1 ng/mg for dust and 0.1 ng/m3 for airborne muramic acid.
Culturing Bacteria from Air Samples
For comparison to QPCR analyses, concentrations of culturable airborne Gram-positive and Gram-negative bacteria were estimated by collecting air samples using two-stage Andersen samplers (Thermo Andersen, Franklin, MA) equipped with Trypticase soy agar (for all bacteria) and MacConkey agar (for Gram-negative bacteria) plates. The counts of Gram-positive bacterial colonies were estimated by subtracting the number of Gram-negative colonies from total number of colonies. The samples were collected at an air flow rate of 28.3 ± 2 l/min for 15 min. The agar plates were subsequently incubated at two incubation temperatures: at 30 ± 2°C for a minimum of three days for mesophilic bacterial species or at 55 ± 2°C for a minimum of seven days for thermophilic bacterial species (US EPA, 2003). The colonies were counted by a using a Quebec darkfield colony counter (Cambridge Instruments, Inc., Buffalo, NY). The counts of mesophilic and thermophilic colonies were combined to obtain the final concentration in colony forming units. Concentrations of culturable bacteria in air samples were described as colony forming units per m3 air (CFU/m3).
Statistical analyses
Independent samples t-test was conducted to examine the differences in means between two ERMI-specific groups for different bacterial variables after log transformation. The non-parametric Spearman’s correlation coefficients were calculated between different bacterial and environmental variables. Non-parametric method was employed because the data were not normally or log-normally distributed in several cases when data for ERMI-specific and all homes were considered. Statistical analyses were performed using SPSS Statistics 17.0 (IBM Corporation, Armonk, NY) and SAS 9.3 software (SAS Institute, Carry, NC).
RESULTS
According to 2011 ERMI assessment, 31 homes belonged to the low ERMI group and 11 homes belonged to the high ERMI group. The average ERMI value over two years (2010 and 2011) was determined for each home to provide a longer term view of the conditions in the home and resulted in 25 low ERMI and 17 high ERMI homes.
The differences between low and high ERMI homes in the Gram-positive and Gram-negative bacterial concentrations for dust and air samples are shown in Table 2a (based on average ERMI) and Table 2b (based on 2011 ERMI values). When average ERMI values were considered, both the concentration and load of Gram-positive bacteria in the dust were significantly greater (t-test p <0.001 and 0.003, respectively) in high ERMI homes than in low ERMI homes (Table 2a). The same trend was observed for the concentration and load of Gram-negative bacteria in dust, but the differences were not statistically significant (Table 2a). Furthermore, none of the air concentrations were significantly different in high versus low ERMI homes separated according to the average ERMI values.
Table 2a.
Geometric mean concentrations of Gram-positive and Gram-negative bacterial populations in low (<5) Environmental Relative Moldiness Index (ERMI) homes and high (>5) ERMI homes considering average ERMI values of two years. Significantly different averages are bolded.
| Category | Gram-positive bacteria | Gram-negative bacteria | ||||
|---|---|---|---|---|---|---|
| Low ERMI homes | High ERMI homes | p-value for t-test | Low ERMI homes | High ERMI homes | p-value for t-test | |
| Dust concentration | ||||||
| Number of cell equivalents/mg dust | 0.5 × 106 | 1.3 × 106 | <0.001 | 1.2 × 105 | 2.3 × 105 | 0.196 |
| Dust loading | ||||||
| Number of cell equivalents/m2 floor | 2.7 × 108 | 4.3 × 108 | 0.003 | 6.0 × 107 | 7.8 × 107 | 0.671 |
| Air concentration | ||||||
| Number of cell equivalents /m3 air | 1.4 × 105 | 1.1 × 105 | 0.467 | 3.8 × 103 | 4.2 × 103 | 0.750 |
| Number of CFU/m3 air* | 8.6 × 102 | 7.7 × 102 | 0.806 | 4 | 7 | 0.30 |
Culturable bacteria were analyzed only in air samples and quantified as colony forming units (CFU).
Note: Average amounts of dust collected from low ERMI and high ERMI homes were 1595 mg and 2158 mg, respectively.
Table 2b.
