Abstract
We used heteroduplex mobility assay (HMA) to determine the genetic variability of 118 respiratory syncytial virus (RSV) field isolates from 19 epidemics occurring in a Japanese urban area between 1980 and 2000. Nucleotides 1 to 584 of the attachment G glycoprotein gene were amplified by reverse transcription-PCR, and the PCR amplicons were analyzed by HMA by using the earliest isolate from 1980 as the reference throughout. We also performed PCR-restriction fragment length polymorphism (RFLP) analysis and phylogenetic analysis on the same nucleotide sequence. PCR-RFLP revealed 9 patterns, whereas HMA produced 31 distinct patterns. The RFLP patterns were divided into two to seven distinct HMA genotypes. Field strains with similar degrees of G gene nucleotide differences from the reference strain often showed distinct HMA types. The RSV genetic heterogeneity detected by direct sequencing of the PCR amplicon was usually identical to HMA analysis. Analysis of the molecular epidemiology of RSV subgroup A isolates obtained by HMA showed that new RSV variants emerged with each epidemic and that previously dominant variants seldom recurred in subsequent epidemics. HMA is useful in detecting genetic variants of RSV subgroup A and has some advantages over other conventional methods.
Human respiratory syncytial virus (RSV) is the most important viral pathogen causing lower respiratory tract infections in infants, immunocompromised hosts, and the elderly (15, 16, 27). Epidemics of RSV occur every winter in temperate climates, during rainy seasons, or year round in tropical regions (28). RSV can infect the same individual repeatedly and infants under 6 months of age who still possess maternal antibodies against the virus (21). These findings prompted one hypothesis that RSV infection may not produce a sufficient immune response against different strains of the virus because of the extensive genetic variability of the strains circulating in a community and worldwide.
RSV was initially found to have two distinct antigenic groups, designated A and B, by their different reactivity with monoclonal antibodies against the viral antigens (1, 20). Epidemiology studies of RSV demonstrated that both antigenic groups circulate concurrently or alternately in a community during epidemics (18, 32). Among viral surface antigens, the attachment G glycoprotein (G protein) has the greatest antigenic diversity between these two groups and among strains within each group. The G-protein variability is concentrated in its ectodomain, which contains two hypervariable regions separated by a conserved 11-amino-acid motif (4, 7, 19, 29). Recent molecular epidemiological studies suggest that many distinct RSV genotypes circulate worldwide and that similar genetic variants are clustered by time rather than by geographic location (6, 8, 17, 26).
Genetic variability of RSV has most often been studied by using restriction fragment length polymorphism (RFLP) analysis and DNA sequencing (4, 30). RFLP analysis is relatively easy to perform, although genotypes obtained by this method provide limited information. DNA sequencing, in contrast, shows the most precise genetic variability, although this method is not available to all laboratories because of costs and equipment.
Heteroduplex mobility assay (HMA) was originally elaborated to detect genetic diversity of the human immunodeficiency virus type 1 in 1994 (10, 11). Since then, it has been widely applied to other viruses for subtyping a viral species (2, 14) and for molecular epidemiology studies (13) and to detect viral genome quasispecies in a single patient (12). The advantages of HMA for molecular epidemiology viral studies are that it is technically simple, it costs less, and it is less time-consuming than the other methods mentioned above. In our study, HMA provided more details of RSV genetic diversity than did RFLP and was compatible with the results of DNA sequencing.
MATERIALS AND METHODS
Virus isolation.
A total of 193 RSV field strains were isolated from infants and children who developed respiratory tract infections and were examined at the Sapporo Medical University Hospital in Sapporo, Japan, during 19 winter seasons from 1980 to 2000. Sample collections were not performed in two successive seasons of 1995 to 1996 and 1996 to 1997. For virus isolation, nasopharyngeal swabs or aspirates were inoculated onto HEp-2 cells (obtained from Cell Resource Center for Biomedical Research Institute of Development, Aging and Cancer, Tohoku University) in 24-well plates and, when the cells showed syncytium formation, the supernatants were snap-frozen in liquid nitrogen and stored at −80°C until further analysis. RSV identification and subgrouping were performed by enzyme-linked immunosorbent assay with subgroup-specific monoclonal antibodies to the F and G proteins of the Long strain of RSV as described previously (34). A total of 118 isolates were categorized as subgroup A (61.1%) and 75 were categorized as subgroup B (38.9%). We used 118 subgroup A strains in the present study. Clinical information was available for 108 of 118 cases with subgroup A infection. Of these, 59 (54.6%) patients were <6 months old, 31 (28.7%) were 6 to <12 months old, and 18 (16.7%) were ≥12 months old: 51 (47.1%), 34 (31.7%), 19 (17.3%), and 4 (3.9%) patients were diagnosed as having bronchiolitis, tracheobronchitis, pneumonia, and upper respiratory tract infection, respectively.
