Abstract
We examined 215 Enterococcus faecalis isolates and found that neither the two-component regulatory locus fsr (E. faecalis regulator) nor gelatinase production was more common in disease-associated isolates than in isolates colonizing healthy individuals (ca. 60 to 65%). The majority of gelatinase-negative isolates, including 14 endocarditis isolates (of 80 isolates tested), contained the previously described 23.9-kb deletion and lacked fsrA and fsrB. While these findings indicate that neither fsr nor gelatinase is required for E. faecalis to cause infection, this study did not address whether fsr or gelatinase affects the severity of disease, as it does in animal models.
Enterococci are normally commensal flora, but they can cause a wide range of diseases, including urinary tract infections, bloodstream infections, wound infections, and endocarditis (12, 13). Although there are over 20 species of enterococci (8), Enterococcus faecalis isolates cause the majority of infections (22). Despite the increasing number of cases of enterococcal infection and the remarkable ability of these organisms to resist antibiotics, relatively little about their pathogenesis is known compared to what is known about some other gram-positive cocci.
Despite its role as a possible virulence factor, gelatinase is not produced by all clinical isolates, and a number of studies have attempted to correlate gelatinase activity with disease caused by E. faecalis (3, 5, 7, 17, 20, 24, 28). Nakayama et al. recently reported that a 23.9-kb chromosomal deletion involving the fsr locus was commonly found in urine isolates and was also found in most gelatinase-negative strains (17). In contrast, another study reported that fsr was present in 100% of 12 endocarditis isolates (20). In the present study, we compared clinical isolates of E. faecalis from patients with endocarditis, urinary tract infections, or blood-borne infections to fecal isolates from healthy volunteers with respect to the presence of fsr and gelatinase production.
A total of 215 E. faecalis isolates from clinical samples and from feces of healthy, community-based volunteers, collected between 1974 and 2003, were used in this study. The identification of all isolates as E. faecalis was initially done by biochemical tests and was confirmed by using an intragenic portion of the ace gene (18) as a probe for colony hybridization (data not shown). E. faecalis OG1RF (15), E. faecalis OG1SSp (6), E. faecalis V583 (23), and Enterococcus faecium TX0016 (2) are well-studied strains and were used in this study as controls. Brain heart infusion agar (Difco Laboratories, Detroit, Mich.) or brain heart infusion broth was used for growth. Gelatinase production was measured as previously described (21) after overnight incubation at 37°C and again after 7 days.
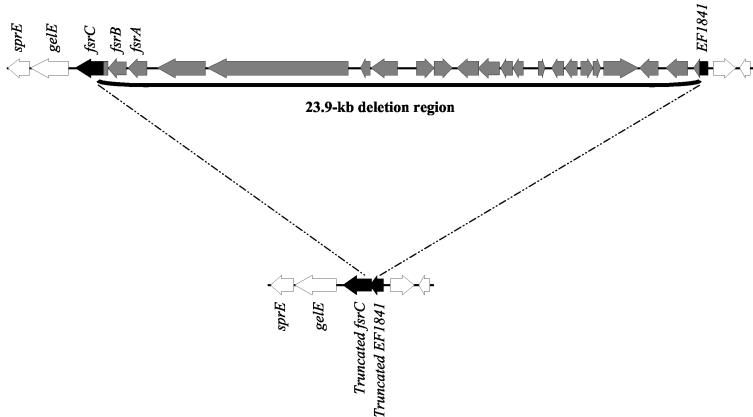
Genomic DNA was isolated as previously described (11, 29), and PCRs were performed with the Optimized Buffer B kit (Invitrogen, San Diego, Calif.) with primers listed in Table 1. The 23.9-kb deletion (17) was detected by using primer pair delsizeF1 and delsizeR1, located in open reading frame EF1841 in the V583 genome database (www.tigr.org) and in the 3′ end of the partially deleted fsrC gene (Fig. 1), respectively.
TABLE 1.
