Abstract
We developed a rapid, sensitive, and reproducible assay to quantify Candida albicans ACT1, CDR1, CDR2, ERG11, and MDR1 mRNA using a two-step reverse transcription and LightCycler real-time PCR (RT-LightCycler PCR) method with sequence-specific hybridization probes. We compared RT-LightCycler PCR with Northern hybridization for quantitative analysis of gene expression in isolates with various fluconazole susceptibilities. Specificity of each LightCycler PCR was verified by LightCycler melting curve analysis and agarose gel electrophoresis of amplified products. Correlation of quantification results between RT-LightCycler PCR and Northern hybridization yielded correlation coefficients of ≥0.91 for all genes except MDR1 (0.74). In this case, reduced correlation was due to the inability of Northern hybridization to accurately quantify the high MDR1 expression in a susceptible dose-dependent isolate which was shown by RT-LightCycler PCR to overexpress MDR1 >200-fold relative to the other isolates tested. In four isolates, low levels of CDR2 mRNA were detected by RT-LightCycler PCR but were undetectable by Northern hybridization. mRNA quantification by RT-LightCycler PCR correlates with Northern hybridization and offers additional advantages, including increased sensitivity and speed of analysis, along with lower RNA concentration requirements and an increased dynamic range of signal detection.
Candida albicans is both a ubiquitous commensal and an opportunistic human pathogen capable of causing mucosal and systemic disease in immunocompromised individuals. Azole antifungals, particularly fluconazole, are often prescribed to treat these infections, and patients may receive extended and/or repeated courses of therapy. As a result, the emergence of azole antifungal drug resistance among C. albicans isolates has been well documented (reviewed in references 16 and 18).
Molecular mechanisms of azole resistance in C. albicans have been identified and include increased expression of the drug target (lanosterol 14-α-demethylase) encoded by ERG11, genetic alteration of ERG11 causing reduced azole affinity, and reduced intracellular accumulation of drug due to increased efflux mediated by multidrug efflux pumps (reviewed in references 18 and 22). Genes encoding drug efflux pumps include CDR1 and CDR2 from the ATP-binding cassette gene family and MDR1 from the major facilitator gene family (18, 22).
Quantification of drug resistance gene expression in C. albicans isolates with reduced fluconazole susceptibility is a valuable tool for understanding the molecular mechanism(s) of fluconazole resistance and monitoring for the emergence of resistance. Traditionally, mRNA quantification by Northern hybridization has been the method of choice for analyzing gene expression in C. albicans (1, 5, 6, 11, 12, 17). However, this method is labor intensive, requires several steps and multiple days to complete, requires relatively large amounts of RNA, and can lack the sensitivity required to detect small changes in gene expression.
More recently, non-real-time reverse transcriptase PCR (RT-PCR) methods for analyzing gene expression have improved sensitivity and increased throughput but still pose limitations with regard to accurate quantification (3, 14, 21). These non-real-time PCR methods measure amplified product at the end of the PCR and are thus subject to the errors caused by the plateauing effect that occurs when reagents become limiting. Moreover, because PCR amplification is exponential in nature, small changes in the amplification efficiency of a given reaction can produce dramatic differences in the amount of final product (3, 21). Therefore, quantification of the final product is not solely dependent upon the initial quantity of target template. Traditional RT-PCR methods also require post-PCR manipulations which take time and increase the risk of laboratory contamination with amplified product.
A further improvement to RT-PCR was accomplished with the introduction of real-time fluorescence PCR technology, which couples PCR with on-line fluorescence detection of amplification products (7, 23). Cycle-by-cycle monitoring of amplification enables discernment of the log-linear phase of amplification for more accurate quantification and eliminates the need for post-PCR sample processing to visualize and analyze products. Quantitative real-time RT-PCR has been successfully used for the measurement of gene expression in a variety of fields, including microbiology (8, 15, 20). The speed of RT-PCR has been further increased by ultra-fast LightCycler PCR, which uses forced air to rapidly heat and cool the chamber along with glass capillaries, which increase sample surface area and serve as natural cuvettes for fluorescence analysis. These features reduce the time to complete 30 cycles of a typical three-step PCR program to 30 min.
The RT-LightCycler PCR method described here couples reverse transcription and real-time PCR to quantify C. albicans ACT1, CDR1, CDR2, ERG11, and MDR1 mRNA in the absence of azole exposure. We utilized fluorescent-labeled, sequence-specific hybridization probes to increase the sensitivity and specificity of detection and quantification of amplified products. This report describes the development and optimization of the method and a comparison of results to those obtained by traditional Northern hybridization.
MATERIALS AND METHODS
Isolates.
Ten isolates of C. albicans (four bloodstream and six mucosal) with varying in vitro fluconazole susceptibilities were selected, including three susceptible, four susceptible dose dependent (SDD), and three resistant isolates. Bloodstream isolates were from a collection derived from active population-based surveillance for candidemia conducted between 1998 and 2000. Mucosal isolates were from human immunodeficiency virus-infected persons with oropharyngeal or vaginal candidiasis. Isolates were stored at −70°C as 30% glycerol stocks in sterile water. Prior to testing, isolates were subcultured onto Sabouraud dextrose agar (SAB) plates (BBL, Cockeysville, Md.) at 35°C.
Broth microdilution susceptibility testing method.
MICs of fluconazole, itraconazole, and voriconazole were determined by the NCCLS M27-A broth dilution method (13). Standard powders of fluconazole and voriconazole were received as gifts from Pfizer Pharmaceuticals Group (Groton, Conn.), and itraconazole drug powder was purchased from Research Diagnostics, Inc. (Flanders, N.J.). The final concentrations of the antifungal agents ranged from 0.125 to 64 μg of fluconazole/ml and 0.015 to 8 μg of itraconazole and voriconazole/ml. The MIC endpoints were read visually following 24 and 48 h of incubation and were defined as the lowest concentration that produced a prominent reduction in growth (∼50%) compared with that of the drug-free growth control. For fluconazole and itraconazole, MIC interpretations were assigned according to the NCCLS criteria (13). No interpretive breakpoints have been established for voriconazole. NCCLS-recommended quality control strains (ATCC 6258 and ATCC 22019) were included in each experiment, and MICs were within the recommended range for each test. Fluconazole MIC determinations were repeated on three different days to determine stability of the susceptibility phenotype.