Geometric mean concentrations of Gram-positive and Gram-negative bacterial populations in low (<5) Environmental Relative Moldiness Index (ERMI) homes (n = 31) and high (>5) ERMI homes (n = 11) considering ERMI values of 2011. Significantly different averages are bolded.
| Category | Gram-positive bacteria | Gram-negative bacteria | ||||
|---|---|---|---|---|---|---|
| Low ERMI homes | High ERMI homes | p-value for t-test | Low ERMI homes | High ERMI homes | p-value for t-test | |
| Dust concentration | ||||||
| Number of cell equivalents/mg dust | 6.68 × 105 | 1.08 × 106 | 0.531 | 1.34 × 105 | 2.27 × 105 | 0.367 |
| Dust loading | ||||||
| Number of cell equivalents/m2 floor | 3.31 × 108 | 3.17 × 108 | 0.953 | 6.66 × 107 | 6.69 × 107 | 0.995 |
| Air concentration | ||||||
| Number of cell equivalents /m3 air | 1.33 × 105 | 1.37 × 105 | 0.915 | 3.28 × 103 | 7.26 × 103 | <0.001 |
| Number of CFU/m3 air* | 4.8 × 102 | 4.7 × 102 | 0.952 | 2 | 3 | 0.413 |
Culturable bacteria were analyzed only in air samples and quantified as colony forming units (CFU).
Note: Average amounts of dust collected from low ERMI and high ERMI homes were 1909 mg and 1578 mg, respectively.
The results were somewhat divergent when high ERMI and low ERMI groups were separated based on 2011 ERMI values (Table 2b). Although increasing trends of Gram-positive and Gram-negative bacterial concentrations were observed in dust samples of high ERMI homes, the differences were not statistically significant. Only total cell concentrations of Gram-negative bacteria in air were significantly higher in high ERMI homes than low ERMI homes.
The total cell concentrations in the air as determined by QPCR were on average about 2,000 (SD=9,600) times higher than the concentrations of culturable bacteria. Furthermore, concentrations of Gram-positive bacteria were higher than Gram-negative bacteria in both high and low ERMI homes in dust as well as in air samples (Tables 2a and 2b). The ratios between average concentration of Gram-positive and Gram-negative cell concentrations in all homes were 2.9 in dust samples and 35.8 in air samples.
Tables 3a and 3b show the differences in the measures of the cell wall components of Gram-positive bacteria (muramic acid) and Gram-negative bacteria (endotoxin) between low versus high ERMI homes, when average ERMI and 2011 ERMI values were considered, respectively. When the judgment was made solely based on average ERMI values, the only significant difference found between the high ERMI and low ERMI groups of homes was for the concentration of dust endotoxin, which was greater in the high ERMI homes. However, when 2011 ERMI values were considered, endotoxin concentration in air and muramic acid concentration in dust were significantly higher in high ERMI homes.
Table 3a.
Comparison of the geometric mean concentrations of endotoxin and muramic acid in dust and air samples from low (<5) Environmental Relative Moldiness Index (ERMI) homes and high (>5) ERMI homes considering average ERMI values of two years. Significantly different averages are bolded.
| Cell wall material | Low ERMI homes | High ERMI homes | p-value for t-test |
|---|---|---|---|
| Endotoxin concentration (EU/mg dust) | 167 | 290 | 0.03 |
| Endotoxin loading (EU/m2 floor) | 1.0 × 105 | 0.98 × 105 | 0.92 |
| Endotoxin air concentration (EU/m3 air) | 5.7 | 4.8 | 0.57 |
| Muramic acid concentration (ng/mg dust) | 7.0 | 9.2 | 0.58 |
| Muramic acid loading (ng/m2 floor) | 3.6 × 103 | 3.1 × 103 | 0.80 |
| Muramic acid air concentration (ng/m3 air) | 0.4 | 0.4 | 0.90 |
Table 3b.
Comparison of the geometric mean concentrations of endotoxin and muramic acid in dust and air samples from low (<5) Environmental Relative Moldiness Index (ERMI) homes (n = 31) and high (>5) ERMI homes (n = 11) considering ERMI values of 2011. Significantly different averages are bolded.
| Cell wall material | Low ERMI homes | High ERMI homes | p-value for t-test |
|---|---|---|---|
| Endotoxin concentration (EU/mg dust) | 211.72 | 203.59 | 0.897 |
| Endotoxin loading (EU/m2 floor) | 1.07 × 105 | 0.84 × 105 | 0.667 |
| Endotoxin air concentration (EU/m3 air) | 4.44 | 8.64 | 0.026 |
| Muramic acid concentration (ng/mg dust) | 5.48 | 21.24 | 0.019 |
| Muramic acid loading (ng/m2 floor) | 2.72 × 103 | 6.25 × 103 | 0.187 |
| Muramic acid air concentration (ng/m3 air) | 0.40 | 0.57 | 0.336 |
When assessing correlations between the various bacterial measures, the data were not separated into high and low ERMI groups because these measures are related to bacteria itself and not likely to be dependent on ERMI categories. The dust concentrations of both Gram-negative and Gram-positive bacteria significantly correlated with dust loading for all homes (Table 4). However, neither the dust concentration nor the dust loading correlated with the respective air concentration.
Table 4.
