Abstract
Many studies have reported the presence of bacterial DNA contamination in commercial Taq DNA polymerase reagents. This is the first report of the presence of phage-like DNA sequences in certain commercial Taq DNA polymerase reagents. Precautions are needed when using amplification reagents with exogenous DNAs.
PCR is a highly sensitive technique widely used for rapid detection of specific DNA sequences, with numerous applications in basic research, clinical diagnosis, and microbial identification. This technique can amplify a single copy of template DNA 106- to 107-fold, causing small amounts of endogenous DNA to be potentially a serious contamination problem (3, 8, 14, 18, 20). The highly conserved bacterial 16S rRNA genes are among the widely used PCR targets for identification and detection of microorganisms (5, 7, 10, 11, 21, 22). Our laboratory has also been utilizing broad-range PCR amplification of the highly conserved 16S rRNA gene sequence to detect the presence of prokaryotic agents in cultures of various clinical specimens. In a study, we observed that PCR amplification using the pA/pH primer pair of the 16S rRNA gene sequence (2, 4) produced several DNA bands with sizes different from the expected target of approximately 1.5 kb. Characterization of these PCR products and further PCR study revealed they were amplified from commercial Taq DNA polymerase reagents apparently contaminated with trace amounts of bacterial phage-like DNAs.
In a routine experiment using the highly conserved 16S rRNA gene primers pA (AGAGTTTGATCCTGGCTCAG) and pH (AAGGAGGTGATCCAGCCGCA) designed by Edwards et al. (4) and used by others (6, 12, 13), we observed a number of unexpected PCR products with sizes ranging from 100 bp to more than 2.0 kb, in addition to the expected 1.5-kb band amplified from the 16S rRNA gene (Fig. 1, band D). PCR amplification conditions were 95°C for 2 min followed by 45 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min and a final 10-min extension step at 72°C. The PCR annealing temperature was set at a stringent parameter to reduce side product formation. Side products or secondary products, not uncommonly seen in PCR studies, were often ignored. However, we selected three PCR products (Fig. 1, bands 3A, 3B, and 7C) for further study. These three unexpected PCR products were gel purified, cloned, and sequenced. The resulting sequences were aligned with sequences in the GenBank database. A small portion of DNA sequence 7C (nucleotides 190 to 2031) showed 81% homology to the tail fiber gene of Pseudomonas phage gh-1. The deduced amino acid sequence of a segment (nucleotides 210 to 2031) of the 7C sequence showed partial homology to the tail fiber protein of enterobacterial phage T7 (88%). Nucleotide sequences 3A and 3B showed no significant match or homology to sequences previously deposited in GenBank. However, deduced amino acid sequences of 3A and 3B showed partial homology to tail tubular protein B (72%) and putative DNA ligase (55%) of Pseudomonas phage gh-1, respectively.
FIG. 1.
PCR products amplified from primary cultures using universal 16S rRNA gene primers pA and pH. PCR products were run on a 1.5% agarose gel with Tris-acetate-EDTA buffer. Lane 1, 100-bp DNA ladder; lanes 2 to 13, PCR products from cultures of clinical samples. The bands of interest, 3A (663 bp), 3B (422 bp), and 7C (2.0 kb), were eluted from the gel, cloned into the pGEM T vector, and sequenced. The size of the expected 16S rRNA gene PCR product was approximately 1.5 kb (band D).
Four different primer pairs were designed from the phage-like DNA sequences 3A, 3B, and 7C; among these four primer sets, two primer pairs (3B-1 and 3B-2) were derived from sequence 3B (Table 1). When these primer sets were used to detect the phage-like DNAs in our cultures by PCR, we found PCR bands of expected sizes were generated from all of our culture samples. However, we noticed PCR amplification against water samples serving as negative controls also produced prominent positive results. All the PCR products were confirmed to be the phage-like DNA sequences by nucleotide sequencing. This repeated finding led us to further study the nature of the phage-like DNA sequences and reexamine our PCR system. We specifically compared, in parallel, the PCR using Taq DNA polymerase reagents from different commercial sources.
TABLE 1.