Sterol quantification method.
Total cellular ergosterol was quantified as previously described (2). The sterol quantification method MIC of fluconazole was defined as the concentration of drug which caused an 80% reduction in the total cellular ergosterol content compared to that in the drug-free control. MICs which fell between two drug concentrations (i.e., less than 80% reduction at one concentration but more than 80% reduction at the next-higher concentration) were mathematically extrapolated based on the amount of reduction by the drug concentration which gave results closest to an 80% reduction end point. Interpretations of fluconazole MICs were assigned according to the NCCLS criteria (13).
Relative quantification of gene expression by LightCycler RT-PCR. (i) RNA extraction.
For each isolate, an overnight culture grown in 10 ml of SAB broth (Difco) was diluted 1:100 in fresh SAB broth and incubated with shaking at 35°C until cells reached the mid-logarithmic phase of growth (optical density at 600 nm = 0.8 to 1.0). Cells were harvested by centrifugation and washed once with sterile distilled water. Cell lysis was performed by resuspending cells in sterile distilled water plus lysis binding solution (Bio 101 Systems, Vista, Calif.), transferring into lysis matrix C tubes (Qbiogene, Carlsbad, Calif.), and vortexing twice for 45 s at speed level 6 in the FastPrep high-speed vortexer (Qbiogene). Total RNA was extracted using an RNAqueous-4PCR kit (Ambion, Austin, Tex.) following the manufacturer's directions. To remove genomic DNA contamination, RNA samples were treated with 2 U of DNase I (Ambion) per 50 μl of RNA at 37°C for 1 h.
(ii) Synthesis of cDNA.
Reverse transcription was performed in a total volume of 20 μl with a 1st Strand cDNA synthesis kit for RT-PCR (Roche Molecular Biochemicals, Indianapolis, Ind.) using 1 μl of RNA, avian myeloblastosis virus RT, and random primer p(dN)6, as recommended by the manufacturer. The cDNA was purified using the QIAquick PCR purification kit (Qiagen, Valencia, Calif.) and quantified by spectrophotometric measurement of the A260 and A280 and standard calculations.
(iii) Quantitative real-time PCR with LightCycler.
Oligonucleotide primers and hybridization probes were designed using LightCycler probe design software (Roche) and are listed in Table 1. In order to optimize the real-time PCR conditions, the optimal MgCl2 concentration and annealing temperature were determined using FastStart DNA Master SYBR Green I (Roche) for LightCycler PCR. In this format, the double-stranded specific, DNA-binding dye, SYBR Green I, was used to detect amplicons instead of fluorescent-labeled hybridization probes. To verify the specificity of the PCR amplification products, LightCycler melting curve analysis was performed using the following thermal cycling profile: 95°C for 0 s, 65°C for 15s, and ramping to 95°C with stepwise signal acquisition. To further verify the specificity of the LightCycler PCR, 5 μl of amplification product was subjected to gel electrophoresis using a 1.2% agarose-TBE (89 mM Tris-borate, 2 mM EDTA; pH 8.3) gel with 1× TBE running buffer. DNA bands were visualized with UV light following ethidium bromide staining.
TABLE 1.
Primers and probes used for RT-LightCycler PCR and Northern hybridization probe synthesis
| Gene | GenBank accession no. | Primer or probed | Sequence (5′ to 3′) | PCR amplicon size (bp) |
|---|---|---|---|---|
| ERG11 | X13296 | LC forward | TGG AGA CGT GAT GCT G | 204 |
| LC reverse | AGT ATG TTG ACC ACC CAT AA | |||
| Donor probea | AAT CTC TGC TAC TTA TAT GAA AGA AAT TAA ACT GAG | |||
| Acceptor probeb | GAG AAC GTG GTG ATA TTG ATC CAA AT | |||
| NH forwardc | ATT GGT ATT CTT ATG GGT GGT CAA CAT AC | |||
| NH reversec | CCC AAT ACA TCT ATG TCT ACC ACC ACC | |||
| CDR1 | X77589 | Forward primer | AAG AGA ACC ATT ACC AGG | 286 |
| Reverse primer | AGG AAT CGA CGG ATC AC | |||
| Donor probe | CAA GAC CAG CAT CTC CAT ATA CTG TAT | |||
| Acceptor probe | ATT CTT TAT GCA AGT GAG GTA TGG TG | |||
| NH forward | GAG ATC TAC CCT TTA AGA TA | |||
| NH reverse | GAA TCG GGA TTC AAT TG | |||
| CDR2 | U63812 | Forward primer | ATG CTG ATG CCC TAG T | 364 |
| Reverse primer | GCT TCC TTA GGA CAT GG | |||
| Donor probe | ATT GTT GTT ATT CTT AGA TGA ACC AAC TTC AG | |||
| Acceptor probe | TTA GAT TCT CAA ACT GCC TGG TCG | |||
| NH forward | GGT ATA TAA ACT GGA CAA C | |||
| NH reverse | GAA TCT GGG TCT AAT TGT | |||
| MDR1 | X53823 | Forward primer | GGA GTT TAG GTG CTG T | 201 |
| Reverse primer | CGG TGA TGG CTC TCA A | |||
| Donor probe | GCC AGT TGG AGA TGG ACT | |||
| Acceptor probe | TTG GTT CAT GTG TAT CAT TTC TG | |||
| NH forwardc | GAG TCG TAG CTA CAT TGC CAT TAA CA | |||
| NH reversec | GGT GAT TTC TAA TGG TCT CCA TAA TGT | |||
| ACT1 | X16377 | Forward primer | CCA GCT TTC TAC GTT TCC | 209 |
| Reverse primer | CTG TAA CCA CGT TCA GAC | |||
| Donor probe | CGG TAT TGT TTT GGA TTC TGG TG | |||
| Acceptor probe | TGG TGT TAC TCA CGT TGT TCC | |||
| NH forwardc | ACC GAA GCT CCA ATG AAT CCA AAA TCC | |||
| NH reversec | GTT TGG TCA ATA CCA GCA GCT TCC AAA |
Donor probes were labeled at the 3′ end with fluorescein.
