Abstract
Seeking a simple disk test for detection of organisms producing plasmid-mediated AmpC β-lactamases, we evaluated the diagnostic utility of the β-lactamase inhibitors 48-1220 (Ro 48-1220) and LN-2-128. Using NCCLS disk methodology, inhibition zone diameters were determined for five β-lactam antibiotics tested alone and in combination with 20 μg of either 48-1220 or LN-2-128. Using an increase of ≥4 mm in zone diameter in the presence of an inhibitor as a positive test, cefotetan with LN-2-128 and 48-1220 was adequate for the detection of organisms producing plasmid-mediated AmpCs (specificity of 90% and sensitivity of 100%).
Organisms producing plasmid-mediated AmpC β-lactamases were first reported in the 1980s (2, 13). These enzymes are derivatives of the chromosomally encoded, clavulanate-resistant AmpC cephalosporinases and have been reported in Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, Salmonella spp., Enterobacter aerogenes, and Proteus mirabilis. The genes are typically encoded on large plasmids containing additional antibiotic resistance genes, leaving few therapeutic options (15). Although it has been over a decade since plasmid-mediated AmpC β-lactamases were discovered, most clinical laboratories and physicians remain unaware of their clinical importance. Current detection methods for organisms producing plasmid-mediated AmpC β-lactamases are technically demanding for clinical laboratories to perform on a routine basis (1, 5, 6, 9, 20). Multiplex PCR is also available as a research tool for detection of plasmid-mediated AmpC β-lactamases but is not yet available for routine use in clinical laboratories (14). As a result, organisms producing these types of β-lactamases often go undetected (15) and therefore have been responsible for several nosocomial outbreaks (3, 10, 11, 13). Community sources of isolates producing plasmid-mediated AmpC β-lactamases have also been involved in outbreaks of infection (7, 8, 16, 19, 21). The detection of organisms producing these β-lactamases is thus important for enhanced infection control and to ensure effective therapeutic options.
Currently, there are no recommendations available from the NCCLS or elsewhere for detection of organisms producing plasmid-mediated AmpC β-lactamases. The NCCLS has established guidelines for the detection of organisms producing extended-spectrum β-lactamases (ESBLs). These include a confirmation test using both cefotaxime and ceftazidime tested alone and in combination with clavulanate (12). We report a study that was conducted to evaluate the diagnostic utility of the AmpC inhibitors 48-1220 (Ro 48-1220) (17) and LN-2-128 (4; J. D. Buynak and L. Vogeti, 24 July 2002, PCT Int. patent appl.) in seeking a simple test for detection of organisms producing plasmid-mediated AmpC β-lactamases, using methodology similar to the NCCLS guidelines for ESBL confirmation disk test. LN-2-128 and 48-1220 have a broad spectrum of inhibition and inhibit both class A (e.g., TEM and SHV) and class C (e.g., AmpC) β-lactamases (4, 18).
In this study, clinical and laboratory strains of K. pneumoniae or E. coli producing the following plasmid-mediated AmpC β-lactamases were used as positive controls: ACT-1, FOX-1, FOX-3, FOX-4, FOX-5, CMY-2, DHA-1, MIR-1, and MOX-1. A strain of Hafnia alvei was used due to the similarity of its chromosomal enzyme to the plasmid-mediated β-lactamase ACC-1. These strains (except for Misc416, PAB-CM30, and CDC2085) are clinical strains with well-characterized enzymes and produce additional β-lactamases, such as SHV-1 and TEM-1. The following clinical and laboratory strains were used as negative controls: K. pneumoniae porin mutant resistant to cefoxitin, K. oxytoca producing high levels of the K1 enzyme, and K. pneumoniae or E. coli producing the β-lactamases SHV-2, SHV-5, TEM-3, TEM-10, CTX-M-1, CTX-M-14, OXA-2, and KPC-1.