Analysis of the correlations between bacterial measures (geometric mean concentrations) in dust and air as determined by QPCR for samples from all homes. Significant correlation values are bolded.
| A. Gram-negative bacteria | |||
|---|---|---|---|
| Bacterial measures in dust and air | Gram-negative dust concentration (number of cell equivalents /mg dust) | Gram-negative dust loading (number of cell equivalents /m2 floor) | |
| Gram-negative dust loading (number of cell equivalents /m2 floor) |
Spearman’s rho p value (n) |
.703 <.001 (42) |
Not applicable |
| Gram-negative in air (number of cell equivalents /m3 air) |
Spearman’s rho p value (n) |
.0001 .998 (38) |
.096 .568 (38) |
| B. Gram-positive bacteria | |||
|---|---|---|---|
| Bacterial measures in dust and air | Gram-positive dust concentration (number of cell equivalents /mg dust) | Gram-positive dust loading (number of cell equivalents /m2 floor) | |
| Gram-positive dust loading (number of cell equivalents /m2 floor) |
Spearman’s rho p value (n) |
.581 <.001 (42) |
Not applicable |
| Gram-positive in air (number of cell equivalents /m3 air) |
Spearman’s rho p value (n) |
.198 .234 (38) |
.231 .163 (38) |
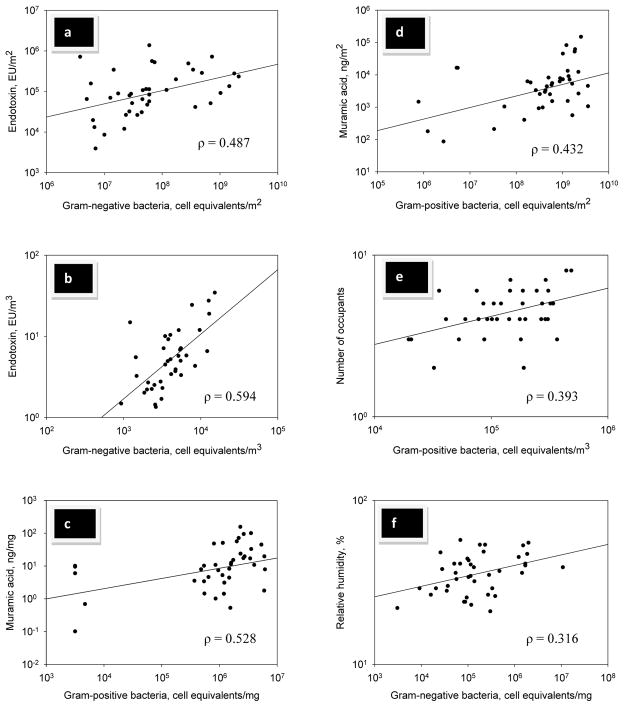
The correlations between bacterial cell wall components and concentration of bacterial cells in dust and air samples for all homes are presented in Table 5. Distributions of data in selected correlations are presented in Figure 1. The dust loading and air concentration of Gram-negative bacteria significantly correlated with respective measure of endotoxin in all homes (Table 5A, Figure 1a, 1b). In addition, there was a significant correlation between Gram-positive cell concentration and muramic acid concentration in dust (Table 5B, Figure 1c) and between Gram-positive cell loading and muramic acid loading (Table 5B, Figure 1d). We also examined the correlations between levels of culturable Gram-positive and Gram-negative bacteria with total Gram-positive and Gram-negative cells measured by QPCR as well as levels of muramic acid and endotoxin (dust concentration, loading, and air). Only muramic acid in air significantly correlated with culturable Gram-positive bacteria in air (ρ = 0.002; p < 0.001, data not shown).
Table 5.
Analysis of the correlation between bacterial measures (geometric mean concentrations) as determined by QPCR and measures of cell components for samples from all homes. Significant correlation values are bolded.
| A. Gram-negative bacteria | ||||
|---|---|---|---|---|
| Bacterial measures in dust and air | Endotoxin dust concentration (EU/mg dust) | Endotoxin dust loading (EU/m2 floor) | Endotoxin in air (EU/m3 air) | |
| Gram-negative dust concentration (number of cell equivalents /mg dust) | Spearman’s rho p value (n) |
.107 .506 (41) |
−.011 .948 (41) |
.002 .990 (42) |
| Gram-negative dust loading (number of cell equivalents /m2 floor) | Spearman’s rho p value (n) |
.002 .988 (41) |
.487 .001 (41) |
.057 .720 (42) |
| Gram-negative in air (number of cell equivalents /m3 air) | Spearman’s rho p value (n) |
−.059 .731 (37) |
.165 .329 (37) |
.594 <.001 (38) |
| B. Gram-positive bacteria | ||||
|---|---|---|---|---|
| Bacterial measures in dust and air | Muramic acid dust concentration (ng/mg dust) | Muramic acid dust loading (ng/m2 floor) | Muramic acid in air (ng/m3 air) | |
| Gram-positive dust concentration (number of cell equivalents /mg dust) | Spearman’s rho p value (n) |
.528 <.001 (42) |
.245 .118 (42) |
−.150 .349 (41) |
| Gram-positive dust loading (number of cell equivalents /m2 floor) | Spearman’s rho p value (n) |
.202 .200 (42) |
.434 .004 (42) |
−.021 .896 (41) |
| Gram-positive in air (number of cell equivalents /m3 air) | Spearman’s rho p value (n) |
.061 .718 (38) |
.216 .192 (38) |
.243 .147 (37) |
Figure 1.