Acceptor probes were labeled at the 5′ end with LightCyler Red 640.
ACT, ERG11, and MDR1 Northern hybridization probe sequences were previously described (5).
LC, LightCycler; NH, Northern hybridization.
Quantification of gene expression by LightCycler was determined relative to a standard curve for each target gene and was included in each LightCycler RT-PCR experiment. The template for the gene-specific standard curves was generated via conventional PCR using 5 ng of genomic DNA from the fluconazole-susceptible C. albicans isolate ATCC 32354, a 0.3 μM concentration of the respective PCR primer (sequences shown in Table 1), 1.5 mM MgCl2, and a 150 μM concentration of each deoxynucleoside triphosphate. Cycling conditions were one cycle at 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by one cycle at 72°C for 7 min. The samples were held at 4°C in the thermal cycler until retrieved.
A standard curve was prepared for each gene (ACT1, CDR1, CDR2, ERG11, and MDR1) by purification and 10-fold serial dilutions of the respective amplicons (generated as described above) (4). For quantification, LightCycler PCR was performed using the FastStart DNA master hybridization probe PCR mix (Roche). The reaction mixture consisted of 2.4 μl of 25 mM MgCl2, a 0.4 μM concentration of each PCR primer, 0.2 μM concentration of each hybridization probe, 2 μl of LightCycler DNA FastStart hybridization mix (Roche), and PCR-grade water up to a final volume of 18 μl. For all samples, a master mix was prepared and 18 μl was transferred into each glass capillary. Two microliters of cDNA from the reverse transcription step (test samples) or PCR amplicons from each dilution (standard curve) were then pipetted into all but one of the LightCycler capillaries. PCR-grade water was added to the latter in place of template to serve as a negative control. Each RNA preparation was tested for genomic DNA contamination in a LightCycler PCR by replacing cDNA template with 2 μl of purified RNA. After 10 min of denaturation at 95°C, 40 to 50 PCR cycles were performed. Cycling conditions were 95°C for 10 s, 62°C for 15 s, and 72°C for 10 s. The concentration of each gene was calculated by reference to the respective standard curve with the aid of the LightCycler software. Relative gene expression was expressed as a ratio of target gene (ERG11, CDR1, CDR2, and MDR1) concentration to housekeeping gene (ACT1) concentration, and values reported represent the mean gene expression from three separate LightCycler PCR experiments using the same RNA preparation ± the standard deviation (SD).
Northern hybridization. (i) Gel electrophoresis and transfer.
Total RNA from each C. albicans strain was obtained as described above. For Northern hybridization, equal amounts (approximately 5 μg) of RNA from each strain were denatured by adding 3 volumes of formaldehyde loading dye (RNAqueous kit; Ambion) and heating at 65°C for 15 min. RNA samples were then subjected to electrophoresis in a denaturing gel containing 1.2% agarose, 2% formaldehyde, and 1× morpholinepropanesulfonic acid (MOPS) buffer (40 mM MOPS, 10 mM sodium acetate, 1 mM EDTA; pH 7.0). Samples were loaded and electrophoresed in 1× MOPS buffer at 30 V until the bromophenol blue had migrated two-thirds of the length of the gel. Northern transfer was performed by capillary transfer overnight onto positively charged, nylon membrane (Roche) with 20× SSC (3 M NaCl, 0.3 M sodium citrate) as transfer buffer. RNA was fixed onto the membrane by UV cross-linking following the manufacturer's recommendations (SpectroLinker, Westbury, N.Y.).
(ii) Probe synthesis and hybridization.
Digoxigenin (DIG)-labeled DNA probes specific for ERG11, CDR1, CDR2, MDR1, and ACT1 were synthesized by PCR using the PCR Dig probe synthesis kit (Roche), genomic DNA from the fluconazole-susceptible C. albicans isolate ATCC 32354 as template, and gene-specific oligonucleotide primers (sequences shown in Table 1). Cycling conditions for ERG11, MDR1, and ACT1 were one cycle at 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by one cycle at 72°C for 7 min. Cycling conditions for CDR1 and CDR2 were the same as above except the annealing temperature was reduced to 52°C. The samples were held at 4°C in the thermal cycler until retrieved. Positive control amplification reactions were included for each gene, using the same DNA template, primers, and cycling conditions but with unlabeled nucleotides. Efficiency of DIG labeling was determined by electrophoresis of 5 μl of DIG-labeled and 5 μl of unlabeled reaction products through a 1.2% agarose-TBE gel with 1× TBE running buffer. A shift in the expected molecular weight between labeled and unlabeled product indicated efficient incorporation of DIG-labeled nucleotides. Separate but identical blots were probed simultaneously with 3 μl of ACT1 probe and 4 μl of a target gene probe (CDR1, CDR2, ERG11, or MDR1) per ml of DIG Easy Hyb solution (Roche) at 42°C, overnight. Posthybridization washes were carried out using the DIG Wash and Block buffer system (Roche) following the manufacturer's directions.
(iii) Detection and quantification of target gene expression.