RNA extraction and cDNA synthesis.
RNA extraction and cDNA synthesis were described previously (26). Briefly, the frozen RSV isolates were inoculated on HEp-2 cells in 24-well plates and total RNA was extracted by adding 0.8 ml of RNAzol B (TEL-TEST, Inc., Friendswood, Tex.) to each well when the extensive cytopathic effect was observed. For cDNA synthesis, 100 ng of the total RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and either random hexamer (Takara) or oligo(dT)12-18 primer (Invitrogen) at 37°C for 1 h.
PCR.
PCR was performed as described previously with minor modifications (26). Primers used in this PCR amplified the sequence between nucleotides (nt) 1 and 584 of the G gene of the Long strain, which covers the first hypervariable region of the G gene. The sequence of each primer was as follows: primer 1 (5′-GGGGCAAATGCAAACATGTCC-3′; nt 1 to 21) and primer 2 (5′-GGTATTCTTTTGCAGATAGC-3′; nt 565 to 584) (6, 35). PCR was carried out in a total volume of 100 μl containing a 1 μM concentration of each primer, a 250 μM deoxynucleoside triphosphate concentration (Takara), 10 mM Tris-HCl (pH 9.0), 500 mM KCl, 1% Triton X-100, 1.5 mM MgCl2, 2 U of Taq DNA polymerase (Promega, Madison, Wis.), and 2 to 4 μl of cDNA template. The mixture was heated at 94°C for 2 min, followed by 30 cycles of denaturing at 94°C for 45 s, annealing at 54 to 57°C for 45 s, and extension at 74°C for 45 s, with a final extension at 74°C for 7 min by using GeneAmp PCR System 9700 (Applied Biosystems, Foster City, Calif.).
RFLP analysis.
The procedure of RFLP was described previously (26). Briefly, 8 μl of each PCR product was digested with AluI, TaqI, and NdeII at 37°C for 90 min. The digested PCR products were analyzed on 2% agarose gels stained with ethidium bromide.
HMA.
To form heteroduplexes, the Sap/645/80 strain of RSV which was the earliest isolate from the study community was used throughout as a reference. An equal volume (4.5 μl) of reference PCR product was mixed with a sample PCR product and 1 μl of 10× annealing buffer (1 M NaCl, 100 mM Tris-HCl [pH 7.8], 20 mM EDTA) (11). The fragment mixture was denatured at 94°C for 2 min and annealed by rapid cooling at 4°C in the thermal cycler. After it cooled, the mixture was loaded onto a 5% polyacrylamide gel. Electrophoresis was performed in a vertical gel apparatus (ATTO, Tokyo, Japan) at 150 V for 3 h. DNA fragments were visualized by ethidium bromide staining.
Nucleotide sequencing and computer analysis.
The PCR products were sequenced directly by a BigDye Terminator v3.0 cycle sequence kit and an ABI Prism 310 Genetic Analyzer (Applied Biosystems). Phylogenetic analysis was done by the neighbor-joining method with CLUSTAL W in DDBJ, and a phylogenetic tree was made by TreeView (version 1.6.6).
RESULTS
Correlation between RFLP analysis and HMA.
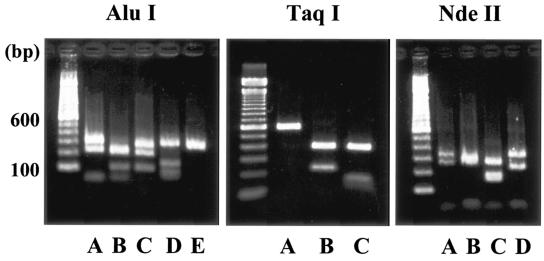
RFLP analysis of 118 isolates revealed five (designated A to E), three (A to C), and four (A to D) restriction patterns by digestion of restriction enzymes AluI, TaqI, and NdeII, respectively (Fig. 1). The combination of these restriction patterns enabled us to classify the isolates into nine patterns (we refer to these patterns as RFLP genotypes), i.e., AAA, ABA, ACB, ABC, BAA, CAA, DBA, EAA, and AAD (Table 1). However, heteroduplex formation between amplicons of the reference (Sap/645/80 strain) and the 118 isolates revealed 31 distinct homoduplex and heteroduplex mobility patterns (HMA genotypes). We assigned HMA genotypes from a to e′ in order of year of emergence (Table 2).