Selected significant (p<0.05) correlations observed in the study: (a) Dust loading of endotoxin vs. Gram-negative bacteria; (b) Airborne concentration of endotoxin vs. Gram-negative bacteria; (c) Dust concentration of muramic acid vs. Gram-positive bacteria; (d) Dust loading of muramic acid vs. Gram-positive bacteria; (e) Number of occupants vs. Gram-positive bacteria in air; (f) Relative humidity vs. dust concentration of Gram-negative bacteria.
The correlations between measures of Gram-negative and Gram-positive bacterial concentrations and some potentially relevant environmental variables in all homes are shown in Table 6. Among the Gram-negative bacterial measures in dust, positive correlations were found between cell concentrations and RH (range: 21 to 57%, average: 36%) (Figure 1f) as well as between endotoxin and age of home (7 to 128 years, average 57 years). Culturable Gram-negative bacteria correlated positively with both RH and temperature (range: 18.6 to 27.4°C, average: 23.3°C). None of the Gram-negative bacterial measures correlated with the number of occupants. Also, none of the bacterial measures of dust loading correlate with environmental variables, except for a significant inverse correlation (ρ = −0.325; p = 0.035) between the loading of Gram-positive bacteria with temperature (data not shown).
Table 6.
Correlations between the selected home environmental factors (temperature, relative humidity, number of occupants, age of the home) and geometric mean concentrations of bacteria, endotoxin, and muramic acid. Significant correlation values are bolded.
| A. Gram-negative bacteria | ||||||
|---|---|---|---|---|---|---|
| Environmental factors/Bacterial measures in dust and air | Gram-negative dust concentration (number of cell equivalents /mg dust) | Endotoxin dust concentration (EU/mg dust) | Gram-negative in air (number of cell equivalents/m3 air) | Culturable Gram-negative in air (CFU/m3 air) | Endotoxin in air (EU/m3 air) | |
| Temperature (°C) | Spearman’s rho p value (n) |
.136 .390 (42) |
.003 .985 (41) |
−.001 .996 (38) |
0.565 <0.001 (42) |
.170 .281 (42) |
| Relative humidity (%) | Spearman’s rho p value (n) |
.316 .041 (42) |
.067 .677 (41) |
.035 .837 (38) |
0.518 <0.001 (42) |
.194 .219 (42) |
| Occupants (number) | Spearman’s rho p value (n) |
.294 .059 (42) |
.036 .825 (41) |
−.167 .315 (38) |
0.125 0.432 (42) |
−.037 .814 (42) |
| Home age (yr.) | Spearman’s rho p value (n) |
.184 .254 (40) |
.440 .005 (39) |
−.076 .655 (36) |
−0.020 0.904 (42) |
.087 .591 (40) |
| B. Gram-positive bacteria | ||||||
|---|---|---|---|---|---|---|
| Environmental factors / Bacterial measures in dust and air | Gram-positive dust concentration (number of cell equivalents /mg dust) | Muramic acid dust concentration (ng/mg dust) | Gram positive in air (number of cell equivalents/m3 air) | Culturable Gram positive in air (CFU /m3 air) | Muramic acid in air (ng/m3 air) | |
| Temperature (°C) | Spearman’s rho p value (n) |
−.364 .018 (42) |
−.332 .032 (42) |
−.219 .187 (38) |
0.429 0.005 (41) |
.221 .165 (41) |
| Relative humidity (%) | Spearman’s rho p value (n) |
−.221 .159 (42) |
.016 .919 (42) |
−.249 .131 (38) |
0.339 0.030 (41) |
.243 .147 (37) |
| Occupants (number) | Spearman’s rho p value (n) |
.029 .855 (42) |
−.353 .022 (42) |
.393 .015 (38) |
0.178 0.266 (41) |
.026 .870 (41) |
| Home age (yr.) | Spearman’s rho p value (n) |
−.144 .377 (40) |
−.214 .183 (40) |
−.142 .408 (36) |
0.034 0.836 (41) |
.054 .740 (39) |
For Gram-positive bacterial measures, concentrations of total Gram-positive bacteria and muramic acid in dust inversely correlated with temperature in the homes. In contrast, culturable airborne bacteria in air positively correlated with temperature and RH. The concentration of Gram-positive bacteria in the air samples positively correlated with the number of occupants in the homes (range: 2 to 9, average: 5) (Figure 1e) but muramic acid concentration in dust inversely correlated with the occupants. Age of home did not seem to be significant factors in controlling the Gram-positive bacterial concentrations (Table 6).