DIG-labeled probe-RNA hybrids were detected using CDP-Star (Roche) following the manufacturer's directions. Chemiluminescent signals were quantified using the LumiImager F1 workstation and LumiAnalyst software (Roche). For each isolate, target gene expression was calculated relative to ACT1 expression to adjust for differences in RNA loading. Two independent Northern hybridization experiments were performed for each gene-isolate combination (using the same RNA preparation), and the average of the results is reported.
Statistical analysis.
Student's t test (two-tailed; two-sample test with unequal variance) was used to determine significant differences in gene expression, as determined by LightCycler RT-PCR, for isolates with different fluconazole susceptibility phenotypes. Pearson's product moment correlation coefficient r was used to determine correlations between Northern hybridization and RT-LightCycler PCR results for each target gene.
RESULTS
Antifungal susceptibility testing.
Table 2 summarizes the in vitro susceptibilities of the 10 C. albicans isolates to fluconazole, itraconazole, and voriconazole as determined by the NCCLS broth microdilution method and their corresponding susceptibility interpretations. Fluconazole MICs were confirmed for all isolates by the sterol quantification method (data not shown). These isolates were selected for evaluation of the RT-LightCycler PCR method because of their stable and unambiguous fluconazole MIC results, i.e., three independent MIC tests, set up on different days and using the same batch of antifungal drug plates, yielded identical results. Trailing growth was not observed in any of the isolates in any azole drug tested.
TABLE 2.
In vitro susceptibilities of the 10 C. albicans isolates used in this study
| Isolate no. | MIC (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Fluconazole
|
Itraconazole
|
Voriconazole
|
||||||
| 24 h | 48 h | Interpretationa | 24 h | 48 h | Interpretationa | 24 h | 48 h | |
| 1 | 64 | ≥64 | R | 0.5 | 0.5 | SDD | 0.5 | 0.5 |
| 2 | 64 | 64 | R | 0.5 | 0.5 | SDD | 0.25 | 0.5 |
| 3 | 64 | ≥64 | R | 0.5 | 0.5 | SDD | 0.25 | 0.25 |
| 4 | 32 | 32 | SDD | 0.25 | 0.25 | SDD | 0.125 | 0.25 |
| 5 | 32 | 32 | SDD | 0.5 | 0.5 | SDD | 0.25 | 0.25 |
| 6 | 16 | 16 | SDD | 0.25 | 0.5 | SDD | 0.125 | 0.25 |
| 7 | 16 | 16 | SDD | 0.125 | 0.25 | SDD | 0.06 | 0.125 |
| 8 | 0.25 | 0.25 | S | 0.06 | 0.06 | S | 0.015 | 0.015 |
| 9 | 0.125 | 0.125 | S | 0.06 | 0.06 | S | ≤0.015 | 0.015 |
| 10 | 0.125 | 0.125 | S | 0.03 | 0.03 | S | ≤0.015 | 0.015 |
R, resistant (fluconazole MIC ≥ 64 μg/ml; itraconazole MIC ≥ 1 μg/ml); SDD, susceptible dose dependent (fluconazole MIC, 16 to 32 μg/ml; itraconazole MIC, 0.25 to 0.5 μg/ml); S, susceptible (fluconazole MIC ≤ 8 μg/ml; itraconazole MIC ≤ 0.125 μg/ml).
Specificity and efficiency of ACT1, CDR1, CDR2, ERG11, and MDR1 LightCycler PCRs.
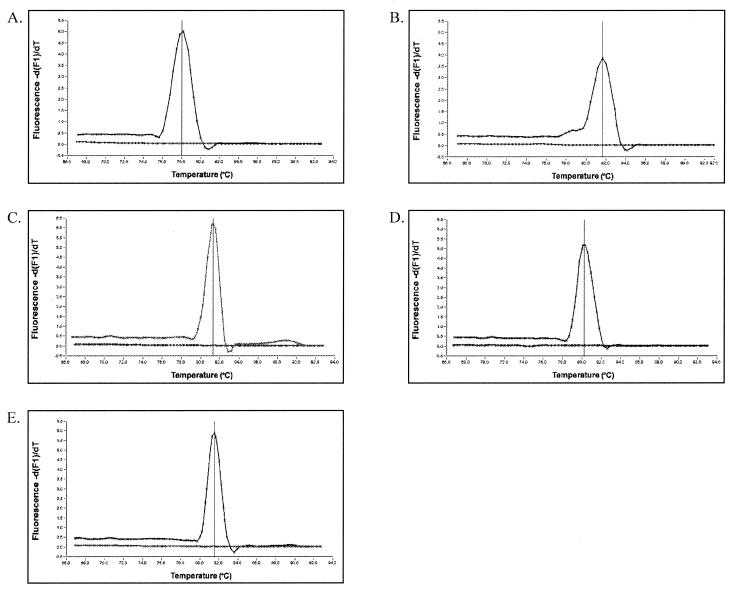
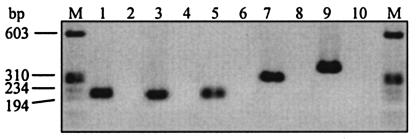
A two-step RT-PCR method was adopted in this study so that both the RT and the LightCycler PCR steps could be optimized for accurate quantification. The specificity and efficiency of each gene-specific LightCycler PCR was determined using the LightCycler melting curve analysis with SYBR Green I dye prior to introducing sequence-specific hybridization probes. Since SYBR Green I binds to double-stranded DNA in a sequence-independent manner and amplification products have sequence-specific melting temperatures (Tm), each unique DNA species can be identified based on its peak fluorescence at a specific temperature. MgCl2 concentration and annealing temperature of each LightCycler PCR were adjusted systematically until the melting curve analysis revealed a single fluorescent peak at the expected temperature and the formation of primer dimers, seen as a low and broad fluorescent peak at temperatures 10 to 15°C lower than that of the specific amplicon, was inhibited. Figure 1 shows the melting curve analysis results for each gene-specific LightCycler PCR. The presence of a single peak at the expected temperature indicates that the primers are specific and nonspecific amplification is not occurring. Specificity of the LightCycler PCR was further verified by agarose gel electrophoresis of the amplification products by observation of a single band of the expected molecular weight (Fig. 2).
