Abstract
We report the evaluation of recombinant severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) nucleocapsid protein enzyme-linked immunosorbent assay (ELISA)-based antibody tests for serodiagnosis of SARS-CoV pneumonia and compare the sensitivities and specificities of this ELISA for detection of immunoglobulin G (IgG), IgM, IgA, and their combinations with serum samples from 149 healthy blood donors who donated blood 3 years ago as controls and 106 SARS-CoV pneumonia patients in Hong Kong. The specificities of the ELISA for IgG, IgM, and IgA detection were 95.3, 96.6, and 96.6%, respectively, with corresponding sensitivities of 94.3, 59.4, and 60.4%, respectively. The present ELISA appears to be a sensitive test for serodiagnosis of SARS-CoV pneumonia, is much more economical and less labor-intensive than the indirect immunofluorescence assay, and does not require cultivation of SARS-CoV.
Severe acute respiratory syndrome (SARS), caused by the SARS coronavirus (SARS-CoV), is a new emerging disease that has affected 30 countries, with more than 8,000 cases and more than 750 deaths (6-10, 12-16, 19). For laboratory diagnosis of SARS-CoV pneumonia, isolation of the virus from clinical specimens is insensitive and requires biosafety level 3 laboratory facilities, while detection of viral RNA by reverse transcription-PCR can achieve a sensitivity of only 50 to 79%, depending on the type and number of clinical specimens collected and the protocol used (22).
At the moment, the most widely used methods for serodiagnosis of SARS-CoV infection in clinical microbiology laboratories are antibody detection in acute- and convalescent-phase serum samples by indirect immunofluorescence assay and enzyme-linked immunosorbent assay (ELISA) with cell culture extract (8, 14). However, antibody detection by indirect immunofluorescence assay and ELISA with cell culture extract may be less reproducible, difficult to standardize, and labor intensive compared with ELISA-based antibody detection tests with recombinant antigens. Furthermore, producing the infected cell lines for coating the ELISA plates and the slides for indirect immunofluorescence require cultivation of SARS-CoV, for which biosafety level 3 laboratory facilities are required. Such facilities are not available in most clinical microbiology laboratories.
ELISA-based antibody detection tests with recombinant antigens are well known to offer higher reproducibilities, are easy to standardize and less labor intensive than antibody detection by indirect immunofluorescence assay and ELISA with cell culture extract, and do not require cultivation of SARS-CoV (1-4, 20, 23). Recently, we have reported the use of a recombinant SARS-CoV nucleocapsid protein ELISA-based immunoglobulin G (IgG) antibody test for the study of the seroprevalence of nonpneumonic SARS-CoV infections (21). In this article, we describe the evaluation of recombinant SARS-CoV nucleocapsid protein ELISA-based IgM and IgA antibody tests for serodiagnosis of SARS-CoV pneumonia. The sensitivities and specificities of antibody tests for detection of IgG, IgM, IgA, and their combinations were compared. The clinical usefulness of these antibody assays for serodiagnosis of SARS-CoV infections is also discussed.
Cloning and purification of (His)6-tagged recombinant nucleocapsid protein were reported previously (21). Serum samples from 149 healthy blood donors who donated blood 3 years ago and 106 SARS-CoV pneumonia patients positive for IgG antibodies against the SARS-CoV as detected by our indirect immunofluorescence assay (14) were used for evaluation of the ELISA-based antibody tests. Serum samples positive for IgG antibodies against SARS-CoV by indirect immunofluorescence assay from the 106 SARS-CoV pneumonia patients were taken at a median of 25 (range, 12 to 43) days from the onset of symptoms. The ELISA-based SARS-CoV antibody tests were modified from our previous publication (21). Twenty, 80, and 30 ng of purified (His)6-tagged recombinant nucleocapsid protein were used for coating the ELISA plates for IgG, IgM, and IgA detection, respectively, and diluted horseradish peroxidase-conjugated goat anti-human IgG (1:4,000), mouse anti-human IgM (1:500), and mouse anti-human IgA (1:1,000) antibodies (Zymed Laboratories Inc., South San Francisco, Calif.) were used as the secondary antibodies.
