Abstract
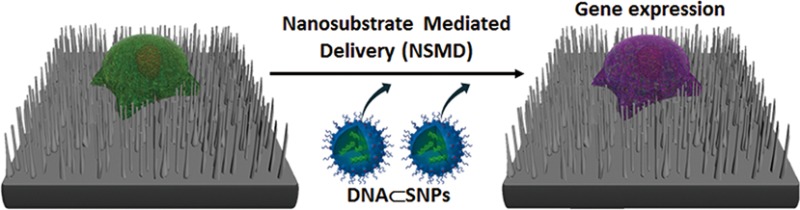
Substrate-mediated gene delivery is a promising method due to its unique ability to preconcentrate exogenous genes onto designated substrates. However, many challenges remain to enable continuous and multiround delivery of the gene using the same substrates without depositing payloads and immobilizing cells in each round of delivery. Herein we introduce a gene delivery system, nanosubstrate-mediated delivery (NSMD) platform, based on two functional components with nanoscale features, including (1) DNA⊂SNPs, supramolecular nanoparticle (SNP) vectors for gene encapsulation, and (2) Ad-SiNWS, adamantane (Ad)-grafted silicon nanowire substrates. The multivalent molecular recognition between the Ad motifs on Ad-SiNWS and the β-cyclodextrin (CD) motifs on DNA⊂SNPs leads to dynamic assembly and local enrichment of DNA⊂SNPs from the surrounding medium onto Ad-SiNWS. Subsequently, once cells settled on the substrate, DNA⊂SNPs enriched on Ad-SiNWS were introduced through the cell membranes by intimate contact with individual nanowires on Ad-SiNWS, resulting in a highly efficient delivery of exogenous genes. Most importantly, sequential delivery of multiple batches of exogenous genes on the same batch cells settled on Ad-SiNWS was realized by sequential additions of the corresponding DNA⊂SNPs with equivalent efficiency. Moreover, using the NSMD platform in vivo, cells recruited on subcutaneously transplanted Ad-SiNWS were also efficiently transfected with exogenous genes loaded into SNPs, validating the in vivo feasibility of this system. We believe that this nanosubstrate-mediated delivery platform will provide a superior system for in vitro and in vivo gene delivery and can be further used for the encapsulation and delivery of other biomolecules.
Keywords: nanosubstrate, self-assembled supramolecular nanoparticles, molecular motif, molecular recognition, gene delivery
Gene delivery constitutes one of the most critical steps in gene manipulation and therapy.1,2 It is generally recognized that an idealized delivery technology must meet certain criteria, including (1) general applicability for delivering a diverse range of gene drugs with high loading efficiency into a wide spectrum of cell types, (2) appropriate stability in a medium or circulation system, (3) high transfection efficiency at low toxicity, and (4) capability of continuous, multiple rounds of delivery while sustaining a steady supply of exogenous genes over the duration of desired biological processes (e.g., cell reprogramming3). The most effective vectors in use are mostly viral vectors owing to their practical advantages such as ease of construction, good production titer, and high transgene expression.4 However, the potential risks of viral vectors such as immunogenicity, inflammatory responses, and potential insertion mutagenesis limit their clinical use.5 By mimicking the size and function of viral vectors, numerous nonviral gene delivery systems based on biocompatible nanostructured materials,6−8 such as inorganic nanoparticles,9−12 carbon nanotubes,13 liposomes,14 cationic polymers,15−18 polypeptides,19 and dendrimers,20 have been developed to provide an alternative approach to the problems encountered in viral gene delivery.
Among the array of delivery approaches, substrate-mediated delivery is a promising method due to its unique ability to preconcentrate one or more exogenous genes onto the designated substrates and to enable localized delivery of these genes into cells that are immobilized on the substrates. Several techniques (e.g., charge interactions,21−23 layer-by-layer deposition,24−28 and self-assembly29−34) have been employed to deposit the genes onto the substrates and have already achieved improved transduction performances compared to those of a solution-based delivery approach. Recently, further improvement on substrate-mediated delivery has been achieved by incorporating nanofeatures (i.e., silicon nanowires, SiNW),35,36 which are capable of penetrating cell membranes, onto the substrates for direct delivery of a variety of exogenous genes already predeposited on the SiNW surfaces. However, due to the depletion of the predeposited payloads throughout the substrate-mediated delivery process, such a method is limited for single use only. Challenges remain to enable continuous and multiround delivery of genes using the same substrates without depositing payloads and immobilizing cells in each round of delivery.