FIG. 1.
Specificity of LightCycler PCR for amplification of C. albicans genes ERG11 (A), CDR1 (B), CDR2 (C), MDR1 (D), and ACT1 (E) as determined by melting curve analysis. Melting peaks were determined by plotting the continuous negative derivative of fluorescence emitted by each sample as PCR products were slowly melted. The no-template control sample for each reaction does not show fluorescence, confirming the absence of primer dimers.
FIG. 2.
Agarose gel electrophoresis of LightCycler PCR products. A single band of the expected molecular weight confirms the specificity of the LightCycler PCR. The absence of a band in the no-template samples indicates the absence of primer dimers. Lanes: M, molecular weight marker IX (Roche); 1, ACT1 amplicon; 2, ACTI no-template control (NTC); 3, MDR1 amplicon; 4, MDR1 NTC; 5, ERG11 amplicon; 6, ERG11 NTC; 7, CDR1 amplicon; 8, CDR1 NTC; 9, CDR2 amplicon; 10, CDR2 NTC.
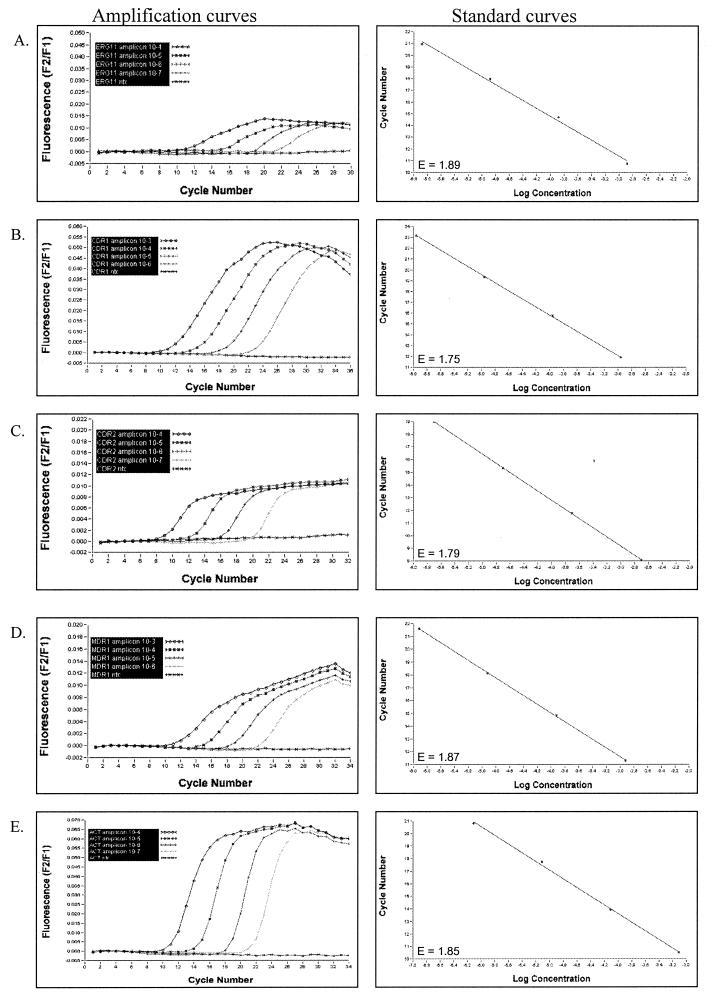
Quantification of LightCycler PCR amplicons is based on extrapolation from a standard curve. The LightCycler software generates a standard curve by plotting the logarithm of fluorescence versus cycle number for each serial dilution of standard template and then identifies the crossing point (cycle number) where the fluorescent signal emerges from background and enters the log-linear phase. Each crossing point is then plotted against the user-defined concentration of that standard to produce a standard curve. Finally, the crossing point for each test sample is measured and the concentration of target DNA in that test sample is calculated based on the standard curve. For accurate quantification, the amplification efficiency (E) of the target gene must be the same as for the housekeeping gene. The E value for a specific LightCycler PCR is determined based on the slope of the standard curve and calculated using the equation E = 10−1/slope. Optimal efficiency is achieved when the slope of the standard curve is approximately −3.3, giving an E of ∼2.0. After optimizing magnesium concentration and annealing temperature and, in some cases, redesigning LightCycler PCR primers, E values for both the drug resistance genes and ACT1 ranged between 1.768 and 1.885. Figure 3 shows representative LightCycler PCR amplification curves and the respective standard curves for ACT1, CDR1, CDR2, ERG11, and MDR1 using four 10-fold serial dilutions of homologous, in vitro-synthesized DNA template.
FIG.3.
LightCycler PCR amplification curves and their respective standard curves from four 10-fold serial dilutions of in vitro-synthesized, C. albicans genes ERG11 (A), CDR1 (B), CDR2 (C), MDR1 (D), and ACT1 (E). For amplification curves, the amount of fluorescence emitted from each sample was measured at the end of each PCR cycle and plotted against the respective PCR cycle number. For the standard curves, log concentration of each standard (as entered by user) was plotted against the cycle number where fluorescence emerged from background during amplification. The E (efficiency) value is shown for each LightCycler standard curve.
Since the crossing point of a given LightCycler PCR refers to the cycle number at which amplicon fluorescence comes out of background and enters the log-linear portion of the amplification curve, the timing of the crossing point correlates with the concentration of template DNA such that the more template DNA, the earlier the crossing point. Template concentrations that are too high result in crossing points that are less than or equal to cycle 5 and must be further diluted for accurate quantification. The initial concentration of template DNA for each standard curve was adjusted such that the crossing point for the highest dilution was greater than cycle 5 and crossing points for all dilutions spanned a range of cycle numbers the crossing point of a given unknown was expected to fall within.
