Abstract
Delta opioid receptors are implicated in a variety of psychiatric and neurological disorders. These receptors play a key role in the reinforcing properties of drugs of abuse, and polymorphisms in OPRD1 (the gene encoding delta opioid receptors) are associated with drug addiction. Delta opioid receptors are also involved in protecting neurons against hypoxic and ischemic stress. Here, we first examined a large sample of 738 elderly participants with neuroimaging and genetic data from the Alzheimer's Disease Neuroimaging Initiative. We hypothesized that common variants in OPRD1 would be associated with differences in brain structure, particularly in regions relevant to addictive and neurodegenerative disorders. One very common variant (rs678849) predicted differences in regional brain volumes. We replicated the association of this single‐nucleotide polymorphism with regional tissue volumes in a large sample of young participants in the Queensland Twin Imaging study. Although the same allele was associated with reduced volumes in both cohorts, the brain regions affected differed between the two samples. In healthy elderly, exploratory analyses suggested that the genotype associated with reduced brain volumes in both cohorts may also predict cerebrospinal fluid levels of neurodegenerative biomarkers, but this requires confirmation. If opiate receptor genetic variants are related to individual differences in brain structure, genotyping of these variants may be helpful when designing clinical trials targeting delta opioid receptors to treat neurological disorders. Hum Brain Mapp 35:1226–1236, 2014. © 2013 Wiley Periodicals, Inc.
Keywords: neuroimaging, genetics, neurodegeneration, drug addiction, opiates
INTRODUCTION
Endogenous opioid peptides are part of the neurohormonal system. They function as hormones, secreted into the blood, and delivered to several distant target tissues. They also act as neuromodulators, influencing how other transmitters act in the central nervous system [Janecka et al., 2004]. Opioid receptors have four major subtypes: mu, delta, kappa, and nociceptin. These G protein‐coupled receptors are encoded by four distinct genes and are stimulated by various endogenous opioid peptides, as well as by opiate drugs.
Opioids may mediate reward by disinhibiting dopamine neurons in the ventral tegmental area, resulting in increased dopamine release in the nucleus accumbens [Volkow et al., 2012a]. Mu opioid receptors are primary modulators of the rewarding effects of drugs of abuse [Volkow et al., 2012a]. Over the last decade, many animal studies have implicated delta opioid receptors in reward [Hirose et al., 2005], opioid tolerance [Zhang et al., 2011] and dependence [Wu et al., 2011], and in several aspects of ethanol addiction, including heavy consumption [Nielsen et al., 2001] and escalation of intake contributing to abuse [Barson et al., 2010]. Chronic ethanol exposure changes delta opioid receptor expression [Van Rijn et al., 2010], and pharmacological manipulations of these receptors modulate ethanol‐related behaviors such as place preference [Van Rijn et al., 2012] and withdrawal‐induced anxiety [Van Rijn et al., 2010]. In addition, several single‐nucleotide polymorphisms (SNPs) in the OPRD1 gene, which encodes delta receptors, contribute to the risk for substance abuse disorders [Levran et al., 2012].
Delta receptors are also involved in neurodegeneration. They promote the processing of amyloid beta (Aβ) precursor protein. Knockdown or antagonization of delta receptors ameliorates Aβ‐related pathology and Aβ‐dependent behavioral deficits in mice [Teng et al., 2012]. One OPRD1 polymorphism (rs1042114) is strongly associated with opioid dependence [Zhang et al., 2004], and changes the evolutionarily conserved phenylalanine to cysteine in the N‐terminus of the receptor. In human cell lines, this change in the amino acid sequence of the delta receptors leads to a detrimental accumulation of one type of Aβ precursor protein, which is degraded less efficiently [Sarajarvi et al., 2004]. Thus, the G allele, which confers a predisposition to addiction [Zhang et al., 2004], also seems to promote neurodegeneration [Sarajarvi et al., 2004].
Commonly carried variants in the OPRD1 gene have been associated with a variety of psychiatric and neurological phenotypes, but to our knowledge, no investigation has assessed whether they influence brain structure. Here, we hypothesized that common variants in OPRD1 might be associated with structural differences in limbic circuits relevant to addiction and in frontotemporal regions vulnerable to neurodegeneration. We tested this hypothesis in a large sample of elderly subjects; to test the generalizability of the finding, we also assessed a replication sample of young adult twins, greatly reducing the risk of false positive findings.
After discovering that the rs678849 locus was associated with local brain volumes in various regions, we performed exploratory tests in the elderly cohort, to assess its effects on other neurodegenerative markers in the cerebrospinal fluid (CSF). We hypothesized that the C allele, which was associated with smaller regional brain volumes, might also be associated with CSF markers of neurodegeneration, including measures of amyloid and tau proteins that abnormally accumulate in Alzheimer's disease.