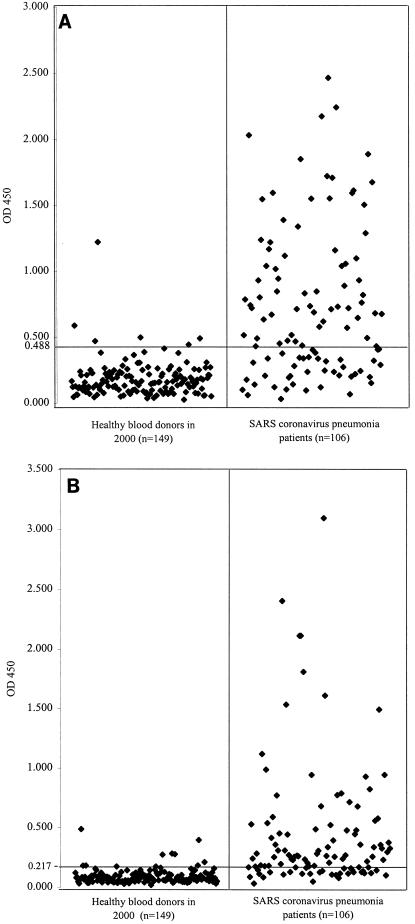
To establish the baseline for the tests, serum samples (all tested negative by the indirect immunofluorescence assay) from 149 healthy blood donors who donated blood 3 years ago were tested in the SARS-CoV antibody ELISA. For the 149 specimens from healthy blood donors, the mean ELISA optical density at 450 nm (OD450) values for IgM and IgA detection were 0.182 and 0.093, respectively, with standard deviations of 0.133 and 0.062, respectively. Absorbance values of 0.488 and 0.217 were selected as the cutoff values (equal to the sum of the mean values from the healthy control and two times the standard deviations) (Fig. 1). With these cutoff values, five of the serum samples obtained from the 149 healthy blood donors had an OD450 of more than 0.488 in the IgM ELISA, and another five had an OD450 of more than 0.217 in the IgA ELISA (Fig. 1). The specificities of the IgM and IgA ELISAs were both 96.6%. These serum samples were also confirmed to be false-positives by Western blot analysis with recombinant nucleocapsid protein and spike protein as the antigens (21).
FIG. 1.
Evaluation of sensitivity and specificity of recombinant nucleocapsid protein-based IgM (A) and IgA (B) antibody ELISA for SARS-CoV pneumonia. Serum specimens were obtained from 106 patients with SARS-CoV pneumonia, and control serum specimens were obtained from 149 healthy blood donors. The test results were plotted as OD450. The cutoff line for positive diagnosis is drawn at a value that equals the sum of the mean value and two times the standard deviation for the healthy blood donors.
The mean OD450 values (IgM and IgA) for the serum samples obtained from the 106 SARS-CoV pneumonia patients that were positive for IgG antibodies against the SARS-CoV in our indirect immunofluorescence assay were 0.739 and 0.451, respectively, with standard deviations of 0.563 and 0.523, respectively. Sixty-three serum samples had an OD450 of over 0.488 in the IgM ELISA, and 64 had an OD450 of over 0.217 in the IgA ELISA (Fig. 1). The sensitivities of the IgM and IgA tests, with the indirect immunofluorescence assay as the gold standard, were hence 59.4 and 60.4%, respectively. Only 38 serum samples (35.8%) had all three antibodies (IgG, IgM, and IgA) against the nucleocapsid protein of SARS-CoV (Table 1). There was no statistically significant difference among the results for serum samples obtained at different times after disease onset (Table 2).
TABLE 1.