Primers used in this study
| Primer | Amplicon size (bp) | Reference | Sequence (5′ to 3′) |
|---|---|---|---|
| Primers used to make probes | |||
| aceF | 1,003 | 18 | GAGCAAAAGTTCAATCGTTGAC |
| aceR | 18 | GTCTGTCTTTTCACTTGTTTCT | |
| aacF | 320 | 4 | GCGGTAGCAGCGCTAGACCAAG |
| aacR | 4 | GCATTTGGTAAGACACCTACG | |
| gelEF | 1,200 | This study | GATGAAGGGAAATAAAATTTTATAC |
| gelER | This study | AGTAAGTAACTGGTCTTGTCTAAG | |
| fsrBF1 | 541 | 21 | GGGAGCTCTGGACAAAGTATTATCTAACCG |
| fsrBR2 | 21 | TTGGTACCCACACCATCACTGACTTTTGC | |
| Primers used to determine size of deletion and for sequencing | |||
| delsizeF1 | 1,800 | This study | GAAACTTCACTTTCTGAATAAACTTG |
| delsizeR1 | This study | GTTGGAGAGTATGCCTATCAACTCG |
FIG. 1.
The 23.9-kb deletion affecting the fsr locus.
Colony hybridization was performed as described (26) with probes labeled by using the RadPrime DNA labeling system (GIBCO/BRL, Gaithersburg, Md.). Probes consisting of intragenic gelE, fsrB (21), and ace (19) PCR products were generated by using OG1RF genomic DNA templates and the primers given in Table 1, while the aac(6′)-Ii probe (4) (to rule out E. faecium) was generated by using TX16 genomic DNA (http://www.hgsc.bcm.tmc.edu/microbial/Efaecium/). Pulsed-field gel electrophoresis was performed as previously described (11, 14).
A number of investigators have studied gelatinase and E. faecalis isolates from clinical and community sources. While some of these studies measured gelatinase activity by plate assay (1, 5, 7, 10, 28), others used DNA-based techniques to determine whether the gelE gene was present (25), and such techniques are now known to not reliably predict the gelatinase phenotype. In our study, we used the gelatinase plate assay, hybridization, and PCR to correlate the observed gelatinase phenotype with the underlying fsr and gelE genotypes. Gelatinase activity was observed for 59.5% of 215 E. faecalis isolates (Table 2). With the exception of the slow-gelatinase-producing mutant OG1SSp (6), all isolates were gelatinase positive after 16 h of incubation. Previously, we reported that 27% of 30 healthy community E. faecalis fecal isolates, identified biochemically, produced gelatinase (5). However, we recently found by genetic methods that some of our original community fecal isolates were actually E. faecium isolates, leading to an underestimation of the percentage of E. faecalis isolates that were gelatinase positive. In the present study, we found that 66.6% of fecal E. faecalis isolates were gelatinase positive, compared to 58.7% of clinical isolates (endocarditis, blood, and urine isolates). Furthermore, we recently tested 15 additional fecal E. faecalis isolates from healthy volunteers and found that 87% were gelatinase producers. Gelatinase production in our clinical isolates correlates with that seen in other studies (1, 7, 16, 28).
TABLE 2.
Summary of results for all isolates tested
| Source of isolates | No. (%) of isolates:
|
Total no. of isolates from source (% of total) | ||||
|---|---|---|---|---|---|---|
| Gelatinasea positive | Gelatinase negative
|
|||||
| fsrB+gelE+ | Lacking fsrB, gelE+b | Lacking fsrB and gelE | fsrB+, lacking gelE | |||
| Endocarditis | 49 (61.2) | 7 (8.7) | 14 (17.5) | 10 (12.5) | 0 | 80 (37.2) |
| Blood | 24 (75) | 1 (3.1) | 5 (15.6) | 2 (6.3) | 0 | 30 (14.8) |
| Urine | 41 (50) | 2 (2.4) | 36 (43.9) | 3 (3.7) | 0 | 82 (38.1) |
| Feces from healthy community-based volunteers | 14 (66.6) | 0 | 5 (23.8) | 1 (4.8) | 1 (4.8) | 21 (9.7) |
| Total | 128 (59.5) | 10 (4.7) | 60 (27.9)b | 16 (7.4) | 1 (0.5) | 215 (100) |
Isolates that produced gelatinase had the fsrB+ gelE+ genotype.
All isolates with this genotype appeared to have the 23.9-kb deletion as determined by PCR (see text).