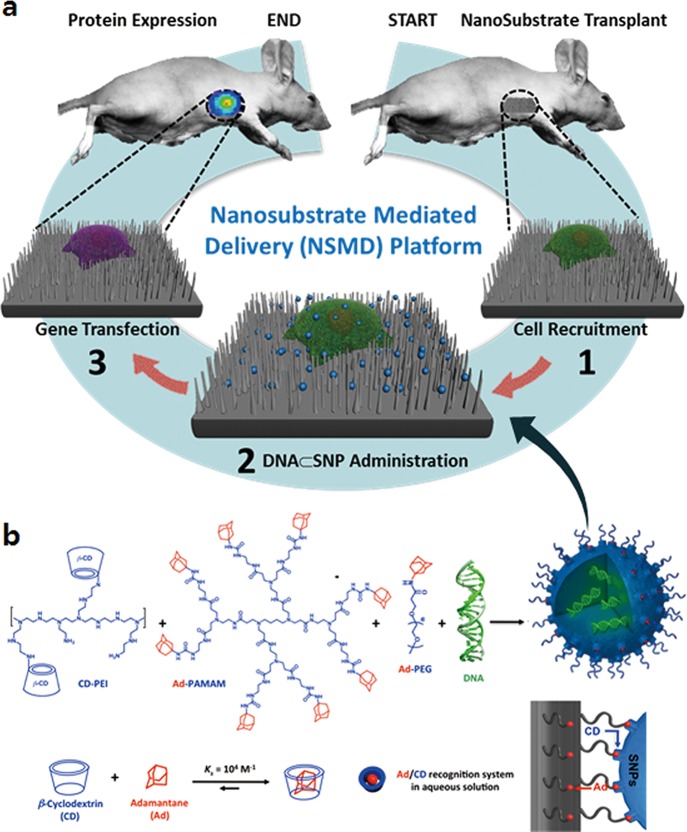
Herein, we introduce a new gene delivery system that centers around the use of two functional components with nanoscale features, including (1) DNA⊂SNPs, supramolecular nanoparticle (SNP) vectors37−43 for encapsulation of plasmid DNA, and (2) Ad-SiNWS, adamantane (Ad)-grafted silicon nanowire substrates44,45 that can mediate the transduction of DNA⊂SNPs from surrounding solution/medium into cells that settle on Ad-SiNWS. Figure 1a illustrates the working mechanism of this nanosubstrate-mediated delivery (NSMD) platform for both in vitro and in vivo settings. Multivalent molecular recognition between the Ad motifs on Ad-SiNWS and the CD motifs on the surfaces of DNA⊂SNPs leads to dynamic assembly and local enrichment of DNA⊂SNPs from the surrounding solution/medium onto Ad-SiNWS. Subsequently, once cells settle on the substrate, DNA⊂SNPs enriched on Ad-SiNWS can be introduced through the cell membranes via intimate contact35,36,46 with the individual nanowires on Ad-SiNWS. After the dynamic dissociation37,39 of DNA⊂SNPs (a reverse process of their initial dynamic assembly onto Ad-SiNWS) into the cytoplasm, highly efficient delivery of exogenous genes can be achieved. In contrast to the conventional substrate-mediated delivery platform, the unique mechanism of our NSMD approach allows for the repeated use of Ad-SiNWS for multiple rounds of delivery. Consequently, sequential delivery of multiple batches of exogenous genes can be accomplished by sequential additions of the corresponding DNA⊂SNPs, thus sustaining a steady supply of exogenous genes over the duration of the desired biological process.
Figure 1.
(a) Schematic illustration of the unique mechanism governing the nanosubstrate-mediated delivery (NSMD) approach for both in vivo and in vitro settings. (b) The multivalent molecular recognition between the Ad motifs on Ad-SiNWS and the β-cyclodextrin (CD) motifs on the surfaces of SNPs leads to dynamic assembly and local enrichment of SNPs onto Ad-SiNWS. The Ad/CD recognition system is also responsible for the supramolecular assembly of DNA⊂SNPs from the three molecular building blocks (i.e., CD-PEI, Ad-PAMAM, and Ad-PEG) and plasmid DNA.