FIG. 1.
Restriction fragment analysis of amplified PCR products derived from cDNAs of the G protein gene between nt 1 and 584. The PCR products were digested with AluI, TaqI, and NdeII and separated by 2% agarose gel electrophoresis. Five, three, and four restriction patterns were obtained with AluI (A to E), TaqI (A to C), and NdeII (A to D), respectively. The molecular weight marker (100-bp DNA ladder) is shown on the left.
TABLE 1.
Distribution of RFLP genotypes of RSV subgroup A isolates during 19 epidemics
| Epidemic season | Total no. of strains | RFLP genotype distribution (no. of strains)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AAA | ABA | ACB | ABC | BAA | CAA | DBA | EAA | AAD | ||
| 1980-1981 | 18 | 17 | 1 | |||||||
| 1981-1982 | 2 | 2 | ||||||||
| 1982-1983 | 5 | 1 | 4 | |||||||
| 1983-1984 | 3 | 3 | ||||||||
| 1984-1985 | 4 | 4 | ||||||||
| 1985-1986 | 5 | 5 | ||||||||
| 1986-1987 | 15 | 2 | 11 | 2 | ||||||
| 1987-1988 | 11 | 10 | 1 | |||||||
| 1988-1989 | 10 | 2 | 8 | |||||||
| 1989-1990 | 4 | 1 | 1 | 2 | ||||||
| 1990-1991 | 16 | 15 | 1 | |||||||
| 1991-1992 | 9 | 8 | 1 | |||||||
| 1992-1993 | 1 | 1 | ||||||||
| 1993-1994 | 1 | 1 | ||||||||
| 1994-1995 | 4 | 1 | 3 | |||||||
| 1997-1998 | 5 | 1 | 1 | 2 | 1 | |||||
| 1998-1999 | 1 | 1 | ||||||||
| 1999-2000 | 3 | 1 | 2 | |||||||
| 2000-2001 | 1 | 1 | ||||||||
| Total | 118 | 29 | 15 | 30 | 2 | 28 | 2 | 5 | 2 | 5 |
TABLE 2.
Distribution of HMA genotypes of RSV subgroup A isolates during 19 epidemics
| Epidemic season | Total no. of strains | Distribution of HMA genotypes (no. of strains)
|
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | e | f | g | h | i | j | k | l | m | n | o | p | q | r | s | t | u | v | w | x | y | z | a′ | b′ | c′ | d′ | e′ | ||
| 1980-1981 | 18 | 17 | 1 | |||||||||||||||||||||||||||||
| 1981-1982 | 2 | 2 | ||||||||||||||||||||||||||||||
| 1982-1983 | 5 | 1 | 3 | 1 | ||||||||||||||||||||||||||||
| 1983-1984 | 3 | 2 | 1 | |||||||||||||||||||||||||||||
| 1984-1985 | 4 | 1 | 2 | 1 | ||||||||||||||||||||||||||||
| 1985-1986 | 5 | 5 | ||||||||||||||||||||||||||||||
| 1986-1987 | 15 | 1 | 8 | 1 | 1 | 1 | 3 | |||||||||||||||||||||||||
| 1987-1988 | 11 | 10 | 1 | |||||||||||||||||||||||||||||
| 1988-1989 | 10 | 6 | 2 | 2 | ||||||||||||||||||||||||||||
| 1989-1990 | 4 | 1 | 1 | 1 | 1 | |||||||||||||||||||||||||||
| 1990-1991 | 16 | 13 | 3 | |||||||||||||||||||||||||||||
| 1991-1992 | 9 | 5 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||
| 1992-1993 | 1 | 1 | ||||||||||||||||||||||||||||||
| 1993-1994 | 1 | 1 | ||||||||||||||||||||||||||||||
| 1994-1995 | 4 | 2 | 1 | 1 | ||||||||||||||||||||||||||||
| 1997-1998 | 5 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||
| 1998-1999 | 1 | 1 | ||||||||||||||||||||||||||||||
| 1999-2000 | 3 | 1 | 2 | |||||||||||||||||||||||||||||
| 2000-2001 | 1 | 1 | ||||||||||||||||||||||||||||||
| Total | 118 | 19 | 1 | 1 | 6 | 1 | 4 | 1 | 5 | 25 | 1 | 1 | 1 | 5 | 20 | 2 | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 2 | 1 | 2 | 3 |
The correlation between RFLP genotypes and HMA genotypes is summarized in Table 3. RFLP genotypes ABA and BAA were further divided into seven HMA genotypes, and RFLP genotype AAA was divided into six HMA genotypes, and RFLP genotype DBA was divided into three HMA genotypes. The other five RFLP genotypes were divided into two distinct HMA genotypes. Each RFLP genotype contained at least two HMA genotypes; however, no HMA genotype belonged to more than one RFLP genotype.