Inhibition zones were determined by NCCLS disk methodology (12) on Mueller-Hinton agar (Oxoid Ltd., Basingstoke, Hampshire, England). Antibiotic disks tested contained 30 μg of cefoxitin, 30 μg of cefotetan, 10 μg of cefpodoxime, 30 μg of ceftazidime, or 30 μg of cefotaxime (Becton Dickinson, Sparks, Md.) alone and in combination with 20 μg of either 48-1220 or LN-2-128. An increase of ≥4 mm in zone diameter in the presence of an inhibitor compared to when the antibiotic was tested alone was considered to be a positive test for the presence of plasmid-mediated AmpC β-lactamases.
Results of tests with the positive and negative control strains are shown in Tables 1 and 2, respectively. In tests with cefotetan, LN-2-128 yielded positive tests with all strains producing AmpC β-lactamases except MOX-1, ACC-1, and FOX-5, while 48-1220 yielded positive tests with all strains producing AmpC β-lactamases except ACT-1 and FOX-1 (Table 1). In tests with cefoxitin, LN-2-128 yielded positive tests with all strains producing AmpC β-lactamases except ACT-1, ACC-1, and MOX-1, while 48-1220 results were the same except the strain producing FOX-5 was also negative (Table 1). Negative control strains yielded negative tests with cefoxitin and cefotetan with both inhibitors except for the strain producing KPC-1 β-lactamase, a carbapenem-hydrolyzing enzyme. This strain yielded a positive test with cefotetan combined with LN-2-128 (Table 2). In tests with ceftazidime, LN-2-128 yielded positive tests with all strains producing AmpCs except ACT-1, FOX-1, and MOX-1, while all negative controls were negative. Cefotaxime and cefpodoxime with both inhibitors and ceftazidime with 48-1220 were too nonspecific for the detection of plasmid-mediated AmpC β-lactamases (Table 2). Representative examples of positive and negative inhibitor-based tests are illustrated in Fig. 1 and 2, respectively. The combination of cefotetan with LN-2-128 and cefotetan with 48-1220 showed a specificity of 90% and a sensitivity of 100% for the detection of organisms producing plasmid-mediated AmpC β-lactamases.
TABLE 1.
Results of inhibitor-based disk tests with positive control strains
| Strain | Organism | Resistance mechanism | Result with druga-inhibitor combination
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTT (LN)b | CTT (48)c | FOX (LN) | FOX (48) | CPD (LN) | CPD (48) | CTX (LN) | CTX (48) | CAZ (LN) | CAZ (48) | |||
| HVAMC39 | K. pneumoniae | ACT-1 | Pd | Ne | N | N | P | P | P | N | N | N |
| Misc304 | K. pneumoniae | MIR-1 | P | P | P | P | P | P | P | P | P | P |
| Misc340 | K. pneumoniae | FOX-1 | P | N | P | P | P | P | N | N | N | N |
| PAB-CM30 | E. coli | FOX-3 | P | P | P | P | P | P | P | P | P | P |
| Misc416 | E. coli | FOX-4 | P | P | P | P | P | P | P | P | P | P |
| CCF52 | K. pneumoniae | FOX-5 | N | P | P | N | P | P | N | N | P | P |
| Kleb249 | K. pneumoniae | CMY-2 | P | P | P | P | P | P | P | P | P | P |
| UMJMH14 | K. pneumoniae | DHA-1 | P | P | P | P | P | P | P | P | P | P |
| CDC2085 | H. alvei | ACC-1 | N | P | N | N | P | P | P | P | P | P |
| Misc339 | K. pneumoniae | MOX-1 | N | P | N | N | N | P | N | P | N | P |
Drugs tested: cefotetan (CTT), cefoxitin (FOX), cefpodoxime (CPD), cefotaxime (CTX), and ceftazidime (CAZ).
Inhibitor LN-2-128.
Inhibitor 48-1220.
P, positive test: increase of ≥4 mm in zone diameter in the presence of the inhibitor.
N, negative test: increase of ≤3 mm in zone diameter in the presence of the inhibitor.
TABLE 2.