Quantification of ACT1, CDR1, CDR2, ERG11, and MDR1 expression in the absence of azole exposure among C. albicans isolates with varying susceptibilities to fluconazole. (i) LightCycler real-time PCR.
Following reverse transcription of mRNA, cDNA was amplified and quantified by LightCycler real-time PCR to determine the expression of ACT1, CDR1, CDR2, ERG11, and MDR1. For each C. albicans isolate, expression of CDR1, CDR2, ERG11, and MDR1 was normalized with ACT1 expression (from the respective isolate) by representing the results as a ratio of resistance gene expression to housekeeping gene expression (ACT1). Results for all 10 isolates are shown in Table 3 and represent the mean expression ± SD for three independent LightCycler PCR runs. ERG11 expression was significantly higher in the fluconazole-resistant isolates compared to the SDD and susceptible isolates, as well as in the SDD isolates compared to the susceptible isolates (P ≤ 0.05). CDR1 and CDR2 overexpression was observed in the fluconazole-resistant isolates compared to the susceptible isolates (P ≤ 0.08). MDR1 expression was less consistent in that only a single fluconazole-SDD isolate expressed high levels while the other nine isolates (including susceptible, SDD, and resistant) expressed very little or no MDR1.
TABLE 3.
LightCycler quantification of C. albicans ERG11, CDR1, CDR2, and MDR1 mRNA
| Isolate no. | Susceptibility interpretationb | Mean expression units ± SDa
|
|||
|---|---|---|---|---|---|
| ERG11c | CDR1d | CDR2d | MDR1 | ||
| 1 | R | 3.98 ± 0.64 | 4.89 ± 0.86 | 1.21 ± 0.16 | 0 |
| 2 | R | 5.39 ± 0.56 | 4.33 ± 0.76 | 1.71 ± 0.29 | 0.20 ± 0.03 |
| 3 | R | 4.78 ± 0.63 | 8.35 ± 0.16 | 0.85 ± 0.09 | 0.63 ± 0.16 |
| 4 | SDD | 3.90 ± 0.45 | 5.16 ± 0.44 | 2.01 ± 0.28 | 0 |
| 5 | SDD | 2.35 ± 0.46 | 13.14 ± 1.43 | 1.01 ± 0.15 | 0.61 ± 0.14 |
| 6 | SDD | 3.97 ± 0.50 | 5.04 ± 0.60 | 2.82 ± 0.45 | 0 |
| 7 | SDD | 2.52 ± 0.40 | 2.42 ± 0.49 | 0.62 ± 0.04 | 156.50 ± 11.63 |
| 8 | S | 2.08 ± 0.32 | 2.43 ± 0.83 | 0.26 ± 0.05 | 0 |
| 9 | S | 1.87 ± 0.08 | 1.43 ± 0.22 | 0.30 ± 0.02 | 0 |
| 10 | S | 1.56 ± 0.27 | 2.06 ± 0.22 | 0.24 ± 0.05 | 0 |
Gene expression normalized to ACT1 expression for each isolate.
Fluconazole susceptibility category (R, resistant; SDD, susceptible dose dependent; S, susceptible).
Overexpression in R compared to S and SDD isolates and in SDD compared to S isolates (P ≤ 0.05).
Overexpression in R compared to S isolates (P ≤ 0.08).
Because LightCycler is more rapid (due to rapid cycling between temperatures and decreased incubation times) and more sensitive (due to quantification during the log-linear phase of amplification and detection with hybridization probes) than conventional PCR, it can be more susceptible to run-to-run variability. Therefore, it was important to validate its reproducibility. For all isolates, the SD among repeated runs was less than or equal to 25% of the mean for all genes analyzed with the exception of isolate 8 and CDR1, which had an SD that was 34% of the mean (Table 3). Although the reproducibility of the LightCycler PCR method was determined using the same RNA preparation from a given isolate, the observed trend in expression was similar when different RNA preparations were analyzed (data not shown).
(ii) Northern hybridization.
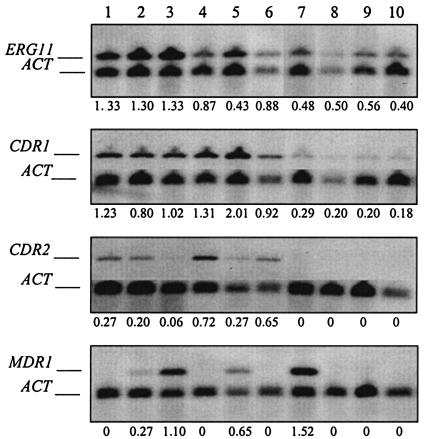
Northern hybridization is currently the most commonly used method for quantifying gene expression in Candida species isolates. Therefore, we compared quantification by this novel RT-LightCycler PCR method with that of Northern hybridization using sequence-specific probes. Figure 4 shows the Northern hybridization results of ACT1, CDR1, CDR2, ERG11, and MDR1 expression for all 10 C. albicans isolates grown in the absence of azole antifungals. Densitometry was used to quantify the Northern blots, and gene expression, relative to ACT1, was determined for each isolate (Fig. 4). The Northern hybridization images in Fig. 4 are from representative experiments, while the densitometry values shown are the average of two independent experiments. Northern hybridization analysis failed to detect low levels of CDR2 expression in one of the SDD isolates (isolate 7) and in all three of the susceptible isolates (isolates 8 to 10). Six of the 10 isolates, including one fluconazole-resistant, two SDD, and all three susceptible isolates (isolates 1, 4, 6, and 8 to 10), lacked MDR1 expression by Northern hybridization (Fig. 4).
FIG. 4.