TABLE 3.
Comparison of 9 RFLP and 31 HMA genotypes of RSV subgroup A isolates during 19 epidemics
| RFLP genotypes (no. of isolates) | HMA genotypes (no. of isolates) |
|---|---|
| AAA (29) | a (19), e (1), h (5), o (2), q (1), b′ (1) |
| ABA (15) | b (1), c (1), d (6), f (4), g (1), l (1), v (1) |
| ACB (30) | i (25), m (5) |
| ABC (2) | j (1), k (1) |
| AAD (5) | d′ (2), e′ (3) |
| BAA (28) | n (19), p (1), r (4), t (1), u (1), y (1), a′ (1) |
| CAA (2) | n (1), s (1) |
| DBA (5) | w (1), x (3), z (1) |
| EAA (2) | b′ (1), c′ (1) |
We could not find any significant difference in clinical categorization of the illnesses between children infected with each genotype.
Genotype distribution determined by RFLP analysis and HMA.
One to four different RFLP genotypes emerged in each epidemic season (Table 1). Six genotypes appeared during three consecutive seasons, and then the genotype changed to another. Genotypes AAA, ABA, and BAA reemerged several years after the first dominance disappeared. Genotype AAA, which is the earliest genotype in the present study, was observed intermittently over a 20-year-period. Likewise, genotypes ABA and BAA also circulated intermittently for 13 and 11 years, respectively. The other six genotypes were detected in only one to four consecutive seasons, and these genotypes never reemerged.
One to six different HMA types occurred in each epidemic season, as was seen in the RFLP genotype distribution (Table 2). However, one HMA genotype lasted only one to three epidemic seasons and was then replaced by another genotype. The maximum duration of circulation of one genotype was 5 years, and individual genotypes usually did not reemerge after the initial circulation period in this observation, although genotype d′ surfaced again exceptionally after a 2-year break.
Correlation between HMA and nucleotide sequencing.
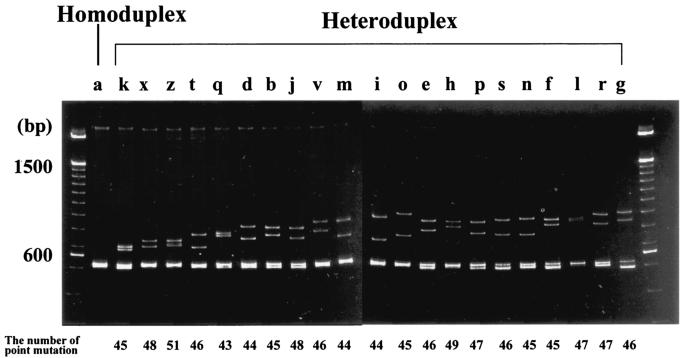
We looked for any correlation between the mobility of heteroduplexes and the degree of nucleotide differences in the corresponding sequence. We selected 21 isolates belonging to different HMA types during the 15 epidemic seasons from 1980 to 1995. Their nucleotide sequences (nt 1 to 584) were compared to that of the reference strain (Sap/645/80) which belongs to genotype a. These strains were aligned in a polyacrylamide gel in order of faster (left) to slower (right) migration (Fig. 2). The nucleotide changes of these isolates compared to those of the reference strain were all within the range of 43 to 51 nt. Therefore, in the present study, a correlation between the mobility of heteroduplexes and the number of point mutations could not be fully evaluated. However, it appeared that strains with a similar amount of G-gene nucleotide differences from the reference strain were often distinct HMA types. For example, the strains belonging to distinct HMA genotypes k, b, o, n, and f had 45- nt changes compared to a reference strain.