METHODS
Subjects
We analyzed two independent samples with neuroimaging and genetic data: the Alzheimer's Disease Neuroimaging Initiative (ADNI) and the Queensland Twin Imaging (QTIM) study. ADNI participants were recruited from 58 sites across North America. The study was conducted according to the Good Clinical Practice guidelines, the Declaration of Helsinki, and U.S. 21 CFR Part 50 (Protection of Human Subjects), and Part 56 (Institutional Review Boards). Written informed consent was obtained from all participants before protocol‐specific procedures were performed. All ADNI data are publicly available (at http://www.loni.ucla.edu/ADNI/). To avoid the known effects of population stratification on genetic analysis [Lander and Schork, 1994], we only included non‐Hispanic Caucasian subjects identified by self‐report and confirmed by multidimensional scaling analysis [Stein et al., 2002]. The ADNI cohort included multiple diagnostic groups: patients with Alzheimer's disease (AD), subjects with mild cognitive impairment (MCI), and healthy elderly (cognitively normal) participants. We used the full ADNI sample, including data from all diagnostic groups, due to the very limited power available when performing any genetic association analysis. Typical effects of candidate genes on the phenotype are often around 1% of the mean value per allele [Stein et al., 2010] so we often pick up effects only when ADNI's full sample is included. Effect sizes for individual genetic variants on brain structure in particular are expected to be small, so the neuroimaging genetics analysis would be underpowered if we further subdivided the sample. Our final analysis comprised 738 individuals (average age ± s.d. = 75.52 ± 6.78 years; 438 men/300 women) including 178 AD, 354 MCI, and 206 healthy participants (Table 1).
Table 1.
Demographic and genetic data for the ADNI cohort (after restricting the genetic analysis to Caucasians)
| ADNI | Males | Females | Total |
|---|---|---|---|
| Total | 438 | 300 | 738 |
| Healthy elderly | 111 | 95 | 206 (28%) |
| MCI | 229 | 125 | 354 (48%) |
| AD | 98 | 80 | 178 (24%) |
| rs678849 0 C alleles | 126 | 82 | 208 (28%) |
| rs678849 1 C alleles | 216 | 157 | 373 (51%) |
| rs678849 2 C alleles | 96 | 61 | 157 (21%) |
| Mean age (±s.d.) | 75.90 (±6.76) | 74.98 (±6.78) | 75.52 (±6.78) |
The QTIM sample has been described in detail elsewhere [Blokland et al., 2011; De Zubicaray et al., 2008; Wright and Martin, 2012b]. Briefly, the QTIM study is a large 5‐year Australian research project to examine genetic effects on brain structure and function. Subjects consist of healthy, young adult monozygotic (MZ) and dizygotic (DZ) twins and their singleton siblings. All twins were screened to exclude pathology known to affect brain structure. No twins reported a history of significant head injury, neurological or psychiatric illness, substance abuse or dependence, or had a first‐degree relative with a psychiatric disorder. The cohort is predominantly Caucasian; 10 individuals were excluded as ancestry outliers, identified by principal component analysis of genome‐wide SNP data (outliers were at least six standard deviations from the mean along the first two principal component axes). Written informed consent was obtained from all subjects before performing protocol‐specific procedures. Our final analysis included 578 individuals; age 23.80 ± 2.24 years, including 224 men and 354 women (Table 2).
Table 2.
Demographic and genetic data in the QTIM cohort
| QTIM | Males | Females | Total |
|---|---|---|---|
| Total | 224 | 354 | 578 |
| rs678849 0 C alleles | 55 | 102 | 157 (27%) |
| rs678849 1 C alleles | 126 | 171 | 297 (51%) |
| rs678849 2 C alleles | 43 | 81 | 124 (22%) |
| Mean age (±s.d.) | 23.54 (±2.22) | 23.96 (±2.25) | 23.80 (±2.24) |
Genotyping and SNP Selection
Both the ADNI and QTIM samples were genotyped using the Illumina 610‐Quad BeadChip (San Diego, CA). Using the Tagger algorithm in Haploview [v4.2; Barrett et al., 2005], tag SNPs were selected to provide coverage of the OPRD1 gene (Fig. 1). These five SNPs spanning OPRD1 were analyzed for association with brain volumes in ADNI. Only the top SNP hit (at the rs678849 locus) was subsequently analyzed for association with CSF biomarkers in a subset of ADNI participants (who volunteered for CSF assessments) and analyzed for association with brain volumes in the QTIM sample for replication purposes.
Figure 1.
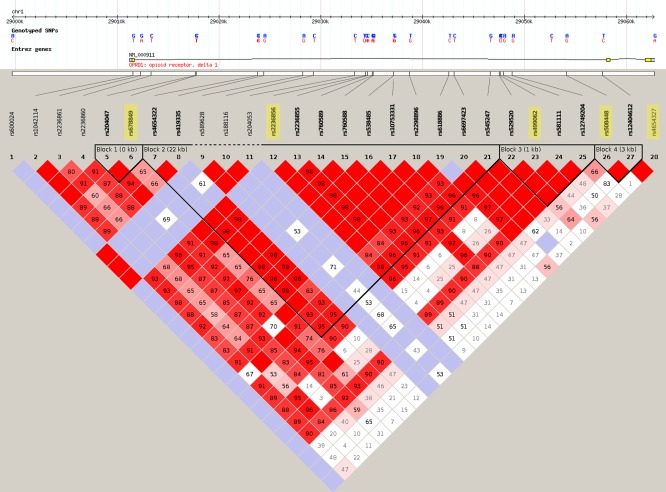
Pattern of linkage disequilibrium (LD) across all OPRD1 SNPs and location of tag SNPs among other OPRD1 SNPs genotyped in the HapMap European (CEU) sample. The pattern of LD across all OPRD1 SNPs genotyped in the HapMap sample is illustrated here. The SNPs are shown on a scale representing their position on chromosome 1. The tag SNPs selected with Haploview (v4.2) are highlighted in yellow. This image is modified from the HapMap website (http://www.hapmap.org/). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Image Acquisition, Correction and Preprocessing
ADNI subjects were scanned with a standardized MRI protocol developed for this cohort [Jack et al., 2008; Leow et al., 2006]. Briefly, high‐resolution structural brain MRI scans were acquired at 58 sites using 1.5 T MRI scanners. A sagittal 3D MP‐RAGE sequence was used, optimized for consistency across sites [Jack et al., 2008; time of repetition (TR)/time of echo (TE) = 2400/1000 ms; flip angle = 8°; field of view = 24 cm; final reconstructed voxel resolution = 0.9375 mm × 0.9375 mm × 1.2 mm]. Image corrections were applied using a processing pipeline at the Mayo Clinic, consisting of: (1) a procedure termed GradWarp to correct geometric distortion due to gradient nonlinearity [Jovicich et al., 2006], (2) a “B1‐correction”, to adjust for image intensity inhomogeneity due to B1 nonuniformity using calibration scans [Jack et al., 2008], (3) “N3” bias field correction, for reducing residual intensity inhomogeneity [Sled et al., 2012], and (4) geometrical scaling, according to a phantom scan acquired for each subject [Jack et al., 2008], to adjust for scanner‐ and session‐specific calibration errors. To adjust for global differences in brain positioning and scale, all subjects' scans were linearly registered to the stereotaxic space defined by the International Consortium for Brain Mapping [ICBM‐53; Mazziotta et al., 2000] using a 9‐parameter (9P) transformation [three translations, three rotations, three scales; Collins et al., 1994]. For ADNI, we used standard trilinear interpolation and resampled the resulting aligned scans to have 1 mm isotropic voxels.