Comparison of sensitivities and specificities of recombinant nucleocapsid protein-based ELISA for detection of IgG, IgM, IgA, and their combinations for SARS-CoV pneumonia
| Specific antibody detected | Sensitivity (%) | Specificity (%) |
|---|---|---|
| IgG | 94.3 | 95.3 |
| IgM | 59.4 | 96.6 |
| IgA | 60.4 | 96.6 |
| IgG and IgM | 54.7 | 99.3 |
| IgG and IgA | 59.4 | 99.3 |
| IgM and IgA | 38.7 | 100 |
| IgG and IgM and IgA | 35.8 | 100 |
| IgG and/or IgM | 96.2 | 93.3 |
| IgG and/or IgA | 95.3 | 92.6 |
| IgM and/or IgA | 80.2 | 94.0 |
| IgG and/or IgM and/or IgA | 96.2 | 90.6 |
TABLE 2.
Sensitivities of recombinant nucleocapsid protein-based ELISA for detection of IgG, IgM, and IgA in serum samples obtained at different periods after disease onset
| Days after disease onset | No. of serum samples | No. (%) positive
|
||
|---|---|---|---|---|
| IgG | IgM | IgA | ||
| 10-20 | 22 | 19 (86) | 13 (59) | 12 (55) |
| 21-30 | 64 | 61 (95) | 36 (56) | 40 (63) |
| 31-40 | 13 | 13 (100) | 8 (62) | 6 (46) |
| >40 | 7 | 7 (100) | 3 (43) | 6 (86) |
Detection of antibody to SARS-CoV nucleocapsid protein appears to be a feasible approach to serodiagnosis of SARS-CoV pneumonia. Previous studies with animal coronaviruses have shown that the nucleocapsid protein is highly immunogenic and abundantly expressed during infection and has been used for serodiagnosis of animal coronavirus infections (5, 17). In one study, recombinant nucleocapsid protein expressed in Escherichia coli with a histidine tag at the amino terminus, the fusion protein purified with a nickel column, was used as the antigen for development of an assay to detect infectious bronchitis virus-specific antibody (11). Recently, Shi et al. described the use of an antigen-capture ELISA for detection of SARS-CoV nucleocapsid protein antibodies. With clinical criteria as the gold standard, this assay had a sensitivity of 89.6% for patients on days 11 to 61 of their illness (18). However, the authors did not show the sensitivities of the assay for IgG, IgM, and IgA detection.
In the present study, with the recombinant SARS-CoV nucleocapsid protein as the antigen for ELISA-based antibody testing with an indirect immunofluorescence assay as the gold standard, detection of IgG antibody was shown to be sensitive, whereas detection of IgM or IgA antibodies had sensitivities of only 59.4 and 60.4%, respectively. The lower sensitivities of IgM and IgA detection could be due to the transient appearance of IgM and IgA antibodies in the patients, in whom the peak levels of IgM and IgA antibodies appeared at different times during the illness. This is in contrast to the high sensitivity of the IgG ELISA, which increased from 86% for patients on days 10 to 20 after disease onset to 95 and 100% for patients on days 21 to 30 and >day 30, respectively, after disease onset (Table 2).
Although six (5.7%) serum samples (all reverse transcription-PCR positive for SARS-CoV) that were seropositive by the indirect immunofluorescence assay were negative by the present recombinant nucleocapsid protein IgG ELISA, of five SARS-CoV pneumonia patients who were “seronegative” by the indirect immunofluorescence assay but reverse transcription-PCR positive for SARS-CoV, two had IgG antibody against the nucleocapsid protein of SARS-CoV as detected by the present ELISA (unpublished data). These results showed that the present recombinant nucleocapsid protein-based IgG ELISA was as sensitive as the indirect immunofluorescence assay for serodiagnosis of SARS-CoV pneumonia. In our experience, one technician who works for 8 h a day can perform the indirect immunofluorescence assay on about 50 serum samples but can perform ELISA on about 300 serum samples. Furthermore, due to the significant variation in the quality of slides used for the indirect immunofluorescence assay, the titer of the same serum sample can differ as much as eightfold when two different batches of slides are used. As ELISA is much less labor intensive, the interpretation of results is more objective, and the ELISA does not require cultivation of SARS-CoV, it could replace the indirect immunofluorescence assay as a test for detection of antibody against SARS-CoV in clinical microbiology laboratories.