FIG. 2.
Heteroduplex mobility assay with amplicons of cDNAs of the G gene (nt 1 to 584) of 21 isolates during 15 epidemic seasons from 1980 to 1995. Heteroduplexes with 21 distinct mobility patterns were aligned from faster (left) and slower (right) migrations, and alphabetical assignment was provided as shown in Table 2. The number of nucleotide changes compared to those of the reference strain are indicated below. The molecular weight marker (100-bp DNA ladder) is shown on both sides.
The nucleotide homology of isolates that belong to the single HMA genotype was also investigated. The nucleotide sequences of several isolates belonging to genotypes f, i, and r were aligned in each genotype, and the alignment revealed 98 to 100% homology (data not shown).
RFLP, HMA, and phylogenetic analysis.
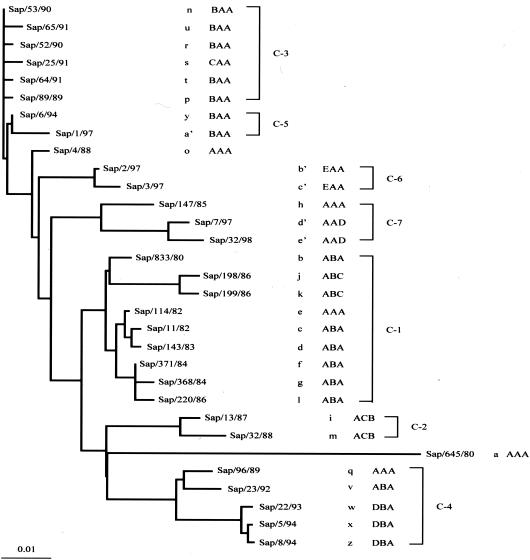
We selected one isolate from each of 31 HMA genotypes and performed phylogenetic analysis using whole sequences from nt 1 to 584. HMA and RFLP genotypes of each strains were also indicated. The phylogenetic tree showed seven clusters (clusters 1 to 7), and each cluster consisted of two to nine isolates that have close isolation times (Fig. 3). Genetic heterogeneity within RSV field strains detected by HMA analysis was confirmed phylogenetically. That is, all 31 strains with individual HMA genotypes were discriminated from each other by nucleotide sequence analysis. Strains within the same branches usually exhibited similar but distinct HMA types (types f, l, and g in C-1; types m and i in C-2; types p, s, and n in C-3; and types x and z in C-4), although there were some exceptions, i.e., types k and j in C-1 and types q and v in C-4. On the other hand, strains belonging to distant clusters sometimes presented similar, although distinct HMA patterns, e.g., types e, h, and p in C-1, C-7, and C-3, respectively (Fig. 2 and 3).
FIG. 3.
Phylogenetic tree of 31 isolates across all HMA genotypes based on sequences of the first variable region of the G gene of RSV. Names of HMA genotypes (a to e′) and RFLP genotypes (three capital letters) are indicated next to the strain names. The phylogenetic clusters (C-1 to C-7) are also indicated.
DISCUSSION
In the present study, we applied HMA to the analysis of G-gene variability of RSV for the first time. HMA is based on the observation that the structural deformations in double-stranded DNA that result from mismatches and nucleotide insertions or deletions reduce the electrophoretic mobility of these fragments in polyacrylamide gels. These mismatches are thought to result from “bubbles” in the heteroduplex that retard the mobility of the fragment through the polyacrylamide matrix. To be detected in ethidium bromide-stained gels, mismatches must amount to at least 1 to 2% of the base pairs in the heteroduplex, whereas 3-nt gaps can be detected in molecules at least as large as 3 kb (11). HMA was originally introduced to detect genetic diversity of the human immunodeficiency virus type 1 (10, 11) and subsequently used to analyze other RNA viruses including lentivirus, hepatitis C and G virus, norovirus, influenza virus, measles virus, and poliovirus (3).
Trincado et al. (31) used HMA for genotyping human cytomegalovirus isolates and compared this method to RFLP, single-stranded conformation polymorphism, and DNA sequencing analyses. These authors concluded that HMA was the most accurate and concise of the four methods used to genotype human cytomegalovirus isolates.