In an earlier paper analyzing fsr (21), it was shown that 62% of 95 isolates produced gelatinase but that 91% of the isolates hybridized to a gelE probe (very similar to the 92% found in the present study), while 71% hybridized to an fsrB probe; an intact fsr locus and gelE are both necessary for gelatinase production (21). Subsequent to our observation of a discrepancy between the presence of gelE and gelatinase production, a study of urine isolates obtained from a single Japanese hospital (17) identified a 23.9-kb deletion affecting the fsr locus, which explains the gelE+, fsrB-lacking genotype (Fig. 1). To expand this observation, we included endocarditis, blood, and community fecal isolates collected from geographically diverse regions over a 29-year period. Using colony hybridization, we found 60 isolates (including 14 endocarditis isolates) that hybridized with the gelE probe but lacked fsrB, suggesting that they lacked the 23.9-kb region (Table 2). In contrast, a previous study involving a small number of endocarditis isolates suggested that fsr was present in all endocarditis isolates (20). The discrepancy between the two studies may have resulted from the small number of isolates used or possibly from the fact that the probe consisted of all three fsr genes, which may have resulted in a positive hybridization result because 1,081 bp of the fsrC gene is still present in strains with the deletion. While the 23.9-kb region was missing in all categories of isolates we tested (Table 2), 16 isolates lacked both gelE and fsrB, similar to the findings presented in the earlier report (21), suggesting an even larger deletion in these isolates, as was also reported by Nakayama et al. (17).
All 60 fsrB-lacking, gelE-containing isolates were confirmed by PCR to lack the 23.9-kb region. Sequencing of the 1.8-kb PCR deletion junctions from 10 isolates (4 endocarditis isolates, 2 other blood isolates, 2 urine isolates, and 2 community fecal isolates) revealed that all were identical for 300 bp on either side of the junction, indicating that the region is highly conserved and also suggesting that the deletion occurred by the same mechanism in all isolates (Fig. 1).
We also examined 35 isolates with the 23.9-kb deletion by pulsed-field gel electrophoresis. The observed banding patterns indicated at least 22 distinct strains (a difference of >7 bands) (27) among these 35 isolates from geographically diverse locations. Several urine isolates collected in the same location were related, while the majority of endocarditis, blood, and community fecal isolates were distinct strains. These results show that the deletion occurred in many distinct clinical strains, as well as in isolates from healthy volunteers.
In conclusion, the percentage of clinical isolates displaying gelatinase activity was similar to the percentage of healthy fecal isolates displaying gelatinase activity. In addition to urine isolates, the previously described 23.9-kb deleted region (17) was also lacking in endocarditis, blood, and healthy stool isolates. While fsr and gelatinase increase the severity of disease in animal models of E. faecalis infection, they are not required for the organism to cause disease (9). Controlled clinical studies will be needed to determine whether the presence of the fsr locus or gelatinase production affects the course of E. faecalis infections in humans or the outcome of the treatment.
Acknowledgments
This work was supported by NIH grant AI 33516 from the Division of Microbiology and Infectious Diseases, NIAID, to B. E. Murray and by FDA grant FD-U-001621-01 to P. C. Okhuysen and NIH grant RR-02558 to the Clinical Research Center.
New community fecal isolates were collected at the Clinical Research Center, University of Texas Medical School. This study was approved by the UTHSC Committee for the Protection of Human Subjects, and informed consent was obtained from participants.
REFERENCES
- 1.Archimbaud, C., N. Shankar, C. Forestier, A. Baghdayan, M. S. Gilmore, F. Charbonne, and B. Joly. 2002. In vitro adhesive properties and virulence factors of Enterococcus faecalis strains. Res. Microbiol. 153:75-80. [DOI] [PubMed] [Google Scholar]
- 2.Arduino, R. C., K. Jacques-Palaz, B. E. Murray, and R. M. Rakita. 1994. Resistance of Enterococcus faecium to neutrophil-mediated phagocytosis. Infect. Immun. 62:5587-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco, A. R., S. La Terra Mule, G. Babini, S. Garbisa, V. Enea, and D. Rusciano. 2003. (-)Epigallocatechin-3-gallate inhibits gelatinase activity of some bacterial isolates from ocular infection, and limits their invasion through gelatine. Biochim. Biophys. Acta 1620:273-281. [DOI] [PubMed] [Google Scholar]
- 4.Coque, T. M., and B. E. Murray. 1995. Identification of Enterococcus faecalis strains by DNA hybridization and pulsed-field gel electrophoresis. J. Clin. Microbiol. 33:3368-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coque, T. M., J. E. Patterson, J. M. Steckelberg, and B. E. Murray. 1995. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J. Infect. Dis. 171:1223-1229. [DOI] [PubMed] [Google Scholar]
- 6.Dunny, G. M., R. A. Craig, R. L. Carron, and D. B. Clewell. 1979. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid 2:454-465. [DOI] [PubMed] [Google Scholar]
- 7.Elsner, H. A., I. Sobottka, D. Mack, M. Claussen, R. Laufs, and R. Wirth. 2000. Virulence factors of Enterococcus faecalis and Enterococcus faecium blood culture isolates. Eur. J. Clin. Microbiol. Infect. Dis. 19:39-42. [DOI] [PubMed] [Google Scholar]
- 8.Facklam, R. R., M. S. Carvalho, and L. M. Teixeira. 2002. History, taxonomy, biochemical characteristics and antibiotic susceptibility testing of enterococci, p. 1-54. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci, pathogenesis, molecular biology and antibiotic resistance. ASM Press, Washington, D.C.
