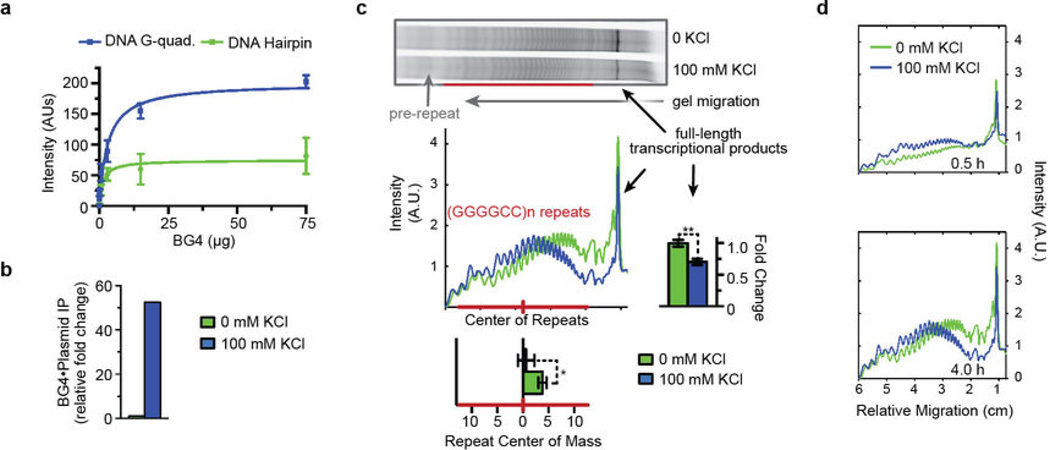
Extended Data Figure 3. G-quadruplexes increase abortive transcripts within the C9orf72 HRE region in vitro.
a) A newly developed BG4 nanobody directly binds the K+-dependent Gquadruplex formed by DNA (GGGGCC)4. BG4 was purified from E. coli, and an ELISA experiment using biotinylated DNA (GGGGCC)4, with or without 100 mM KCl during annealing, was used to determine the specificity of BG4, essentially as previously described28. Data show the mean ± S.D. n = 3. b) The ΔCt graph for 100 mM KCl vs 0 mM KCl shows that BG4 pulled down >50-fold more plasmid when the plasmid containing (GGGGCC)21 repeats was annealed in the presence of 100 mM KCl, directly demonstrating the presence of Gquadruplexes formed by the sequence. The DNA pulldown with BG4 is described in the Supplementary Methods. n = 1. c) Formation of G-quadruplexes in the coding region prior to transcription causes a shift to an earlier termination of abortive transcripts and further loss of the full-length products. The shift is represented below the densitometric plot profile by a significant change in the center of mass for the repeat region (n = 5). The further loss of full-length transcriptional product is shown on the graph to the right of the transcription profile (n = 3). P-values were calculated using a paired fit for each trial assuming a parametric distribution. Data are means ± s.e.m. *P < 0.05; **P < 0.01. d) Accumulation of the abortive transcripts indicates that the repeat-dependent impairment of transcription is not transient but persists over time ± KCl. Aliquots were removed from the in vitro transcription reactions at different time points and then visualized and analyzed as in (c).