In the QTIM cohort, all subjects were imaged on the same scanner using structural whole brain MRI at 4 T (Bruker Medspec). T1‐weighted images were acquired with an inversion recovery rapid gradient echo sequence (inversion time/TR/TE = 700/1, 500/3.35 ms; flip angle = 8°; slice thickness = 0.9 mm, with a 256 × 256 acquisition matrix). All images were initially corrected for intensity nonuniformity [Sled et al., 2012] within an automatically delineated mask of the brain [Smith, 1998]. Images were then spatially normalized to the ICBM‐152 template [Mazziotta et al., 2000] using a 9P transformation that rotated and scaled each image to minimize a normalized mutual information cost function [Jenkinson et al., 2002]. For the QTIM cohort, we did not use standard trilinear interpolation. Instead, images were then resampled in the space of the template using sinc interpolation to yield 1 mm3 isotropic voxels. In this way, each brain was globally matched in size and mutually aligned, but local differences in shape and size remained intact. Subjects' brain images were not skull‐stripped during preprocessing for either cohort.
Minimal Deformation Target and Tensor‐Based Morphometry
For each cohort, we created a minimal deformation target (MDT), which serves as an unbiased average template image for automated image registration, and to reduce statistical bias. For the ADNI sample, an MDT was created from the MRI scans of 40 randomly selected healthy elderly subjects, as detailed elsewhere [Hua et al., 2008a, 2008b]. For the QTIM cohort, an MDT was created from the T1‐weighted images of 40 (20 female/20 male) randomly selected, nonrelated participants [Gutman et al., 2010]. The MDT image was calculated from a geometric composite averaging of source moves from the sKLMI to an average affine‐registered target image following the procedure detailed elsewhere [Hua et al., 2008a, 2008b].
To quantify 3D patterns of volumetric tissue variations, all individual T1‐weighted images (N = 1,316) were nonlinearly aligned to their cohort‐specific template with an inverse‐consistent 3D elastic warping technique using a mutual information cost function [Leow et al., 2005]. For each subject, a separate Jacobian matrix field was derived from the gradients of the deformation field that aligned that individual brain to the MDT template. The determinant of the local Jacobian matrix was derived from the forward deformation field to characterize local volume differences. Color‐coded Jacobian determinants were used to illustrate regions of volume expansion, that is, those with det J(r) > 1, or contraction, that is, det J(r) < 1 [Chung et al., 2001; Freeborough and Fox, 1998; Riddle et al., 2011; Thompson et al., 2009] relative to the group template. As all images within each cohort were registered to the same study‐specific template, these Jacobian maps shared common anatomical coordinates, defined by the normal template. Individual Jacobian maps were retained for further statistical analyses.
Regression of Structural Brain Differences with the Tag SNPs
In the ADNI cohort, we investigated the effects of five tag SNPs on local brain volumes. For each SNP, we used univariate linear regression to associate the number of minor alleles (0, 1, or 2) with the Jacobian values (describing the amount of brain tissue deficit or excess relative to the standard template) at each voxel in the brain, after covarying for age and sex. In the QTIM cohort, kinship structure was also taken into account using mixed‐effects modeling. We performed univariate associations with the top SNP hit identified in ADNI (rs678849), using a mixed model approach controlling for age and sex [emmaX: http://genetics.cs.ucla.edu/emmax/news.html; Kang et al., 2010], to account for the familial relatedness between subjects. A kinship matrix was used to describe the expected proportion of common genetic variation between subjects. As detailed elsewhere [Jahanshad et al., 2012], a “0” in the kinship matrix represents the relation between unrelated individuals, “1” denotes the relationship of a pair of MZ twins (with identical genomes), and DZ twins and nontwin siblings within the same family are denoted by “0.5” (as they are expected to share approximately half of their SNPs).