DISCUSSION
The results suggest that both short-term and long-term mold contamination in homes could be linked with the bacterial concentrations in house dust; however, only the current mold status was associated with bacterial concentrations in air. This was likely due to different levels of environmental tolerance among molds and bacteria when they are growing in aerosolizable dust in moisture damaged home environments. Diverging associations were observed between the home moldiness and different measures of bacteria. Long-term mold contamination was associated with Gram-positive bacteria when measured with QPCR, but not when measured as a cell wall component (muramic acid), and with Gram-negative bacteria when measured as cell-wall component (endotoxin), but not when measured by QPCR. This contradictory observation indicates that cell concentrations obtained by QPCR and concentrations of cell wall components may not represent the same bacterial identity and represent too broad categories to understand the bacterial composition and sources of the home microbiota. Another possible explanation is that molds and bacteria growing in water damaged building materials could have different levels of tolerance for environmental stresses. Short-term water damage in building materials can support both mold and bacteria, but with time, the level of bacterial contamination may change faster than the mold contamination. Gram-positive bacteria can grow in parallel with molds because they are more tolerant to dry conditions (Møretrø et al., 2010; Janning and in’t Veld, 1994) than Gram-negative bacteria, whose growth could be diminished when building material is drying. This explanation seems feasible because we found that current moldiness was associated with airborne Gram-negative bacterial concentration.
Although the dust samples from high ERMI homes contained significantly greater concentrations of endotoxin, we did not find a significant difference in the concentration of Gram-negative bacteria in the dust samples between high and low ERMI homes. Similarly, dust concentrations of endotoxin and gram-negative bacteria did not correlate with each other. One reason for this finding may be that different species of Gram-negative bacteria produce different amounts of endotoxin (Weber-Frick and Schmidt-Lorenz, 1988). The amount of endotoxin might depend on the species present in low versus high ERMI homes and some Gram-negative species could be present in higher concentration in moldy homes. For example, Kettleson et al. (2013) found that the Gram-negative bacterium, Stenotrophomonas maltophilia, occurred in significantly higher concentrations in high ERMI homes versus low ERMI homes. Interestingly, in air samples, endotoxin and Gram-negative bacteria correlated and both were also higher in high ERMI homes. These observations support the previous studies indicating that exposure assessment based on dust versus air samples could provide different perspectives (Adhikari et al., 2010). Another observation of our study was that the age of a home was significantly correlated with the endotoxin concentration in dust but not with the other bacterial measurements. This observation is consistent with the previous reports on endotoxin levels in homes, for example, a large study in German homes demonstrated that endotoxin concentrations were higher in old buildings (means ratio = 1.52, 95% C.I.: 1.14; 2.04; Bischof et al., 2002). Older homes may favor Gram-negative bacteria (Kettleson et al., 2013) due to accumulation of dust serving as growth substrate. Endotoxin can remain in homes for longer period because it is a stable molecule.
Good correlations were found in both dust concentration and dust loading between muramic acid and Gram-positive bacteria. Therefore, somewhat surprisingly, differing associations were found between Gram-positive bacterial measures and occupancy. As expected, occupancy was positively correlated with Gram-positive bacteria in air, possibly due to contributions of Gram-positive bacteria from skin, but negatively with muramic acid in dust because Gram-positive bacterial community could be largely different in dust. Furthermore, differences in growth rate and developmental state of the Gram-positive bacteria between human body and dust can also affect cell wall composition of Gram-positive bacteria (Chien et al., 2012) including muramic acid.
Home temperature was inversely correlated with the dust concentrations of Gram-positive bacteria and muramic acid. How typical temperature range in home environments affect Gram-positive bacterial growth in house dust is quite unknown. Our observations suggest that home temperature may have a negative influence on muramic acid content of Gram-positive bacteria in dust. On the other hand, when culturable bacteria were considered, both Gram-positive and Gram-negative bacteria in air were positively correlated with home temperature, which is consistent with observations reported earlier (Obbard et al., 2000).
Home RH positively influenced Gram-negative, but not Gram-positive bacteria, analyzed by QPCR. However, for airborne culturable bacterial concentrations, both Gram-positive and Gram-negative bacteria demonstrated positive correlations with RH, similar to previous report (Aydogdu et al., 2005). Although both Gram-negative and Gram-positive bacteria have been shown to persist under desiccating conditions, previous studies suggest that Gram-positive bacteria exhibit enhanced tolerance to dry conditions compared to Gram-negative bacteria (Møretrø et al., 2010; Janning and in’t Veld, 1994). This may explain why we found a greater portion of Gram-negative bacteria in dust than in air samples. However, all of these observations should be confirmed in larger studies because of some acknowledged limitations of the present study.