Results of inhibitor-based disk tests with negative control strains
| Strain | Organism | Resistance mechanism | Result with druga-inhibitor combination
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTT (LN)b | CTT (48)c | FOX (LN) | FOX (48) | CPD (LN) | CPD (48) | CTX (LN) | CTX (48) | CAZ (LN) | CAZ (48) | |||
| Kleb67 | K. oxytoca | Hyper K1 | Nd | N | N | N | Pe | P | P | P | N | N |
| Kleb196 | K. pneumoniae | OMP | N | N | N | N | N | N | N | N | N | N |
| PAB-C14 | E. coli | SHV-2 | N | N | N | N | N | P | N | P | N | P |
| Kleb116 | K. pneumoniae | SHV-5 | N | N | N | N | N | P | P | P | N | P |
| PAB-C3 | E. coli | TEM-3 | N | N | N | N | N | P | P | P | N | P |
| PAB-C10 | E. coli | TEM-10 | N | N | N | N | N | P | N | P | N | P |
| Misc337 | E. coli | CTX-M-1 | N | N | N | N | N | P | P | P | N | N |
| Misc419 | E. coli | CTX-M-14 | N | N | N | N | P | P | N | P | N | N |
| PAB-C19 | E. coli | OXA-2 | N | N | N | N | N | N | N | N | N | P |
| Kleb265 | K. pneumoniae | KPC-1 | P | N | N | N | P | N | P | N | N | N |
Drugs tested: cefotetan (CTT), cefoxitin (FOX), cefpodoxime (CPD), cefotaxime (CTX), and ceftazidime (CAZ).
Inhibitor LN-2-128.
Inhibitor 48-1220.
N, negative test: increase of ≤3 mm in zone diameter in the presence of the inhibitor.
P, positive test: increase of ≥4 mm in zone diameter in the presence of the inhibitor.
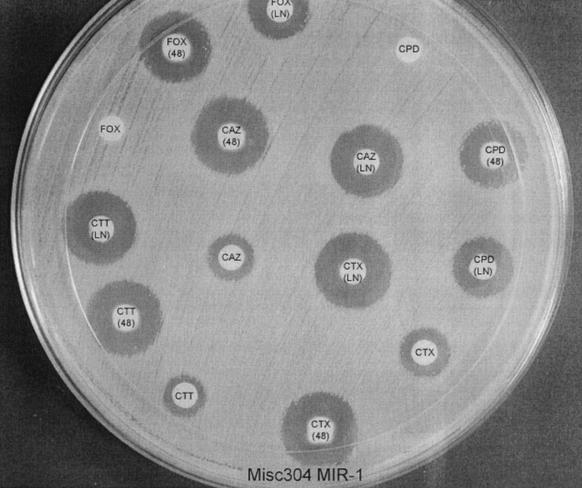
FIG. 1.
Inhibitor-based disk test with Misc 304 producing the β-lactamase MIR-1. CTT, cefotetan; CTT(LN), CTT plus LN-2-128; CTT(48), CTT plus 48-1220; FOX, cefoxitin; FOX(LN), FOX plus LN-2-128; FOX(48), FOX plus 48-1220; CTX, cefotaxime; CTX(LN), CTX plus LN-2-128; CTX(48), CTX plus 48-1220; CPD, cefpodoxime; CPD(LN), CPD plus LN-2-128; CPD(48), CPD plus 48-1220; CAZ, ceftazidime; CAZ(LN), CAZ plus LN-2-128; CAZ(48), CAZ plus 48-1220.
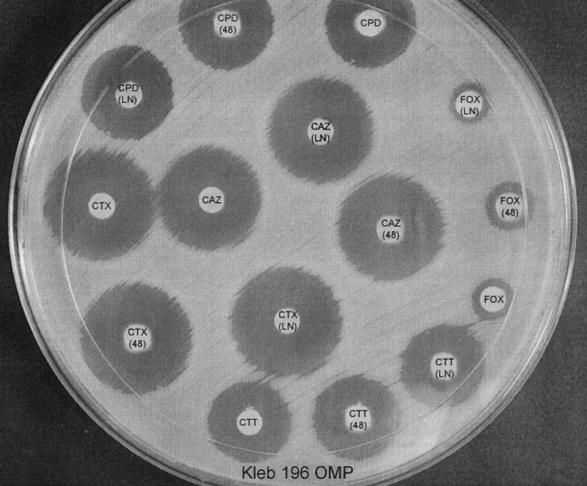
FIG. 2.