Multiple Comparisons Correction
Computing thousands of association tests across the brain can introduce a high Type I (false positive) error rate in neuroimaging studies, if not controlled. To control these errors, we used a searchlight method for false discovery rate (FDR) correction [Langers et al., 2007] which controls the FDR, where possible, in any reported statistical maps. We implemented this searchlight method to correct the map of statistical associations between the image phenotype (morphometry) and genotype at the rs678849 locus. All maps shown are thresholded at the appropriate corrected P‐value, after performing searchlight FDR (q = 0.05), to show only regions of significance that passed the multiple comparisons correction.
Post Hoc Analyses: CSF Biomarkers in ADNI
For 387 of our subjects (75.07 ± 7.0 years, 223 men, and 164 women), CSF data was available from the ADNI public database (http://www.loni.ucla.edu/ADNI/Data/). Baseline values of two CSF biomarkers of neurodegeneration (total t‐tau and the beta amyloid isoform, Aβ42) were analyzed. CSF samples were obtained through lumbar puncture, after an overnight fast. Samples from various sites were transferred, on dry ice, to the ADNI Biomarker Core Laboratory at the University of Pennsylvania Medical Center, where the levels of t‐tau and Aβ42 are measured with a multiplex immunoassay platform under the direction of Drs. Leslie Shaw and John Trojanowski [Trojanowski et al., 2010].
We first tested the rs678849 polymorphism for association with CSF biomarker levels using an additive model, and the results failed to reach statistical significance. In ADNI, CSF is available for only around half the subjects who underwent neuroimaging, which makes it more difficult to detect the effects of a single SNP in a three‐group analysis. As so few subjects were included in the CSF analyses, we also tested a dominant model. We compared mean biomarker levels between C homozygotes and carriers of 1 or 2 T alleles. We used multiple regression to test how well the genotype at the rs678849 locus might predict CSF biomarker levels, after adjusting for age and sex.
RESULTS
Neuroimaging
In the ADNI sample, one very common variant, the intronic OPRD1 polymorphism rs678849 (C/T, minor allele frequency: T = 0.462), predicted differences in regional brain volumes, after covarying for age and sex. Smaller volumes in frontal (anterior cingulate gyrus, right middle frontal and precentral gyrus, left inferior frontal gyrus, and frontal pole), temporal (inferior and middle temporal gyri, and temporal fusiform gyri, especially on the left) and occipital brain regions (lateral occipital and occipital fusiform gyri) were statistically related to carrying at least one C allele (Fig. 2, critical P = 7.74 × 10−5). None of the other four tag SNPs (rs2236856, rs499062, rs508448, and rs4654327) was significantly associated with regional brain volumes, after multiple comparisons correction.
Figure 2.
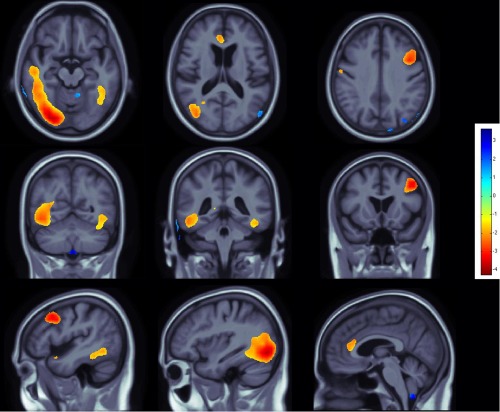
Effects of C allele at the rs678849 locus in the delta opioid receptor gene, on regional brain volumes in the ADNI cohort. The intronic OPRD1 polymorphism rs678849 was significantly associated with regional frontal, temporal, and occipital brain volumes. Negative beta values (warm colors) show regions where C allele carriers had lower tissue volumes. Tests for associations are adjusted for age and sex; maps are corrected for multiple comparisons with the searchlight false discovery rate (FDR) method. Images are in radiological convention (left side of the brain shown on the right). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
The top SNP hit (at the rs678849 locus) also predicted differences in regional brain volumes in the QTIM cohort, after covarying for age and sex and after multiple comparisons correction. Smaller volumes in the orbitofrontal cortex, in superior and posterior parietal regions, and in the right cerebellum were statistically related to carrying at least one C allele (Fig. 3). On all significance maps, the color bar encodes the average percentage of volume difference relative to the template for the subjects carrying at least one C allele, versus noncarriers (Figs. 2 and 3).
Figure 3.
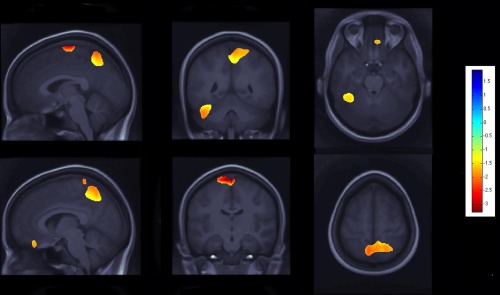
Effects of C allele at the rs678849 locus in the delta opioid receptor gene, on regional brain volumes in the QTIM cohort. The same OPRD1 polymorphism, rs678849, was significantly associated with regional frontal, temporal, and occipital brain volumes. Negative beta values (warm colors) show regions where C allele carriers had lower tissue volumes. Tests for associations are adjusted for age and sex; maps are corrected for multiple comparisons with the searchlight false discovery rate (FDR) method. Images are in radiological convention (left side of the brain shown on the right). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Post Hoc Analyses
In 387 of our ADNI subjects, CSF biomarkers of neurodegeneration (total t‐tau and the beta amyloid isoform Aβ42) were obtained. Based on our finding that the rs678849 predicted differences in regional brain volumes, we tested how well genotype at the rs678849 locus predicted CSF measures of t‐tau and Aβ42, adjusting for age and sex. These post hoc analyses revealed trends (0.05 < P < 0.10) for a main effect of genotype on CSF levels of biomarkers associated with neurodegeneration (P = 0.079 for t‐tau/Aβ42 ratio, P = 0.066 for t‐tau levels), and trend level evidence for a possible genotype by diagnosis interaction (P = 0.096 for t‐tau/Aβ42 ratio, P = 0.077 for t‐tau), with AD coded as “2”, MCI coded as “1”, and healthy elderly coded as “0”.