One limitation of the study is associated with changing of the ERMI status for 17 homes over the year. The participating families in this study were mainly low-income, living in older, often rental, properties (Reponen et al 2013). If water problems develop in the home, the family may not have resources for or control over timely repairs. In 13 homes, the ERMI values had changed to low ERMI status in 2011 indicating that an intervention, e.g. repairs or carpet replacement may have occurred. However, in four homes, the ERMI values were found to have changed into high ERMI status by the second sampling in 2011, suggesting increased home moldiness. The long-term associations between various bacterial contaminants might be clearer if the families were more stable and lived in well maintained homes. Another limitation of our study is that the quantification of Gram-positive and Gram-negative bacteria was conducted using only B. subtilis and E. coli as standards, respectively. This is a common problem with studies that try to quantify by QPCR large and diverse groups of microorganisms. This investigation was also limited to only a few environmental factors that might affect bacterial populations and statistical adjustment with confounding factors was unattainable due to the limited number of homes sampled.
Although the dust-based ERMI values in infant’s homes have been found to be predictive of the development of asthma (Reponen et al., 2011), it is unclear what dust bacterial concentrations mean in terms of bacterial exposure and health outcomes. However, cell products may be important in understanding these outcomes. Further long-term studies on ecology of mold and bacteria with increased number of samples are required to obtain more clarification. For example, enrichment of different bacterial species in dust and air samples and relationship between diversity of molds and bacteria could be investigated by applying high-throughput DNA sequencing methods.
In conclusion, multiple measures of bacterial populations may be needed to evaluate their impact on human exposures. It may also be useful to target specific species of bacteria for assessment rather than large categories of bacteria, like Gram staining status.
Highlights.
High ERMI and low ERMI homes differ in bacterial contaminant levels.
Only the current mold status is associated with bacterial concentrations in air.
Dusts versus air samples provide different perspectives of bacterial contamination.
Occupancy in homes correlates with total Gram-positive bacterial cells in air.
Correlations of temperature and RH with culturable and total bacteria are unalike.
Acknowledgments
This study was supported by Grant No. OHLHH0199-09 from the Healthy Homes Technical Studies Program of the U.S. Department of Housing and Urban Development (HUD). We also acknowledge a partial support from the HUD Grant OHLHH0162-07 and the National Institute of Environmental Health Sciences (NIEHS) Grant No. T32ES010957-11 awarded to the University of Cincinnati. The CCAAPS birth cohort study was supported by NIEHS Grant ES11170. There are no financial interests to disclose. Technical assistance from Dr. Umesh Singh, Mrs. Sewwandi Rathnayake, Mrs. Moumita Ghosh, Mrs. Sonam Gupta, and Mr. Lev Lazinskiy during air and dust sampling is graciously acknowledged. The authors are also thankful for Drs. William G. Lindsley and Bean T. Chen at NIOSH for providing the NIOSH two-stage cyclones.
Footnotes
NOTICE
The U.S. Environmental Protection Agency (EPA) through its Office of Research and Development partially funded and collaborated in the research described here. It has been subjected to the Agency’s peer review and approved as an EPA publication. Mention of trade names or commercial products does not constitute endorsement or recommendation by the EPA for use. Commercial use of the ERMI technology can provide royalties to the EPA.