- 9.Jones, R. N., and L. M. Deshpande. 2003. Distribution of fsr among Enterococcus faecalis isolates from the SENTRY antimicrobial surveillance program. J. Clin. Microbiol. 41:4004-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanemitsu, K., T. Nishino, H. Kunishima, N. Okamura, H. Takemura, H. Yamamoto, and M. Kaku. 2001. Quantitative determination of gelatinase activity among enterococci. J. Microbiol. Methods 47:11-16. [DOI] [PubMed] [Google Scholar]
- 11.Malathum, K., K. V. Singh, G. M. Weinstock, and B. E. Murray. 1998. Repetitive sequence-based PCR versus pulsed-field gel electrophoresis for typing of Enterococcus faecalis at the subspecies level. J. Clin. Microbiol. 36:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moellering, R. C., Jr. 1992. Emergence of Enterococcus as a significant pathogen. Clin. Infect. Dis. 14:1173-1176. [DOI] [PubMed] [Google Scholar]
- 13.Murray, B. E. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28:2059-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray, B. E., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. Akkermans, W. M. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama, J., R. Kariyama, and H. Kumon. 2002. Description of a 23.9-kilobase chromosomal deletion containing a region encoding fsr genes which mainly determines the gelatinase-negative phenotype of clinical isolates of Enterococcus faecalis in urine. Appl. Environ. Microbiol. 68:3152-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nallapareddy, S. R., R. W. Duh, K. V. Singh, and B. E. Murray. 2002. Molecular typing of selected Enterococcus faecalis isolates: pilot study using multilocus sequence typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nallapareddy, S. R., X. Qin, G. M. Weinstock, M. Hook, and B. E. Murray. 2000. Enterococcus faecalis adhesin, ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect. Immun. 68:5218-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillai, S. K., G. Sakoulas, H. S. Gold, C. Wennersten, G. M. Eliopoulos, R. C. Moellering, Jr., and R. T. Inouye. 2002. Prevalence of the fsr locus in Enterococcus faecalis infections. J. Clin. Microbiol. 40:2651-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruoff, K. L., L. de la Maza, M. J. Murtagh, J. D. Spargo, and M. J. Ferraro. 1990. Species identities of enterococci isolated from clinical specimens. J. Clin. Microbiol. 28:435-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidtchen, A., I. M. Frick, E. Andersson, H. Tapper, and L. Bjorck. 2002. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 46:157-168. [DOI] [PubMed] [Google Scholar]
- 25.Shepard, B. D., and M. S. Gilmore. 2002. Differential expression of virulence-related genes in Enterococcus faecalis in response to biological cues in serum and urine. Infect. Immun. 70:4344-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh, K. V., T. M. Coque, G. M. Weinstock, and B. E. Murray. 1998. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol. Med. Microbiol. 21:323-331. [DOI] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vergis, E. N., N. Shankar, J. W. Chow, M. K. Hayden, D. R. Snydman, M. J. Zervos, P. K. Linden, M. M. Wagener, and R. R. Muder. 2002. Association between the presence of enterococcal virulence factors gelatinase, hemolysin, and enterococcal surface protein and mortality among patients with bacteremia due to Enterococcus faecalis. Clin. Infect. Dis. 35:570-575. [DOI] [PubMed] [Google Scholar]
- 29.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.2. In F. M. Ausubel, R. Brent, R. E. Kingston, D. M. David, J. G. Scidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Green Publishing Associates, Brooklyn, N.Y. [DOI] [PubMed]