Quantification of C. albicans CDR1, CDR2, ERG11, and MDR1 mRNA by Northern hybridization. Quantification of target gene expression was accomplished using densitometry of chemiluminescent signals. Expression of drug resistance genes was normalized to that of ACT1 expression in the same isolate. Densitometry values shown below each image are the average of two independent experiments.
Correlation of LightCycler PCR and Northern blotting methods for quantification of gene expression.
Correlation between RT-LightCycler PCR and Northern hybridization results for quantifying CDR1, CDR2, ERG11, and MDR1 expression, relative to ACT1 expression, from 10 C. albicans isolates with varying susceptibilities to fluconazole was determined. Excellent correlation (Pearson's r = 0.97 for ERG11, 0.94 for CDR1, and 0.91 for CDR2) was observed for all genes except MDR1, which showed lower correlation between methods (Pearson's r = 0.74). Reduced correlation for MDR1 was due to the decreased dynamic range of the Northern hybridization procedure relative to that of LightCycler PCR. Even though direct quantification of Northern hybridization chemiluminescent signals with the LumiImager increases the dynamic range of signal detection 100-fold over that of X-ray film, the high levels of MDR1 expression observed in isolate 7 could not be accurately quantified due to signal saturation. Although the Northern hybridization method demonstrated that isolate 7 expressed more MDR1 than any of the other nine isolates, the magnitude of overexpression could not be accurately quantified by this method. In the case of CDR2, Northern hybridization failed to detect low levels of expression in susceptible isolates 8 to 10 and the SDD isolate 7. CDR2 expression was detectable by the RT-LightCycler PCR method in these same isolates and was shown to be, on average, 12-fold lower than that in isolates which were positive for CDR2 expression by Northern hybridization.
DISCUSSION
The ability to detect and quantify mRNA transcripts is a powerful tool for studying gene expression and regulation. In fungi, primary as well as secondary azole resistance has been associated with overexpression of CDR1, CDR2, ERG11, and MDR1 (18, 22). Furthermore, it has been demonstrated that overexpression of CDR1 and CDR2 can lead to cross-resistance of the same isolate to multiple azole antifungals, whereas overexpression of MDR1 has been associated with fluconazole resistance only (1).
Northern hybridization is a well-established method for RNA analysis and has been useful for characterizing drug resistance gene expression in clinical isolates of C. albicans (1, 5, 6, 11, 12, 17). However, in the era of PCR and specifically with the advent of RT-PCR, quantification of mRNA using these modern approaches offers significant advantages over Northern hybridization. These include increased sensitivity for detection of low-abundance mRNAs, increased dynamic range of detection for more accurate quantification, a “micro” format to support high-throughput screening applications, and the ability to convert unstable RNA template into cDNA, which is less susceptible to degradation. The LightCycler system, which couples high-speed PCR with on-line fluorescent detection of amplification products, adds additional advantages to RT-PCR, including quantification during the log-linear phase of amplification, increased speed of analysis, and no need for post-PCR sample processing.
To date, a few reports have described the application of LightCycler PCR to the detection, identification, or quantification of fungal cells (9, 10, 15, 19). However, none have applied this technology to quantify the expression of the known drug resistance genes in C. albicans. Loeffler et al. (10) described a quantitative LightCycler PCR assay to detect C. albicans and Aspergillus fumigatus DNA in human blood specimens with high sensitivity and specificity. Although these authors' previously published PCR-enzyme-linked immunosorbent assay was equally sensitive, the LightCycler assay decreased the time to results by 2 h (10). Another report described the use of LightCycler PCR to detect point mutations in C. albicans and Candida tropicalis ERG11 from isolates with decreased fluconazole susceptibility (9). This approach took advantage of the LightCycler melting curve function to detect differences in Tm among clinical isolates with different ERG11 genotypes. When compared to conventional cycle sequencing, the LightCycler method performed equally well in the case of C. albicans. For C. tropicalis, the LightCycler method detected a heterozygous genotype in five isolates that was not detected in the same five isolates by conventional cycle sequencing. Excellent sensitivity and specificity of the LightCycler assay were demonstrated by these investigators and, coupled with the ability to analyze 32 samples per 45-min run, the potential for rapid screening was noted (9). Selvarangan et al. (19) described a different application of the LightCycler melting curve function to differentiate between the germ-tube-positive yeasts C. albicans and Candida dubliniensis. This method utilized a single fluorescent-labeled nucleic acid probe which bound equally well to LightCycler PCR products from both species but disassociated from each at a characteristic temperature. Differentiation between the two species was accomplished in 2 h or less by detection of peak fluorescence at the species-specific Tm (19). The fourth report described the use of LightCycler real-time RT-PCR to quantify C. albicans actin mRNA as a means of assessing viability in a model of cutaneous candidiasis (15). Results demonstrated the sensitivity of the method in detecting 1 fg of the plasmid DNA containing a 192-bp fragment of the ACT1 exon (pACT) in real time, with a reproducibility of 100% among five replicates (15). Each of these assays illustrates a unique application of the LightCycler technology, while all share the common features of excellent sensitivity, specificity, and speed of analysis. Furthermore, these studies demonstrate the flexibility of the LightCycler system and its usefulness in both the research laboratory as well as the routine clinical laboratory.
The novel RT-LightCycler PCR assay described here for the relative quantification of mRNA is yet another application of real-time PCR. To increase sensitivity and specificity of the assay, we added sequence-specific hybridization probes to detect C. albicans ACT1, CDR1, CDR2, and MDR1 LightCycler PCR products. This detection format uses two oligonucleotide probes, labeled with different fluorescent dyes, which bind very closely to each other on the target sequence. It is only when the dyes are in close proximity to each other, as when bound to the target sequence, that a fluorescent light is emitted. Therefore, in the absence of target sequence, no fluorescence is detected. This approach, unlike the SYBR Green I dye, which binds nonspecifically to all double-stranded DNA, eliminates the contribution of nonspecific amplification products, such as primer dimers, to the quantification of the target sequence.