Primers designed from the cloned DNA sequences (3A, 3B, and 7C) and their respective PCR product sizes
| Primer | Sequence | Expected size (bp) of PCR product |
|---|---|---|
| 3A F | 5′ AGGACCCCTTCGTCACGC 3′ | 264 |
| 3A R | 5′ CGCTGAGGACATCTCGGC 3′ | 264 |
| 3B-1 F | 5′ ACCGCCGACCTGCTGAAACG 3′ | 270 |
| 3B-1 R | 5′ CAACCGTCTTCCTTGACC 3′ | 270 |
| 3B-2 F | 5′ AGCCTAAGATGAGCACCATTCG 3′ | 188 |
| 3B-2 R | 5′ TTGTAGACCCACTTGTCGCCCTG 3′ | 188 |
| 7C F | 5′ AAGCACGGGACCTTACTACG 3′ | 207 |
| 7C R | 5′ ACCAGCAGATGCCTCACAGG 3′ | 207 |
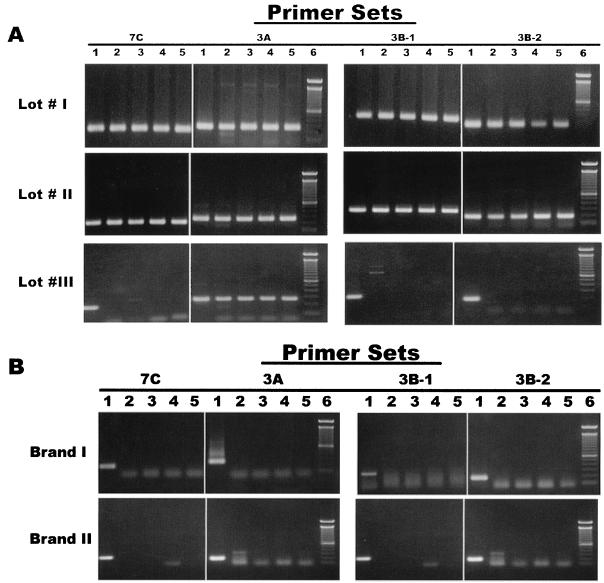
Taq DNA polymerases from five different commercial lots were tested. Three lots were from one company (company A), and the other two lots were two different brands from a second company (company B). All five lots of polymerase enzymes together with the accompanying reagents were tested against the four primer pairs derived from phage-like DNA sequences. PCR was performed against deionized water samples with or without UV treatment, which has been shown to reduce false-positive PCR signals (15, 17). Cloned 3A, 3B, or 7C DNA was added into one of each group of reaction tubes to serve as the positive controls for the respective primer set. No exogenous DNA template was added in any other PCR tubes. The reaction cocktail consisted of 1× buffer (100 mM Tris-HCl, pH 8.3), 200 μM deoxynucleoside triphosphate, 2.5 mM MgCl2, 25 pmol of each primer, and 1.25 U of Taq DNA polymerase. In the PCR study, the same PCR cocktails were used with different Taq polymerases. PCR amplification conditions were 95°C for 10 min, 45 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min followed by a final 10-min extension step at 72°C. The phage-like DNAs were amplified from all three lots of DNA polymerase from company A by the 3A primer set (Fig. 2A). Moreover, two lots of enzyme reagents from this company tested positive by all four primer sets examined (Fig. 2A). In comparison, Taq DNA polymerase of two brands from company B did not amplify the phage-like DNAs in these water samples when tested against the same four primer pairs (Fig. 2B). Only the tubes of positive controls with cloned 3A, 3B, or 7C DNA for each of the respective primer sets had positive PCR products. Since the companies provided their own buffer and MgCl2 solutions, we also tested the buffer solutions that had the same ingredients and similar pH as well as MgCl2 solutions by swapping them with other companies' polymerase reagents in the PCR study. The buffer and MgCl2 solutions from each company made no difference in the results produced by each of the polymerase reagents tested.
FIG. 2.
PCR amplification using four sets of primers (3A, 3B-1, 3B-2, and 7C) derived from bacteriophage-like DNA using commercial Taq polymerase reagents from company A (A) and company B (B). PCR products were run on a 1.5% agarose gel with Tris-acetate-EDTA buffer. Lane 1, UV-treated double-distilled H2O (ddH2O; Gibco-Invitrogen) with cloned DNA 3A, 3B, or 7C for the respective primer sets tested; lane 2, UV-treated laboratory ddH2O; lane 3, laboratory ddH2O without UV treatment; lane 4, UV-treated ddH2O (Gibco-Invitrogen); lane 5, ddH2O (Gibco-Invitrogen) without UV treatment; lane 6, 100-bp DNA ladder size marker.
Different sequences of the primer sets designed from different gene fragments often exhibited different sensitivities in detecting the respective genes by PCR. The 3A primer set appeared to be the most sensitive primer set in detecting the trace amount of contaminating phage-like DNAs among the four primer sets tested in this study. On the other hand, each lot of the commercial Taq DNA polymerase could be contaminated by different amounts of the phage-like DNAs. Compared to lots 1 and 2, lot 3 was apparently contaminated with less of the phage-like DNAs that could be detected only by PCR with the more sensitive 3A primer set.
The presence of DNA contamination surrounding Taq DNA polymerase was previously reported in many PCR studies (1, 9, 16, 19, 20). Most of the contamination reported was exogenous bacterial DNA. Precautions were called for in PCR detection of bacterial ribosomal DNA (rDNA) sequences in samples using the highly preserved 16S rDNA primer sets for broad-range amplification. Our study is the first report of finding bacterial phage-like DNA in some commercial Taq DNA polymerase reagents. Interestingly, the phage-like DNA was accidentally discovered in studies that used a previously reported broad-range primer set for amplifying 16S rDNA. Initial amplification of the phage like DNAs was likely due to, by chance, partial annealing between the “mismatched” primer set and the trace amount of phage-like DNAs in certain reaction tubes. However, presence of these contaminating DNAs in the Taq DNA polymerase became evident after PCR amplification with certain phage-specific primers. The phage-like DNAs were amplified in all specimens when the contaminated reagents were used. It is important to note that, although our study showed the prevalence of contamination by bacterial phage-related DNAs in the Taq DNA polymerase reagents from one particular company, it does not exclude the possibility of contamination by other kinds of DNAs or genes in the reagents of other commercial brands.