Detection of specific antibodies against the SARS-CoV nucleocapsid protein by the present ELISA would be useful for serodiagnosis of SARS-CoV pneumonia, particularly during a SARS epidemic. It is well known that in the presence of possible cross-reactions, the positive predictive value of serological tests depends on the prevalence of the infection in a particular location at a particular moment. When the prevalence of an infection is low, the positive predictive value of a serological test would be low despite its high specificity. On the other hand, the positive predictive value of the serological test would be increased if the prevalence of the infection is high, such as during an epidemic and when applied in clinically compatible cases. Although the amino acid identity between the nucleocapsid protein of SARS-CoV and the nucleocapsid protein of the human coronaviruses OC43 and 229E was only 32.7 and 21.3%, respectively, and it has been shown, in one study, that there were no cross-reactions between 13 pairs of OC43 and 14 pairs of 229E human coronavirus-infected serum samples and SARS-CoV by cell culture extract-based ELISA (8), our findings revealed that about 80% of the “seropositive” serum samples from healthy blood donors and nonpneumonic hospitalized patients, as detected by the recombinant nucleocapsid protein IgG ELISA, were false-positives when the Western blot assay with the recombinant spike protein of SARS-CoV was used for confirmation (21). As a small proportion of false-positive reactions do appear with the present ELISA, all positive results, especially those obtained in the context of a low disease prevalence or from clinically incompatible cases, should be confirmed by Western blot analysis with recombinant nucleocapsid protein and spike protein as the antigens (21).
Acknowledgments
This study was supported by Research Grant Council grant HKU 7532/03M, the SARS Research Fund, the University SARS Donation Fund, University of Hong Kong, and the Research Fund for the Control of Infectious Diseases.
REFERENCES
- 1.Briese, T., C. G. Hatalski, S. Kliche, Y. S. Park, and W. I. Lipkin. 1995. Enzyme-linked immunosorbent assay for detecting antibodies to Borna disease virus-specific proteins. J. Clin. Microbiol. 33:348-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao, L., C. M. Chan, C. Lee, S. S. Y. Wong, and K. Y. Yuen. 1998. MP1 encodes an abundant and highly antigenic cell wall mannoprotein in the pathogenic fungus Penicillium marneffei. Infect. Immun. 66:966-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao, L., D. L. Chen, C. Lee, C. M. Chan, K. M. Chan, N. Vanittanakom, D. N. C. Tsang, and K. Y. Yuen. 1998. Detection of specific antibodies to an antigenic mannoprotein for diagnosis of Penicillium marneffei penicilliosis. J. Clin. Microbiol. 36:3028-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, C. M., P. C. Y. Woo, A. S. P. Leung, S. K. P. Lau, X. Y. Che, L. Cao, and K. Y. Yuen. 2002. Detection of specific antibodies to an antigenic cell wall galactomannoprotein for serodiagnosis of Aspergillus fumigatus aspergillosis. J. Clin. Microbiol. 40:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denac, H., C. Moser, J. D. Tratschin, and M. A. Hofmann. 1997. An indirect ELISA for the detection of antibodies against porcine reproductive and respiratory syndrome virus using recombinant nucleocapsid protein as antigen. J. Virol. Methods 65:169-181. [DOI] [PubMed] [Google Scholar]
- 6.Drosten, C., S. Günther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. M. Fouchier, A. Berger, A. M. Burguière, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Müller, V. Rickerts, M. Stürmer, S. Vieth, H. D. Klenk, A. D. M. E. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 7.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 8.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, and The SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 9.Lee, N., D. Hui, A. Wu, P. Chan, P. Cameron, G. M. Joynt, A. Ahuja, M. Y. Yung, C. B. Leung, K. F. To, S. F. Lui, C. C. Szeto, S. Chung, and J. J. Y. Sung. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1986-1994. [DOI] [PubMed] [Google Scholar]