Quantification of mRNA from the clinical C. albicans isolates was achieved by extrapolation from standard curves generated from known concentrations of standard DNA templates. Therefore, careful consideration was given to designing a standard curve to ensure that amplification efficiencies of target sequences were the same for both the test samples and the standards. For this method, we prepared external standards of in vitro-synthesized ACT1, CDR1, CDR2, ERG11, and MDR1 sequences and included the appropriate standard curves in every LightCycler PCR experiment. Therefore, the concentration of target in the test samples was calculated based on its specific standard curve that was generated during the same LightCycler PCR run. Although this conservative approach added five additional reaction tubes per target gene per LightCycler PCR and thereby reduced the number of test samples that could be analyzed in a given run, it eliminated any concern of variation that may occur between individual LightCycler PCR runs. Upon analysis of our results from multiple LightCycler PCR runs, we found run-to-run variation to be minor and the practice of using a previously generated and stored standard curve to quantify target sequence concentration in test samples acceptable.
In conclusion, the RT-LightCycler PCR method for C. albicans mRNA quantification correlated well with traditional Northern hybridization while offering several advantages, including increased sensitivity and specificity, accurate quantification of very low and very high mRNA quantities, increased throughput and speed of analysis, and a closed-tube system that does not require post-PCR sample analysis. The use of this novel RT-LightCycler PCR assay to rapidly screen clinical C. albicans isolates and quantify the expression of known drug resistance genes could be a valuable complement to in vitro antifungal susceptibility testing for detecting and monitoring the emergence of azole antifungal drug resistance. Adaptation of this method to screen non-C. albicans species of Candida is under way.
Acknowledgments
J.P.F. was supported by a postgraduate fellowship, SFRH/BD5315/01, from the Portuguese Science and Technology Foundation.
REFERENCES
- 1.Albertson, G. D., M. Niimi, R. D. Cannon, and H. F. Jenkinson. 1996. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob. Agents Chemother. 40:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthington-Skaggs, B. A., D. W. Warnock, and C. J. Morrison. 2000. Quantitation of Candida albicans ergosterol content improves the correlation between in vitro antifungal susceptibility test results and in vivo outcome after fluconazole treatment in a murine model of invasive candidiasis. Antimicrob. Agents Chemother. 44:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freeman, W. M., S. J. Walker, and K. E. Vrana. 1999. Quantitative RT-PCR: pitfalls and potential. BioTechniques 26:112-125. [DOI] [PubMed] [Google Scholar]
- 4.Fronhoffs, S., G. Totzke, S. Stier, N. Wernert, M. Rothe, T. Bruning, B. Koch, A. Sachinidis, H. Vetter, and Y. Ko. 2002. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Mol. Cell. Probes 16:99-110. [DOI] [PubMed] [Google Scholar]
- 5.Henry, K. W., J. T. Nickels, and T. D. Edlind. 2000. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 44:2693-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry, K. W., M. C. Cruz, S. K. Katiyar, and T. D. Edlind. 1999. Antagonism of azole activity against Candida albicans following induction of multidrug resistance genes by selected antimicrobial agents. Antimicrob. Agents Chemother. 43:1968-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 8.Kreuzer, K. A., U. Lass, A. Bohn, O. Landt, and C. A. Schmidt. 1999. LightCycler technology for the quantitation of bcr/abl fusion transcripts. Cancer Res. 59:3171-3174. [PubMed] [Google Scholar]
- 9.Loeffler, J., L. Hagmeyer, H. Hebart, N. Henke, U. Schumacher, and H. Einsele. 2000. Rapid detection of point mutations by fluorescence resonance energy transfer and probe melting curves in Candida species. Clin. Chem. 46:631-635. [PubMed] [Google Scholar]
- 10.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and LightCycler system. J. Clin. Microbiol. 38:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Ribot, J. L., R. K. McAtee, L. N. Lee, W. R. Kirkpatrick, T. C. White, D. Sanglard, and T. F. Patterson. 1998. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 42:2932-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marr, K. A., C. N. Lyons, T. Rustad, R. A. Bowden, and T. C. White. 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob. Agents Chemother. 42:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Okeke, C. N., R. Tsuboi, M. Kawai, M. Yamazaki, S. Reangchainam, and H. Ogawa. 2000. Reverse transcription-3′ rapid amplification of cDNA ends-nested PCR of ACT1 and SAP2 as a means of detecting viable Candida albicans in an in vitro cutaneous candidiasis model. J. Investig. Dermatol. 114:95-100. [DOI] [PubMed] [Google Scholar]
- 15.Okeke, C. N., R. Tsuboi, and H. Ogawa. 2001. Quantification of Candida albicans actin mRNA by the LightCycler system as a means of assessing viability in a model of cutaneous candidiasis. J. Clin. Microbiol. 39:3491-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perea, S., and T. F. Patterson. 2002. Antifungal resistance in pathogenic fungi. Clin. Infect. Dis. 35:1073-1080. [DOI] [PubMed] [Google Scholar]
- 17.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 19.Selvarangan, R., A. P. Limaye, and B. T. Cookson. 2002. Rapid identification and differentiation of Candida albicans and Candida dubliniensis by capillary-based amplification and fluorescent probe hybridization. J. Clin. Microbiol. 40:4308-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, R. D., J. Li, C. T. Noguchi, and A. N. Schechter. 2000. Quantitative PCR analysis of HbF inducers in primary human adult erythroid cells. Blood 95:863-869. [PubMed] [Google Scholar]
- 21.Souaze, F., A. Ntodou-Thome, C. Y. Tran, W. Rostene, and P. Forgez. 1996. Quantitative RT-PCR: limits and accuracy. BioTechniques 21:280-285. [DOI] [PubMed] [Google Scholar]
- 22.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-138. [DOI] [PubMed] [Google Scholar]