We performed further exploratory tests to examine results within diagnostic groups, to determine if any one diagnostic group was responsible for carrying the effect. Genotype predicted levels of neurodegenerative biomarkers in the CSF for healthy elderly (n = 105) but not for individuals with MCI (n = 182) or AD (n = 100). Healthy homozygous C allele carriers had higher t‐tau/Aβ42 ratios (suggesting a greater risk for neurodegenerative diseases) than participants with at least one T‐allele (P = 0.027, Fig. 4). In addition, healthy homozygous C allele carriers showed a trend for higher mean t‐tau levels (perhaps suggesting a higher risk for neurodegenerative disorders) than participants with at least one T‐allele (P = 0.063, Fig. 5).
Figure 4.
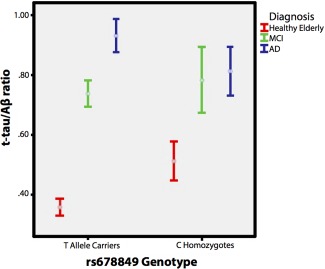
Effects of genotype at the rs678849 locus on t‐tau/Aβ42 ratios Significant main effect of genotype on t‐tau/Aβ42 ratios in healthy elderly (n = 105, represented in red). Homozygous C allele carriers had higher ratios (suggesting a greater risk for neurodegenerative diseases) than participants with at least one T allele (P = 0.027). AD: Alzheimer's disease patients (blue) and MCI: participants with mild cognitive impairment (green). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Figure 5.
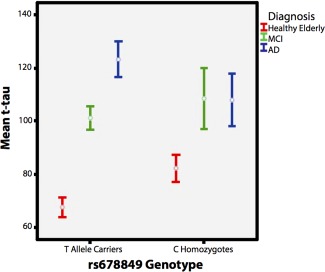
Effects of genotype at rs678849 locus on mean t‐tau levels. Trend for main effect of genotype on mean t‐tau levels in healthy elderly (n = 105, represented in red). Homozygous C allele carriers had higher mean t‐tau levels (suggesting a greater risk for neurodegenerative diseases) than participants with at least one T allele (P = 0.063). AD: Alzheimer's disease patients (blue), MCI: participants with mild cognitive impairment (green), and t‐tau levels are reported in pg/ml. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
DISCUSSION
This study is the first to report an association between brain structure and a commonly carried opioid receptor genetic variant. We identified a SNP in OPRD1, the gene encoding delta opioid receptors, which predicts tissue volumes in various brain regions. In much previous research, investigating gene–brain relationships, replication of initially promising results has proven very difficult. For instance, our group recently found that several candidate genes reportedly related to hippocampal volumes (including BDNF, DISC1, and COMT) were not significantly associated with hippocampal volumes in genome‐wide association meta‐analyses from a large multinational consortium [Stein et al., 2010]. Here, we were able to replicate the genetic association of the C‐allele at the rs678849 locus with smaller regional brain volumes in samples from two continents (United States and Australia), separated in mean age by ∼ 50 years, and using data collected on scanners with different field strengths (1.5 vs. 4 T), suggesting that the association of this OPRD1 SNP with brain volumes may be robust and persist throughout life.
Although the same allele was associated with reduced tissue volumes in both samples, none of the significant voxels after correction for multiple comparisons in the QTIM sample overlapped with the voxels that survived statistical thresholding in the original ADNI sample. In the QTIM sample, we performed an unbiased search across each voxel of the full brain without incorporating previous information from our earlier tests in the ADNI dataset. The reason for this is that voxels that are significant after correction for multiple comparisons do not necessarily represent the only voxels where this OPRD1 SNP may have an effect. We wanted to perform an analysis that allowed for the possibility to observe an effect anywhere in the brain. As such, the most appropriate test is a brain‐wide search, as it asserts only that the gene has some effect in the brain, rather than making a stronger assertion about its specific localization.
From a biological point of view, subjects in the QTIM study are, on average, around 50 years younger than ADNI participants. Thus, a gene may have regional effects on the brain that either spread out or become more regionally specific over the human lifespan. Or, perhaps more likely, it could be that a very weak effect is spread over the entire brain in both samples, but due to noise and biological variability in the two cohorts, different locations in the brain provide the highest effect sizes to the statistical maps in each cohort, with the result that the maps are significant in aggregate. As an alternative more stringent approach, a conjunction test [Jovicich et al., 2006] could be performed to directly identify voxels where a gene effect is statistically significant in several cohorts at once. Such tests have relatively low power, as they require that the gene effect be found consistently in the same voxels in all cohorts, rather than across the brain in aggregate in each cohort. Different formulations of the null hypothesis may therefore greatly affect the power and scope of the inferences. In addition, there may be gene effects that are detectable at only one part of the lifespan, so many biologically important gene effects may not be detectable in both young and old cohorts.