- 10.Marra, M. A., S. J. M. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. N. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 11.Ndifuna, A., A. K. Waters, M. Zhou, and E. W. Collisson. 1998. Recombinant nucleocapsid protein is potentially an inexpensive, effective serodiagnostic reagent for infectious bronchitis virus. J. Virol. Methods 70:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholls, J. M., L. L. Poon, K. C. Lee, W. F. Ng, S. T. Lai, C. Y. Leung, C. M. Chu, P. K. Hui, K. L. Mak, W. Lim, K. W. Yan, K. H. Chan, N. C. Tsang, Y. Guan, K. Y. Yuen, and J. S. Peiris. 2003. Lung pathology of fatal severe acute respiratory syndrome. Lancet 361:1773-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peiris, J. S. M., C. M. Chu, V. C. C. Cheng, K. S. Chan, I. F. N. Hung, L. L. M. Poon, K. I. Law, B. S. F. Tang, T. Y. W. Hon, C. S. Chan, K. H. Chan, J. S. C. Ng, B. J. Zheng, W. L. Ng, R. W. M. Lai, Y. Guan, K. Y. Yuen, Members of the HKU/UCH, and the SARS Study Group. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia—a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peiris, J. S. M., S. T. Lai, L. L. M. Poon, Y. Guan, L. Y. C. Yam, W. Lim, J. Nicholls, W. K. S. Yee, W. W. Yan, M. T. Cheung, V. C. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, K. Y. Yuen, and Members of the SARS Study Group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Brunham, A. J. McGeer, the National Microbiology Laboratory, Canada, and the Canadian Severe Acute Respiratory Syndrome Study Team. 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348:1995-2005. [DOI] [PubMed] [Google Scholar]
- 16.Rota, P. A., M. S. Obserste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Peñaranda, B. Bankamp, K. Maher, M. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Günther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 17.Seah, J. N., L. Yu, and J. Kwang. 2000. Localization of linear B-cell epitopes on infectious bronchitis virus nucleocapsid protein. Vet. Microbiol. 75:11-16. [DOI] [PubMed] [Google Scholar]
- 18.Shi, Y., Y. Yi, P. Li, T. Kuang, L. Li, M. Dong, Q. Ma, and C. Cao. 2003. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J. Clin. Microbiol. 41:5781-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsang, K. W., P. L. Ho, G. C. Ooi, W. K. Yee, T. Wang, M. Chan-Yeung, W. K. Lam, W. H. Seto, L. Y. Yam, T. M. Cheung, P. C. Wong, B. Lam, M. S. Ip, J. Chan, K. Y. Yuen, and K. N. Lai. 2003. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1977-1985. [DOI] [PubMed] [Google Scholar]
- 20.Woo, P. C. Y., K. T. K. Chong, A. S. P. Leung, S. S. Y. Wong, S. K. P. Lau, and K. Y. Yuen. 2003. AFLMP1 encodes an antigenic cell wall protein in Aspergillus flavus. J. Clin. Microbiol. 41:845-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woo, P. C. Y., S. K. P. Lau, H. W. Tsoi, K. H. Chan, B. H. L. Wong, X. Y. Che, V. K. P. Tam, S. C. F. Tam, V. C. C. Cheng, I. F. N. Hung, S. S. Y. Wong, B. J. Zheng, Y. Guan, and K. Y. Yuen. 2004.. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet 57:281-285. [DOI] [PMC free article] [PubMed]
- 22.Yam, W. C., K. H. Chan, L. L. M. Poon, Y. Guan, K. Y. Yuen, W. H. Seto, and J. S. M. Peiris. 2003. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 41:4521-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuen, K. Y., C. M. Chan, K. M. Chan, P. C. Y. Woo, X. Y. Che, A. S. P. Leung, and L. Cao. 2001. Characterization of AFMP1, a novel target for serodiagnosis of aspergillosis. J. Clin. Microbiol. 39:3830-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]