Genetic variability of the G gene of RSV has been investigated during the past 10 years in both developed and developing countries by using RFLP and nucleotide sequencing (5, 8, 9, 22-25, 30, 34). These investigations revealed that multiple genotypes cocirculated in each epidemic and that the dominant genotype changes year by year. The present study demonstrates that the results of HMA for the G-gene variability correlate well with those of RFLP analysis and, moreover, that HMA can detect more detailed genotype manifestations. We and others (8) found that some RFLP genotypes emerged for very long periods. In a study of the molecular epidemiology of RSV in Korea, one genotype (GP-A2) of subgroup A was detected over eight consecutive epidemics, and one subgroup B genotype (GP-B1) was also found for six epidemics (8). In our study, the genotype AAA that was epidemic in the 1980 and 1981 season emerged repeatedly over 20 years. However, the genotype AAA strains isolated after these two seasons were never assigned to the same HMA genotype as that of the former AAA strains. The same observation was found in RFLP genotype ABA, which had appeared over 13 years. The genotype ABA corresponded to seven HMA genotypes and two of them, genotypes d and f, appeared for only 3 and 4 years, respectively. All strains with different HMA genotypes were confirmed as discrete strains by nucleotide sequence analysis. Our findings show that HMA is precise and can detect more subtle genetic changes in RSV isolates than can RFLP. In this regard, HMA is more discriminatory than RFLP. Our observations also indicate that in RSV epidemics new genotypes are continuously emerging, and these new genotypes prevail for only a limited period, at most 4 to 5 years.
We confirmed that RSV field strains with similar sequence divergence from the reference strain could often display distinct HMA patterns. However, it also appeared that the locations of nucleotide changes in each strain compared to the reference strain varied, although the total number of mutations was the same or very similar between the strains. Thus, our results suggest that the mobility of heteroduplexes is not only based on the number of point mutations but also correlates with the location of the mismatch, i.e., structural differences in heteroduplexes reduce the electrophoretic mobility. Therefore, HMA may be more sensitive with respect to structural deformations in double-stranded DNA than nucleotide sequence. The nucleotide sequences of isolates in the same HMA genotype had at least 98% homology. This was comparable to the previous observation that a sequence divergence of more than 1 to 2% was necessary for detectable heteroduplex retardation (3, 10).
The HMA had some correlation with phylogenetic analysis of the G gene by nucleotide sequencing. In the present study, the phylogenetic tree showed seven clusters, and each cluster consisted of two to nine HMA genotypes. The strains within each branch represent similar but distinct HMA types. However, the degree of genetic divergence evident in the phylogenetic analysis as distinct clusters could not be represented precisely by HMA analysis. Therefore, the HMA analysis in the present study could distinguish genetic divergence among strains with a higher sensitivity, but it might not be possible to express these quantitatively.
In a study of the differentiation of influenza A viruses from different animal species, such as avian and swine viruses, there was an excellent correlation between HMA results and nucleotide divergence, as determined by direct sequencing and phylogenetic analysis (14). In our study, the RSV field strains with various sequence divergence from reference DNA could not be evaluated; however, we confirmed that HMA could differentiate between strains with a similar sequence divergence from reference DNA.
In conclusion, it appears that HMA is useful for elucidating the genetic variability of RSV field strains. This method can demonstrate a clear distribution of many RSV genotypes and provides more precise molecular epidemiology than RFLP analysis. In addition, the HMA genotypes correlated to some extent with the clustering of strains in a phylogenetic tree.
HMA is technically easy and inexpensive compared to other conventional methods for the assessment of genetic diversity of viral strains. In particular, in developing countries, the molecular epidemiology of RSV epidemics is becoming more important, but equipment for DNA sequencing is not always available in many laboratories. For this reason, HMA is an alternative and important method for identifying genetic variants and investigating the molecular epidemiology of RSV infection. Further prospective studies are needed to support and confirm these observations.
Acknowledgments
We thank Peter M. Olley (University of Alberta) for language advice.