Conflict of interest
All authors have no actual or potential conflict of interest including any financial, personal or other relationships with other people or organizations within three years of beginning the submitted work that could inappropriately influence, or be perceived to influence, their work. This study was supported by Grant No. OHLHH0199-09 from the Healthy Homes Technical Studies Program of the U.S. Department of Housing and Urban Development (HUD). We also acknowledge a partial support from the HUD Grant OHLHH0162-07 and the National Institute of Environmental Health Sciences (NIEHS) Grant No. T32ES010957-11 awarded to the University of Cincinnati. The CCAAPS birth cohort study was supported by NIEHS Grant ES11170. There are no financial interests to disclose. The U.S. Environmental Protection Agency (EPA) through its Office of Research and Development partially funded and collaborated in the research described here. It has been subjected to the Agency’s peer review and approved as an EPA publication. Mention of trade names or commercial products does not constitute endorsement or recommendation by the EPA for use. Commercial use of the ERMI technology can provide royalties to the EPA.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adhikari A, Jung J, Reponen T, Lewis JS, DeGrasse EC, Grimsley LF, et al. Aerosolization of fungi, (1→3)-β-D glucan, and endotoxin from flood-affected materials collected in New Orleans homes. Environ Res. 2009;109:215–24. doi: 10.1016/j.envres.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari A, Lewis JS, Reponen T, Degrasse EC, Grimsley LF, Chew GL, et al. Exposure matrices of endotoxin, (1→3)-β-D-glucan, fungi, and dust mite allergens in flood-affected homes of New Orleans. Sci Total Environ. 2010;408:5489–98. doi: 10.1016/j.scitotenv.2010.07.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MA, Nikulin M, Koljalg U, Andersson MC, Rainey F, Reijula K, et al. Bacteria, molds, and toxins in water-damaged building materials. Appl Environ Microbiol. 1997;63:387–93. doi: 10.1128/aem.63.2.387-393.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydogdu H, Asan A, Otkun MT, Ture M. Monitoring of fungi and bacteria in the indoor air of primary schools in Edirne city, Turkey. Indoor Built Environ. 2005;14:411–25. [Google Scholar]
- Bischof W, Koch A, Gehring U, Fahlbusch B, Wichmann HE, Heinrich J, et al. Predictors of high endotoxin concentrations in the settled dust of German homes. Indoor Air. 2002;12:2–9. doi: 10.1034/j.1600-0668.2002.120102.x. [DOI] [PubMed] [Google Scholar]
- Chien AC, Hill NS, Levin PA. Cell size control in bacteria. Curr Biol. 2012;22:R340–9. doi: 10.1016/j.cub.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Reponen T, Bernstein DI, Olds R, Levin L, Liu X, et al. The effect of home characteristics on dust antigen concentrations and loads in homes. Sci Total Environ. 2006;371:31–43. doi: 10.1016/j.scitotenv.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douwes J, Thorne P, Pearce N, Heederik D. Bioaerosol health effects and exposure assessment: progress and prospects. Ann Occup Hyg. 2003;47:187–200. doi: 10.1093/annhyg/meg032. [DOI] [PubMed] [Google Scholar]
- Dunn RR, Fierer N, Henley JB, Leff JW, Menninger HL. Home life: factors structuring the bacterial diversity found within and between homes. PLoS One. 2013;8:e64133. doi: 10.1371/journal.pone.0064133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feezor RJ, Oberholzer C, Baker HV, Novick D, Rubinstein M, Moldawer LL, et al. Molecular characterization of the acute inflammatory response to infections with Gram-negative versus Gram-positive bacteria. Infect Immun. 2003;7:5803–13. doi: 10.1128/IAI.71.10.5803-5813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland RA, Brinkman NE, Vesper SJ. Evaluation of rapid DNA extraction methods for the quantitative detection of fungal cells using real time PCR analysis. J Microbiol Meth. 2002;50:319–23. doi: 10.1016/s0167-7012(02)00037-4. [DOI] [PubMed] [Google Scholar]
- Haugland RA, Varma M, Wymer LJ, Vesper SJ. Quantitative PCR analysis of selected Aspergillus, Penicillium and Paecilomyces species. Syst Appl Microbiol. 2004;27:198–210. doi: 10.1078/072320204322881826. [DOI] [PubMed] [Google Scholar]
- Heederik D, von Mutius E. Does diversity of environmental microbial exposure matter for the occurrence of allergy and asthma? J Allergy Clin Immunol. 2012;130:44–50. doi: 10.1016/j.jaci.2012.01.067. [DOI] [PubMed] [Google Scholar]
- Janning B, in’t Veld PH. Susceptibility of bacterial strains to desiccation: a simple method to test their stability in microbiological reference materials. Anal Chim Acta. 1994;286:469–76. [Google Scholar]
- Kärkkäinen PM, Valkonen M, Hyvärinen A, Nevalainen A, Rintala H. Determination of bacterial load in house dust using qPCR, chemical markers and culture. J Environ Monit. 2010;12:759–68. doi: 10.1039/b917937b. [DOI] [PubMed] [Google Scholar]