The association of this OPRD1 SNP with tissue volumes in both samples is an important finding for several reasons. Degenerative disorders are typically diagnosed only after severe brain changes have occurred, so it is vital to discover genetic variants that may influence brain volumetry or atrophy in the general population. Animal and human cell line studies implicate delta receptors in neurodegeneration and neuroprotection [Feng et al., 2009], strengthening the case that these receptors may affect brain structure at the macroscopic level. For the first time, we show a direct association between a variant in the gene encoding delta receptors and regional brain volumes, in several systems where cellular and molecular processes are mediated by these receptors.
The genotype associated with reduced volumes (the C allele at the rs678849 locus) also predicted CSF levels of classical neurodegenerative biomarkers in healthy elderly people, suggesting a possible link to physiological measures of neurodegenerative diseases. This association was only significant in healthy elderly, perhaps because, in the MCI and AD subjects, other genes such as ApoE [Schellenberg and Montine, 2011] may be a primary driver factor affecting CSF levels of neurodegenerative biomarkers. We were unable to replicate these results due to the lack of comparable data from a different cohort. In addition, one could argue that some multiple comparisons correction would be appropriate, as two biomarkers were assessed. Although a Bonferroni correction may be overly conservative as the total tau and t‐tau/Aβ42 ratio are correlated measures, this would increase the P‐values well above the trend range of significance. These limitations make the CSF findings less convincing than the imaging results. Nevertheless, if replicated, this SNP, in conjunction with others [Kohannim et al., 2012], may eventually help predict who among the asymptomatic elderly may be at greater risk for developing neurodegenerative disorders. Such a genetic profile may help to target interventions to those with greater inherited predisposition for such diseases.
Interestingly, an association between rs678849 and cocaine addiction was reported in a recent study [Crist et al., 2012]. The authors found that the T allele was overrepresented in the nonaddicted control population, suggesting a “protective” effect. Therefore, similar to the rs1042114 G allele which is associated both with opiate addiction [Zhang et al., 2004] and with a detrimental accumulation of Aβ precursor protein [Sarajarvi et al., 2004], the C allele at the rs678849 locus appears to confer a predisposition to not only cocaine addiction, but also to higher CSF levels of neurodegenerative biomarkers and smaller regional brain volumes. We cannot exclude the possibility that individuals genetically predisposed to cocaine addiction and other risky behaviors or addictive substances, may show brain changes due to downstream effects of their lifestyle choices, rather than as a direct influence of a specific SNP. This may be an alternative explanation for the different brain structure effects in the elderly cohort. Even so, we observed volumetric differences in young and healthy subjects screened to exclude neurological or psychiatric illnesses and substance abuse or dependence, so the structural brain differences may be directly associated with their genotype.
Interestingly, in the QTIM sample, we observed a statistical association between the C allele at this locus and lower volumes in superior and posterior parietal cortices, and in the orbitofrontal cortex, all of which have been implicated in studies of drug abuse. Reduced superior parietal volumes were associated with alcohol use disorders [Van Holst et al., 2000] and stimulant abuse was linked to abnormal functional activation in posterior parietal regions [Aron and Paulus, 2007]. Volume reductions in the orbitofrontal cortex are reported in drug abusers [Tanabe et al., 2010], who also show functional and metabolic abnormalities in this region [London et al., 2010]. A single DNA variant may be associated with diverse phenotypes (pleiotropy), so future studies are needed to determine whether the rs678849 polymorphism confers a higher risk for addiction and neurodegeneration via overlapping or different biological mechanisms.
We identified a polymorphism in OPRD1 associated with regional brain volumes, but we are not able to provide mechanistic evidence for how this SNP might affect brain structure. Unlike the rs1042114 SNP, which leads to an amino acid substitution affecting the functioning of delta receptors, rs678849 is an intronic SNP that is less coherently linked with clear changes in function. The mechanisms that relate this polymorphism to brain volumes, levels of neurodegeneration markers, and addiction in other studies remain largely unknown. SNPs in noncoding regions can affect gene splicing, transcription factor binding, mRNA degradation, and other molecular genetic processes. We cannot conclude with certainty that the rs678849 SNP is driving the observed association with brain volumes. Untyped variants with high linkage disequilibrium to this SNP may be responsible for the signal.
Delta opioid receptors are an evolving target for the treatment of many neurological and psychiatric diseases [Pradhan et al., 2012]. The genetic association of an OPRD1 variant with neuroimaging and biomarker phenotypes, along with the known involvement of the OPRD1 gene in drug abuse, provide additional support for the relevance of these receptors as possible therapeutic targets in both neurodegenerative and addictive disorders. Although neuroimaging changes are not the primary outcome measure of most drug trials, genotyping of the rs678849 variant may be of interest in drug trials targeting delta receptors to treat brain disorders, as patients could, in principle, have therapeutic outcomes that depend on their genotype. Clearly, greater understanding of how OPRD1 and other genes affect the brain is necessary before we can contemplate clinical trials using it as a pertinent genotype. Nevertheless, this study may eventually inform novel and targeted pharmacological interventions for preventing and managing neurological disorders.
ACKNOWLEDGMENTS
We acknowledge the investigators who participated in twin recruitment and data collection in the QTIM study, in particular research nurses Marlene Grace and Ann Eldridge from the Queensland Institute of Medical Research.
Conflicts of interest: M.W.W. received commercial support unrelated to the topic of this article.
The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles.