Scientists in a wide variety of fields use the highly sensitive PCR technique. Identification of exogenous DNA contamination in Taq DNA polymerase reagents as well as notification of the potential problems due to contamination of the reagents should be important to the scientific community. The tainted amplification reagents, even with only trace amounts of DNA, can produce artificial results that can be extremely confusing and misleading. It could also be very time-consuming for the scientist to follow up on the artificial products.
Nucleotide sequence accession numbers. The DNA sequences determined in this study were deposited in the GenBank database under the following accession numbers: 3A, AY587125; 3B, AY587126; 7C, AY587127.
Acknowledgments
We thank Douglas J. Wear and Shimin Zhang for their constructive discussion and kind help in manuscript preparation.
REFERENCES
- 1.Böttger, E. C. 1990. Frequent contamination of Taq DNA polymerase with DNA. Clin. Chem. 36:1258-1259. [PubMed] [Google Scholar]
- 2.Bruce, K. D., W. D. Hiorns, J. L. Hobman, A. M. Osborn, P. Strike, and D. A. Ritchie. 1992. Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl. Environ. Microbiol. 58:3413-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, E. B. Kaczmarski, and A. J. Fox. 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol. 38:1747-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao, S. J., and P. S. Moore. 1996. Molecular approaches to the identification of unculturable infectious agents. Emerg. Infect. Dis. 2:159-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godreuil, S., M. N. Didelot, C. Perez, A. Lefléche, P. Boiron, J. Reynes, F. Laurent, H. Jean-Pierre, and H. Marchandin. 2003. Nocardia veterana isolated from ascitic fluid of a patient with human immunodeficiency virus infection. J. Clin. Microbiol. 41:2768-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greisen, K., M. Loeffelholz, A. Purohit, and D. Leong. 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32:335-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilali, F., P. Saulnier, E. Chachaty, and A. Andremont. 1997. Decontamination of polymerase chain reaction reagents for detection of low concentrations of 16S rRNA genes. Mol. Biotechnol. 7:207-216. [DOI] [PubMed] [Google Scholar]
- 9.Hughes, M. S., L. A. Beck, and R. A. Skuce. 1994. Identification and elimination of DNA sequences in Taq DNA polymerase. J. Clin. Microbiol. 32:2007-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klausegger, A., M. Hell, A. Berger, K. Zinober, S. Baier, N. Jones, W. Sperl, and B. Kofler. 1999. Gram type-specific broad-range PCR amplification for rapid detection of 62 pathogenic bacteria. J. Clin. Microbiol. 37:464-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolbert, C. P., and D. H. Persing. 1999. Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr. Opin. Microbiol. 2:299-305. [DOI] [PubMed] [Google Scholar]
- 12.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14541-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaughlin, R. W., H. Vali, P. C. K. Lau, R. G. E. Palfree, A. De Ciccio, M. Sirois, D. Ahmad, R. Villemur, M. Desrosiers, and E. C. S. Chan. 2002. Are there naturally occurring pleomorphic bacteria in the blood of healthy humans? J. Clin. Microbiol. 12:4771-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier, A., D. Persing, M. Finken, and E. C. Böttger. 1993. Elimination of contaminating DNA within polymerase chain reaction reagents: implication for a general approach to detection of uncultured pathogens. J. Clin. Microbiol. 31:646-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ou, C. Y., J. L. Moore, and G. Schochetman. 1991. Use of UV irradiation to reduce false positivity in polymerase chain reaction. BioTechniques 10:442, 444, 446. [PubMed] [Google Scholar]
- 16.Rand, K. H., and H. Houck. 1990. Taq polymerase contains bacterial DNA of unknown origin. Mol. Cell. Probes 4:445-450. [DOI] [PubMed] [Google Scholar]
- 17.Rochelle, P. A., A. J. Weightman, and J. C. Fry. 1992. DNase I treatment of Taq DNA polymerase for complete PCR decontamination. BioTechniques 13:520. [PubMed] [Google Scholar]
- 18.Saiki, R. K. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 19.Sarkar, G., and S. S. Sommer. 1993. Removal of DNA contamination in polymerase chain reaction reagents by ultraviolet irradiation. Methods Enzymol. 218:381-388. [DOI] [PubMed] [Google Scholar]
- 20.Sharma, J. K., V. Gopalkrishna, and B. C. Das. 1992. A simple method for elimination of unspecific amplifications in polymerase chain reaction. Nucleic Acids Res. 20:6117-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson, K. H., R. B. Blitchington, and R. C. Greene. 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]