REFERENCES
- 1.Anderson, L. J., J. C. Hierhorzer, C. Tsou, R. M. Hendry, B. F. Fernie, Y. Stone, and K. McIntosh. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151:626-633. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann, M. H., E. L. Delwart, E. G. Shpaer, P. Lingenfelter, R. Singal, and J. I. Mullins. 1994. Rapid genetic characterization of HIV type 1 strains from World Health Organization-sponsored vaccine evaluation sites using a heteroduplex mobility assay. AIDS Res. Hum. Retrovir. 10:1345-1353. [DOI] [PubMed] [Google Scholar]
- 3.Barlow, K. L., J. Green, and J. P. Clewley. 2000. Viral genome characterization by the heteroduplex mobility and heteroduplex tracking assays. Rev. Med. Virol. 10:321-335. [DOI] [PubMed] [Google Scholar]
- 4.Cane, P. A., D. A. Matthews, and C. R. Pringle. 1991. Identification of variable domains of the attachment (G) protein of subgroup A respiratory syncytial viruses. J. Gen. Virol. 72:2091-2096. [DOI] [PubMed] [Google Scholar]
- 5.Cane, P. A., and C. R. Pringle. 1992. Molecular epidemiology of respiratory syncytial virus: rapid identification of subgroup A lineages. J. Virol. Methods 40:297-306. [DOI] [PubMed] [Google Scholar]
- 6.Cane, P. A., and C. R. Pringle. 1995. Evolution of subgroup A respiratory syncytial virus: evidence for progressive accumulation of amino acid changes in the attachment protein. J. Virol. 69:2918-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cane, P. A. 1997. Analysis of linear epitopes recognized by the primary human antibody response to a variable region of the attachment (G) protein of respiratory syncytial virus. J. Med. Virol. 51:297-304. [DOI] [PubMed] [Google Scholar]
- 8.Choi, E. H., and H. J. Lee. 2000. Genetic diversity and molecular epidemiology of the G protein of subgroup A and B of respiratory syncytial viruses isolated over 9 consecutive epidemics in Korea. J. Infect. Dis. 181:1547-1556. [DOI] [PubMed] [Google Scholar]
- 9.Coggins, W. B., E. J. Lefkowitz, and W. M. Sullender. 1998. Genetic variability among group A and group B respiratory syncytial viruses in a children's hospital. J. Clin. Microbiol. 36:3552-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 11.Delwart. E. L., E. G. Shpaer, and J. I. Mullins. 1995. Heteroduplex mobility assay for phylogenetic analysis, p. 154-160, In D. H. Gelfand, J. J. Sninsky, and M. Innis (ed.), PCR strategies. Academic Press, Inc., San Diego, Calif.
- 12.Delwart, E. L., H. Pan, H. W. Sheppard, D. Wolpert, A. U. Neumann, B. Korber, and J. I. Mullins. 1997. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J. Virol. 71:7498-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz, R. S., C. F. De Oliveira, R. Pardini, E. Operskalski, A. J. Mayer, and M. P. Busch. 1999. HIV type 1 tat gene heteroduplex mobility assay as a tool to establish epidemiologic relationships among HIV type 1-infected individuals. AIDS Res. Hum. Retrovir. 15:1151-1156. [DOI] [PubMed] [Google Scholar]
- 14.Ellis, J. S., and M. C. Zambon. 2001. Combined PCR-heteroduplex mobility assay for detection and differentiation of influenza A viruses from different animal species. J. Clin. Microbiol. 39:4097-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falsey, A. R., C. K. Cunningham, W. H. Barker, R. W. Kouides, J. B. Yuen, M. Menegus, L. B. Weiner, C. A. Bonville, and R. F. Betts. 1995. Respiratory syncytial virus and influenza A infections in the hospitalized elderly. J. Infect. Dis. 172:389-394. [DOI] [PubMed] [Google Scholar]
- 16.Fouillard, L., L. Mouthon, J. P. Laporte, F. Isnard, J. Stachowiak, M. Aoudjhane, J. C. Lucet, M. Wolf, F. Bricourt, and L. Douay. 1992. Severe respiratory syncytial virus pneumonia after autologous bone marrow transplantation: a report of three cases and review. Bone Marrow Transplant. 9:97-100. [PubMed] [Google Scholar]