REFERENCES
- Aron JL, Paulus MP (2007): Location, location: Using functional magnetic resonance imaging to pinpoint brain differences relevant to stimulant use. Addiction 102(Suppl 1):33–43. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2005): Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. [DOI] [PubMed] [Google Scholar]
- Barson JR, Carr AJ, Soun JE, Sobhani NC, Rada P, Leibowitz SF, Hoebel BG (2010): Opioids in the hypothalamic paraventricular nucleus stimulate ethanol intake. Alcohol Clin Exp Res 34:214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokland GA, McMahon KL, Thompson PM, Martin NG, De Zubicaray GI, Wright MJ (2011): Heritability of working memory brain activation. J Neurosci 31:10882–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MK, Worsley KJ, Paus T, Cherif C, Collins DL, Giedd JN, Rapoport JL, Evans AC (2001): A unified statistical approach to deformation‐based morphometry. Neuroimage 14:595–606. [DOI] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC (1994): Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr 18:192–205. [PubMed] [Google Scholar]
- Crist RC, Ambrose‐Lanci LM, Vaswani M, Clarke TK, Zeng A, Yuan C, Ferraro TN, Hakonarson H, Kampman KM, Dackis CA, Pettinati HM, O'Brien CP, Oslin DW, Doyle GA, Lohoff FW, Berrettini WH(2012): Case–control association analysis of polymorphisms in the delta‐opioid receptor, OPRD1, with cocaine and opioid addicted populations. Drug Alcohol Depend. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Zubicaray GI, Chiang MC, McMahon KL, Shattuck DW, Toga AW, Martin NG, Wright MJ, Thompson PM (2008): Meeting the challenges of neuroimaging genetics. Brain Imaging Behav 2:258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Chao D, He X, Yang Y, Kang X, Lazarus LH, Xia Y (2009): A novel insight into neuroprotection against hypoxic/ischemic stress. Sheng Li Xue Bao 61:585–592. [PMC free article] [PubMed] [Google Scholar]
- Freeborough PA, Fox NC (1998): Modeling brain deformations in Alzheimer disease by fluid registration of serial 3D MR images. J Comput Assist Tomogr 22:838–843. [DOI] [PubMed] [Google Scholar]
- Gutman B, Svarer C, Leow AD, Yanovsky I, Toga AW, Thompson PM (2010):Creating Unbiased Minimal Deformation Templates for Brain Volume Registration. Organization For Human Brain Mapping, Barcelona, Barcelona, Spain, June 2010. [Google Scholar]
- Hirose N, Murakawa K, Takada K, Oi Y, Suzuki T, Nagase H, Cools AR, Koshikawa N (2005): Interactions among mu‐ and delta‐opioid receptors, especially putative delta1‐ and delta2‐opioid receptors, promote dopamine release in the nucleus accumbens. Neuroscience 135:213–225. [DOI] [PubMed] [Google Scholar]
- Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, Jack CRJr, Weiner MW, Thompson PM (2008a): Tensor‐based morphometry as a neuroimaging biomarker for Alzheimer's disease: An MRI study of 676 AD, MCI, and normal subjects. Neuroimage 43:458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Lee S, Klunder AD, Toga AW, Lepore N, Chou YY, Brun C, Chiang MC, Barysheva M, Jack CRJr Bernstein MA, Britson PJ, Ward CP, Whitwell JL, Borowski B, Fleisher AS, Fox NC, Boyes RG, Barnes J, Harvey D, Kornak J, Schuff N, Boreta L, Alexander GE, Weiner MW, Thompson PM and the Alzheimer's Disease Neuroimaging, Initiative (2008b): 3D characterization of brain atrophy in Alzheimer's disease and mild cognitive impairment using tensor‐based morphometry. Neuroimage 41:19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CRJr Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, LWhitwell J, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox‐Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW (2008): The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 27:685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Kohannim O, Hibar DP, Stein JL, McMahon KL, de Zubicaray GI, Medland SE, Montgomery GW, Whitfield JB, Martin NG, Wright MJ, Toga AW, Thompson PM (2012): Brain structure in healthy adults is related to serum transferrin and the H63D polymorphism in the HFE gene. Proc Natl Acad Sci USA 109:E851–E859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janecka A, Fichna J, Janecki T (2004): Opioid receptors and their ligands. Curr Top Med Chem 4:1–17. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S (2002): Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17:825–841. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A (2006): Reliability in multi‐site structural MRI studies: Effects of gradient non‐linearity correction on phantom and human data. Neuroimage 30:436–443. [DOI] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E (2010): Variance component model to account for sample structure in genome‐wide association studies. Nat Genet 42:348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohannim O, Jahanshad N, Braskie MN, Stein JL, Chiang MC, Reese AH, Hibar DP, Toga AW, McMahon KL, de Zubicaray GI, Medland SE, Montgomery GW, Martin NG, Wright MJ, Thompson PM (2012): Predicting white matter integrity from multiple common genetic variants. Neuropsychopharmacology 37:2012–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Schork NJ (1994): Genetic dissection of complex traits. Science 265:2037–2048. [DOI] [PubMed] [Google Scholar]
- Langers DR, Jansen JF, Backes WH (2007): Enhanced signal detection in neuroimaging by means of regional control of the global false discovery rate. Neuroimage 38:43–56. [DOI] [PubMed] [Google Scholar]
- Leow A, Huang SC, Geng A, Becker J, Davis S, Toga A, Thompson P (2005): Inverse consistent mapping in 3D deformable image registration: Its construction and statistical properties. Inf Process Med Imaging 19:493–503. [DOI] [PubMed] [Google Scholar]