- Kettleson E, Kumar S, Reponen T, Vesper S, Méheust D, Grinspun SA, et al. Stenotrophomonas, Mycobacterium, and Streptomyces in home dust and air: associations with moldiness and other home/family characteristics. Indoor Air. 2013;23:387–96. doi: 10.1111/ina.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMasters GK, Wilson K, Levin L, Biagini J, Ryan P, Lockey JE, et al. High prevalence of aeroallergen sensitization among infants of atopic parents. J Pediatr. 2006;149:505–11. doi: 10.1016/j.jpeds.2006.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley WG, Schmechel D, Chen BT. A two-stage cyclone using microcentrifuge tubes for personal bioaerosol sampling. J Environ Monit. 2006;8:1136–42. doi: 10.1039/b609083d. [DOI] [PubMed] [Google Scholar]
- Matera G, Barreca GS, Puccio R, Quirino A, Liberto MC, De Rosa M, et al. Stenotrophomonas maltophilia lipopolysaccharide (LPS) and antibiotics: “in vitro” effects on inflammatory mediators. Infez Med. 2004;12:227–38. [PubMed] [Google Scholar]
- Møretrø T, Heir E, Mo KR, Habimana O, Abdelgani A, Langsrud S. Factors affecting survival of Shigatoxin-producing Escherichia coli on abiotic surfaces. Int J Food Microbiol. 2010;138:71–7. doi: 10.1016/j.ijfoodmicro.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Obbard GI, Viswanathan S, Huan Y. Airborne bacteria and fungal spores in the indoor environment: a case study in Singapore. Acta Biotechnol. 2000;20:67–73. [Google Scholar]
- Peltola JS, Andersson MA, Kampfer P, Auling G, Kroppenstedt RM, Busse HJ, et al. Isolation of toxigenic Nocardiopsis strains from indoor environments and description of two new Nocardiopsis Species, N. exhalans sp. nov. and N umidischolae sp Nov. Appl Environ Microbiol. 2001;67:4293–304. doi: 10.1128/AEM.67.9.4293-4304.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkäranta M, Toivola M, Paulin L, Nevalainen A. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiol. 2008;8:56. doi: 10.1186/1471-2180-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reponen T, Vesper S, Levin L, Johansson E, Ryan P, Burkle J, et al. High Environmental Relative Moldiness Index during infancy as predictor of age seven asthma. Ann Allergy Asthma Immun. 2011;107:120–6. doi: 10.1016/j.anai.2011.04.018. [DOI] [PubMed] [Google Scholar]
- Reponen T, Lockey J, Berstein DI, Vesper SJ, Levin L, Zheng S, et al. Infant origins of childhood asthma associated with specific molds. J Allergy Clin Immunol. 2012;130:639–44. doi: 10.1016/j.jaci.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reponen T, Levin L, Zheng S, Vesper S, Ryan P, Grinspun SA, LeMasters G. Family and home characteristics correlate with mold in homes. Environ Research. 2013;124:67–70. doi: 10.1016/j.envres.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintala H, Pitkäranta M, Toivola M, Paulin L, Nevalainen A. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiol. 2008;8:56. doi: 10.1186/1471-2180-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PH, Lemasters GK, Biswas P, Levin L, Hu S, Lindsey M, et al. A comparison of proximity and land use regression traffic exposure models and wheezing in infants. Environ Health Perspec. 2007;115:278–84. doi: 10.1289/ehp.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U, Levin L, Grinshpun SA, Schaffer C, Adhikari A, Reponen T. Influence of home characteristics on airborne and dustborne endotoxin and β-D-glucan. J Environ Monitor. 2011;13:3246–53. doi: 10.1039/c1em10446b. [DOI] [PubMed] [Google Scholar]
- Suihko ML, Priha O, Alakomi HL, Thompson P, Mälarstig B, Stott R, et al. Detection and molecular characterization of filamentous actinobacteria and thermoactinomycetes present in water-damaged building materials. Indoor Air. 2009;19:268–77. doi: 10.1111/j.1600-0668.2009.00591.x. [DOI] [PubMed] [Google Scholar]
- Torvinen E, Meklin T, Torkko P, Suomalainen S, Reiman M, Katila ML, et al. Mycobacteria and fungi in moisture-damaged building materials. Appl Environ Microbiol. 2006;72:6822–4. doi: 10.1128/AEM.00588-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Environmental Protection Agency. A Standardized EPA Protocol for Characterizing Indoor Air Quality in Large Office Buildings. Washington, DC, USA: US Environmental Protection Agency; 2003. [Google Scholar]
- Vesper SJ, McKinstry C, Haugland RA, Wymer L, Ashley P, Cox D, et al. Development of an environmental relative moldiness index for homes in the U. S J Occu Environ Med. 2007;49:829–33. doi: 10.1097/JOM.0b013e3181255e98. [DOI] [PubMed] [Google Scholar]
- Vesper S. Traditional mould analysis compared to a DNA-based method of mould analysis. Crit Rev Microbiol. 2011;37:15–24. doi: 10.3109/1040841X.2010.506177. [DOI] [PubMed] [Google Scholar]
- Weber-Frick C, Schmidt-Lorenz W. The effect of temperature on the growth and lipopolysaccharide production of Gram-negative bacteria. Zentralbl Bakteriol Mikrobiol Hyg B. 1988;187:56–69. [PubMed] [Google Scholar]
- World Health Organization (WHO) Europe. WHO guidelines for indoor air quality: Dampness and mould. Copenhagen; Denmark: 2009. [PubMed] [Google Scholar]