Inhibitor-based disk test with Kleb 196 OMP, a porin mutant resistant to cefoxitin. CTT, cefotetan; CTT(LN), CTT + LN-2-128; CTT(48), CTT plus 48-1220; FOX, cefoxitin; FOX(LN), FOX plus LN-2-128; FOX(48), FOX plus 48-1220; CTX, cefotaxime; CTX(LN), CTX plus LN-2-128; CTX(48), CTX plus 48-1220; CPD, cefpodoxime; CPD(LN), CPD plus LN-2-128; CPD(48), CPD plus 48-1220; CAZ, ceftazidime; CAZ(LN), CAZ plus LN-2-128; CAZ(48), CAZ plus 48-1220.
Current methods for detection of plasmid-mediated AmpC β-lactamases are technically demanding and time consuming and are therefore unsuitable for clinical laboratories to perform on a routine basis (1, 5, 6, 9, 20). For example, the three-dimensional method involves the use of a sterile scalpel blade to cut a slit in the agar, followed by the inoculation of a prepared enzyme extract into the slit (5, 6, 9), and the test can be difficult and subjective to interpret. Other detection methods are easier to perform but are still difficult to interpret (1, 20). The inhibitor-based tests are simple to perform and easy to interpret, having clear-cut guidelines for interpretation. This test is very similar to the NCCLS ESBL confirmation test (12). Therefore, it can be easily introduced into the routine workflow of the clinical laboratory.
It is important that none of the phenotypic tests (including the inhibitor-based test) described for the detection of AmpC β-lactamases can distinguish between E. coli strains producing plasmid-mediated β-lactamases and strains with chromosomal AmpC alterations (8). The inhibitor-based test does have the ability to distinguish K. pneumoniae strains that are cefoxitin resistant due to plasmid-mediated AmpC β-lactamases from strains with porin mutations (Fig. 1 and 2). This does have important infection control consequences (15).
Plasmid-mediated AmpC β-lactamases are a heterogeneous group of enzymes that originated from the chromosomal genes of bacteria such as Enterobacter, Citrobacter freundii, Morganella morganii, Aeromonas spp., and H. alvei (15). Therefore, it is very possible that they will behave differently to different β-lactamase inhibitors. Very limited research has been undertaken in this regard. This is most likely the reason for the negative results with some of the positive control strains. We included a variety of control strains with different types of plasmid-mediated AmpC β-lactamases in our initial study to evaluate if β-lactamase inhibitors will be able to inhibit most of these enzymes. In our study, the detection of organisms producing plasmid-mediated AmpC β-lactamases with inhibitor-based tests showed potential and should be further investigated.
Acknowledgments
We thank John D. Buynak (Southern Methodist University, Dallas, Tex.) and Pierre Weber (F. Hoffman-La Roche Ltd., Basel, Switzerland), respectively, for kindly providing the samples of LN-2-128 and 48-1220 that made this study possible.