- Leow AD, Klunder AD, Jack CRJr Toga AW, Dale AM, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Whitwell JL, Borowski BJ, Fleisher AS, Fox NC, Harvey D, Kornak J, Schuff N, Studholme C, Alexander GE, Weiner MW, Thompson PM (2006): Longitudinal stability of MRI for mapping brain change using tensor‐based morphometry. Neuroimage 31:627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levran O, Yuferov V, Kreek MJ (2012): The genetics of the opioid system and specific drug addictions. Hum Genet 131:823–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London ED, Ernst M, Grant S, Bonson K, Weinstein A (2000): Orbitofrontal cortex and human drug abuse: functional imaging. Cereb Cortex 10:334–342. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero‐Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B (2001): A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci 356:1293–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CK, Simms JA, Li R, Mill D, Yi H, Feduccia AA, Santos N, Bartlett SE (2012): Delta‐opioid receptor function in the dorsal striatum plays a role in high levels of ethanol consumption in rats. J Neurosci 32:4540–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan AA, Befort K, Nozaki C, Gaveriaux‐Ruff C, Kieffer BL (2011): The delta opioid receptor: An evolving target for the treatment of brain disorders. Trends Pharmacol Sci 32:581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle WR, Li R, Fitzpatrick JM, Donlevy SC, Dawant BM, Price RR (2004): Characterizing changes in MR images with color‐coded Jacobians. Magn Reson Imaging 22:769–777. [DOI] [PubMed] [Google Scholar]
- Sarajarvi T, Tuusa JT, Haapasalo A, Lackman JJ, Sormunen R, Helisalmi S, Roehr JT, Parrado AR, Makinen P, Bertram L, Soininen H, Tanzi RE, Petaja‐Repo UE, Hiltunen M (2011): Cysteine 27 variant of the delta‐opioid receptor affects amyloid precursor protein processing through altered endocytic trafficking. Mol Cell Biol 31:2326–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg GD, Montine TJ (2012): The genetics and neuropathology of Alzheimer's disease. Acta Neuropathol 124:305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC (1998): A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17:87–97. [DOI] [PubMed] [Google Scholar]
- Smith SM (2002): Fast robust automated brain extraction. Hum Brain Mapp 17:143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Hua X, Morra JH, Lee S, Hibar DP, Ho AJ, Leow AD, Toga AW, Sul JH, Kang HM, Eskin E, Saykin AJ, Shen L, Foroud T, Pankratz N, Huentelman MJ, Craig DW, Gerber JD, Allen AN, Corneveaux JJ, Stephan DA, Webster J, DeChairo BM, Potkin SG, Jack CRJr, Weiner MW, Thompson PM (2010): Genome‐wide analysis reveals novel genes influencing temporal lobe structure with relevance to neurodegeneration in Alzheimer's disease. Neuroimage 51:542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann O, Bernard M, Brown AA, Cannon DM, Chakravarty MM, Christoforou A, and others (2012): Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 44:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe J, Tregellas JR, Dalwani M, Thompson L, Owens E, Crowley T, Banich M (2009): Medial orbitofrontal cortex gray matter is reduced in abstinent substance‐dependent individuals. Biol Psychiatry 65:160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L, Zhao J, Wang F, Ma L, Pei G (2010): A GPCR/secretase complex regulates beta‐ and gamma‐secretase specificity for abeta production and contributes to AD pathogenesis. Cell Res 20:138–153. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Giedd JN, Woods RP, Macdonald D, Evans AC, Toga AW (2000): Growth patterns in the developing brain detected by using continuum mechanical tensor maps. Nature 404:190–193. [DOI] [PubMed] [Google Scholar]
- Trojanowski JQ, Vandeerstichele H, Korecka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter WZ, Weiner MW, Jack CRJr Jagust W, Toga AW, Lee VM, Shaw LM (2010): Update on the biomarker core of the Alzheimer's Disease Neuroimaging Initiative subjects. Alzheimer's Dement 6:230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holst RJ, De Ruiter MB, Van Den Brink W, Veltman DJ, Goudriaan AE (2012): A voxel‐based morphometry study comparing problem gamblers, alcohol abusers, and healthy controls. Drug Alcohol Depend 124:142–148. [DOI] [PubMed] [Google Scholar]
- Van Rijn RM, Brissett DI, Whistler JL (2010): Dual efficacy of delta opioid receptor‐selective ligands for ethanol drinking and anxiety. J Pharmacol Exp Ther 335:133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rijn RM, Brissett DI, Whistler JL (2012a): Emergence of functional spinal delta opioid receptors after chronic ethanol exposure. Biol Psychiatry 71:232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rijn RM, Brissett DI, Whistler JL (2012b): Distinctive modulation of ethanol place preference by delta opioid receptor‐selective agonists. Drug Alcohol Depend 122:156–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fowler JS, Tomasi D, Baler R (2011): Food and drug reward: Overlapping circuits in human obesity and addiction. Curr Top Behav Neurosci 1:1–24. [DOI] [PubMed] [Google Scholar]
- Wright MJ, Martin NG (2004): Brisbane adolescent twin study: Outline of study methods and research projects. Aust J Psychol 56:65–78. [Google Scholar]
- Wu Y, Chen R, Zhao X, Li A, Li G, Zhou J (2011): JWA regulates chronic morphine dependence via the delta opioid receptor. Biochem Biophys Res Commun 409:520–525. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J (2008): The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry 13:531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bao L, Guan JS (2006): Role of delivery and trafficking of delta‐opioid peptide receptors in opioid analgesia and tolerance. Trends Pharmacol Sci 27:324–329. [DOI] [PubMed] [Google Scholar]