- 17.Garcia, O., M. Martin, J. Dopazo, J. Arbiza, S. Frabasile, J. Russi, M. Hortal, P. Perez-Brena, I. Martinez, B. Garcia-Barreno, and J. A. Melero. 1994. Evolutionary pattern of human respiratory syncytial virus (subgroup A): co-circulating lineages and correlation of genetic and antigenetic changes in the G glycoprotein. J. Virol. 68:5448-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendry, R. M., A. L. Talis, E. Godfray, L. J. Anderson, B. F. Fernie, and K. McIntosh. 1986. Concurrent circulation of antigenically distinct strains of respiratory syncytial virus during community outbreaks. J. Infect. Dis. 153:291-297. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. USA 84:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mufson, M. A., C. Orvell, B. Rafnar, and E. Norrby. 1985. Two distinct subtypes of human respiratory syncytial virus. J. Gen. Virol. 66:2111-2124. [DOI] [PubMed] [Google Scholar]
- 21.Mufson, M. A., R. B. Belshe, C. Orvell, and E. Norrby. 1987. Subgroup characteristics of respiratory syncytial virus strains recovered from children with two consecutive infections. J. Clin. Microbiol. 25:1536-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peret, T. C. T., C. B. Hall, K. C. Schnabel, J. A. Golub, and L. J. Anderson. 1998. Circulation pattern of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J. Gen. Virol. 79:2221-2229. [DOI] [PubMed] [Google Scholar]
- 23.Peret, T. C. T., C. B. Hall, G. W. Hammond, P. A. Piedra, G. A. Storch, W. M. Sullender, C. Tsou, and L. J. Anderson. 2000. Circulation pattern of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J. Infect. Dis. 181:1891-1896. [DOI] [PubMed] [Google Scholar]
- 24.Rajala, M. S., W. M. Sullender, A. K. Prasad, L. Dar, and S. Broor. 2003. Genetic variability among group A and B respiratory syncytial virus isolated from a large referral hospital in New Delhi, India. J. Clin. Microbiol. 41:2311-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roca, A., M.-P. Loscertales, L. Quinto, P. Perez-Brena, N. Vaz, P.-L. Alonso, and J.-C. Saiz. 2001. Genetic variability among group A and B respiratory syncytial viruses in Mozambique: identification of a new cluster of group B isolates. J. Gen. Virol. 82:103-111. [DOI] [PubMed] [Google Scholar]
- 26.Seki, K., H. Tsutsumi, M. Ohsaki, H. Kamasaki, and S. Chiba. 2001. Genetic variability of respiratory syncytial virus subgroup A strain in 15 successive epidemics in one city. J. Med. Virol. 64:374-380. [DOI] [PubMed] [Google Scholar]
- 27.Simoes, E. A. F., and X. Carbonell-Estrany. 2003. Impact of severe diseases caused by respiratory syncytial virus in children living in developed countries. Pediatr. Infect. Dis. J. 22:S13-S20. [DOI] [PubMed] [Google Scholar]
- 28.Stensballe, L. G., J. K. Devasundaram, and E. A. F. Simoes. 2003. Respiratory syncytial virus epidemics: the ups and downs of a seasonal virus. Pediatr. Infect. Dis. J. 22:S21-S32. [DOI] [PubMed] [Google Scholar]
- 29.Sullender, W. M., M. A. Mufson, L. J. Anderson, and G. W. Wertz. 1991. Genetic diversity of the attachment protein of subgroup B respiratory syncytial viruses. J. Virol. 65:5425-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullender, W. M., L. Sun, and L. J. Anderson. 1993. Analysis of respiratory syncytial virus genetic variability with amplified cDNAs. J. Clin. Microbiol. 31:1224-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trincado, D. E., G. M. Scott, P. A. White, C. Hunt, L. Rasmussen, and W. D. Rawlinson. 2000. Human cytomegalovirus strains associated with congenital and perinatal infections. J. Med. Virol. 61:481-487. [DOI] [PubMed] [Google Scholar]
- 32.Tsutsumi, H., M. Onuma, K. Suga, T. Honjo, Y. Chiba, S. Chiba, and P. L. Ogra. 1988. Occurrence of respiratory syncytial virus subgroup A and B strains in Japan, 1980 to 1987. J. Clin. Microbiol. 26:1171-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsutsumi, H. K. Nagai, K. Suga, and S. Chiba. 1989. Antigenetic variation of human RSV strains isolated in Japan. J. Med. Virol. 27:124-130. [DOI] [PubMed] [Google Scholar]
- 34.Venter, M., S. A. Madhi, C. T. Tiemessen, and B. D. Schoub. 2001. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J. Gen. Virol. 82:2117-2124. [DOI] [PubMed] [Google Scholar]
- 35.Wertz, G. W., P. L. Collins, Y. Huang, C. Gruber, S. Levine, and A. L. Ball. 1985. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc. Natl. Acad. Sci. USA 82:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]