REFERENCES
- 1.Barnaud, G., G. Arlet, C. Verdet, O. Gaillot, P. H. Lagrange, and A. Philippon. 1998. Salmonella enteritidis: AmpC plasmid-mediated inducible β-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob. Agents Chemother. 42:2352-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind, A., Y. Chong, and S. Schweighart. 1989. Extended broad-spectrum β-lactamase in Klebsiella pneumoniae including resistance to cephamycins. Infection 17:316-321. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC beta-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buynak, J. D., L. Vogeti, V. R. Doppalapudi, G. M. Solomon, and H. Chen. 2002. Cephalosporin-derived inhibitors of β-lactamase. Part 4: the C3 substituent. Bioorg. Med. Chem. Lett. 12:1663-1666. [DOI] [PubMed] [Google Scholar]
- 5.Coudron, P. E., E. S. Moland, and K. S. Thomson. 2000. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J. Clin. Microbiol. 38:1791-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coudron, P. E., N. D. Hanson, and M. W. Climo. 2003. Occurrence of extended-spectrum and AmpC beta-lactamases in bloodstream isolates of Klebsiella pneumoniae: isolates harbor plasmid-mediated FOX-5 and ACT-1 AmpC beta-lactamases. J. Clin. Microbiol. 41:772-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fey, P. D., T. J. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N. Engl. J. Med. 27:1242-1249. [DOI] [PubMed] [Google Scholar]
- 8.Hanson, N. D. 2003. AmpC β-lactamases: what do we need to know for the future? J. Antimicrob. Chemother. 52:2-4. [DOI] [PubMed] [Google Scholar]
- 9.Manchanda, V., and N. P. Singh. 2003. Occurrence and detection of AmpC β-lactamases among gram-negative clinical isolates using a modified three-dimensional test at Guru Tegh Bahadur Hospital, Delhi, India. J. Antimicrob. Chemother. 51:415-418. [DOI] [PubMed] [Google Scholar]
- 10.M'Zali, F. H., J. Heritage, D. M. Gascoyne-Binzi, M. Denton, N. J. Todd, and P. M. Hawkey. 1997. Transcontinental importation into the UK of Escherichia coli expressing a plasmid-mediated AmpC-type beta-lactamase exposed during an outbreak of SHV-5 extended-spectrum beta-lactamase in a Leeds hospital. J. Antimicrob. Chemother. 40:823-831. [DOI] [PubMed] [Google Scholar]
- 11.Nadjar, D., M. Rouveau, C. Verdet, J. Donay, J. Herrmann, P. H. Lagrange, A. Philippon, and G. Arlet. 2000. Outbreak of Klebsiella pneumoniae producing transferable AmpC-type beta-lactamase (ACC-1) originating from Hafnia alvei. FEMS Microbiol. Lett. 187:35-40. [DOI] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing; twelfth informational supplement. M100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Papanicolaou, G. A., A. A. Medeiros, and G. A. Jacoby. 1990. Novel plasmid-mediated beta-lactamase (MIR-1) conferring resistance to oxyimino- and alpha-methoxy beta-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 34:2200-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pérez-Pérez, F. J., and N. D. Hanson. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pitout, J. D., M. D. Reisbig, M. Mulvey, L. Chui, M. Louie, L. Crowe, D. L. Church, S. Elsayed, D. Gregson, R. Ahmed, P. Tilley, and N. D. Hanson. 2003. Association between handling of pet treats and infection with Salmonella enterica serotype Newport expressing the AmpC beta-lactamase, CMY-2. J. Clin. Microbiol. 41:4578-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richter, H. G. F., P. Angehrn, C. Hubschwerlen, M. Kania, M. G. P. Page, J. L. Specklin, and F. K. Winkler. 1996. Design, synthesis, and evaluation of 2 beta-alkenyl penam sulfone acids as inhibitors of beta-lactamases. J. Med. Chem. 39:3712-3722. [DOI] [PubMed] [Google Scholar]
- 18.Tzouvelekis, L. S., M. Gazouli, E. E. Prinarakis, E. Tzelepi, and N. J. Legakis. 1997. Comparative evaluation of the inhibitory activities of the novel pencillamic acid sulfone Ro 48-1220 against β-lactamases that belong to groups 1, 2b, and 2be. Antimicrob. Agents Chemother. 41:475-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yong, D., R. Park, J. H. Yum, K. Lee, E. C. Choi, and Y. Chong. 2002. Further modification of the Hodge test to screen AmpC β-lactamase (CMY-1)-producing strains of Escherichia coli and Klebsiella pneumoniae. J. Microbiol. Methods 51:407-410. [DOI] [PubMed] [Google Scholar]
- 21.Zansky, S., B. Wallace, D. Schoonmaker-Bopp, P. Smith, F. Ramsey, J. Painter, A. Gupta, P. Kalluri, and S. Noviello. 2002. From the Centers for Disease Control and Prevention. Outbreak of multidrug resistant Salmonella Newport—United States, January-April 2002. JAMA 288:951-953. [PubMed] [Google Scholar]