Unlike other artificial delivery vectors, DNA⊂SNPs can be prepared by a convenient, flexible, and modular self-assembled synthetic approach from a collection of molecular building blocks [i.e., cyclodextrin (CD)-grafted branched polyethylenimine (CD-PEI), adamantane-grafted polyamidoamine dendrimer (Ad-PAMAM), and Ad-grafted poly(ethylene glycol) (Ad-PEG), as well as plasmid DNA] via a multivalent molecular recognition between Ad and CD motifs.36 This self-assembled synthetic strategy enables control over the sizes, surface chemistry, and payloads of SNP vectors for a wide range of diagnostic and therapeutic applications such as positron emission tomography (PET) imaging,37 magnetic resonance imaging (MRI),43 photothermal treatment,38 on-demand release of a drugs,47 and highly efficient delivery of genes,40,48,49 transcription factors,50 and drug-polymer conjugates.42 As for DNA-encapsulated SNPs (DNA⊂SNPs), Figure 1b illustrates the mechanism of the supramolecular synthetic strategy.37−43 CD-PEI and Ad-PAMAM first self-assemble via the cooperation of Ad/CD recognition motifs into cationic hydrogel networks that can encapsulate DNA plasmids, thus creating the cores of SNPs.39,40 The ligand module (Ad-PEG) then acts as a capping/solvation reagent that constrains continuous growth of the DNA-encapsulated hydrogel networks and simultaneously confers desired solubility, structural stability, and stealth (in circulatory system) to the resulting DNA⊂SNPs with controllable sizes of ca. 100 nm. Again, upon exposure of DNA⊂SNPs to Ad-SiNWS, the multivalent Ad/CD molecular recognition (Figures 1b and S2) induces local enrichment of DNA⊂SNPs onto Ad-SiNWS via dynamic exchange39,40 and results in the highly efficient delivery of exogenous genes.
Results and Discussion
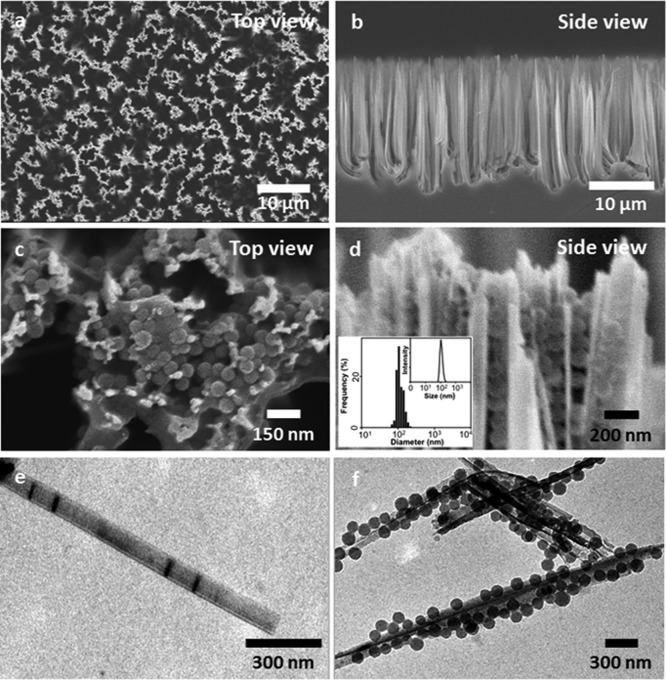
Scanning electron microscope (SEM) images of the Ad-SiNWS, which were prepared from wet-etching followed by covalent functionalization of Ad, showed that the diameters and lengths of Ad-SiNWS are ca. 100–200 nm and 15–20 μm, respectively (Figure 2a and b). The molecular mechanism that drives the local enrichment of DNA⊂SNPs from the solution onto Ad-SiNWS is unambiguously verified by SEM images of Ad-SiNWS and enhanced green fluorescent protein (EGFP) plasmid-encapsulated SNPs34,35 (pEGFP⊂SNPs)-grafted Ad-SiNWS (Figure 2c and d). The narrow size distribution (106 ± 5 nm) of the immobilized pEGFP⊂SNPs (characterized by SEM) agrees with that observed by dynamic light scattering (DLS, inset in Figure 2d) measurements. Further, Ad-SiNWS and pEGFP⊂SNPs-grafted Ad-SiNWS were released from the silicon substrates via sonication for transmission electron microscope (TEM) studies. Their morphology and sizes (Figure 2e and f) are consistent with those observed by SEM. The interactions between cells and Ad-SiNWS were also verified by SEM images (Figure S3, Supporting Information). Regardless of cell type, every cell successfully interacts on Ad-SiNWS as a result of gravitation and Brownian motion. In addition, no detectable difference in terms of cell morphologies was found before or after DNA⊂SNP treatment.
Figure 2.
(a and b) SEM images of the Ad-SiNWS, which were prepared from wet-etching followed by covalent functionalization of Ad. The diameters and lengths of Ad-SiNWS are ca. 100–200 nm and 15–20 μm, respectively. (c and d) Upon exposure of 100 nm pEGFP⊂SNPs in the solution/medium to Ad-SiNWS, the resulting pEGFP⊂SNPs-grafted Ad-SiNWS were examined by SEM. The narrow size distribution (106 ± 5 nm) of pEGFP⊂SNPs on Ad-SiNWS agrees with that observed by DLS measurements (inset d). (e and f) Free Ad-SiNWS and pEGFP⊂SNPs-grafted Ad-SiNWS were released from the substrates, and their morphology and sizes were further examined by TEM.
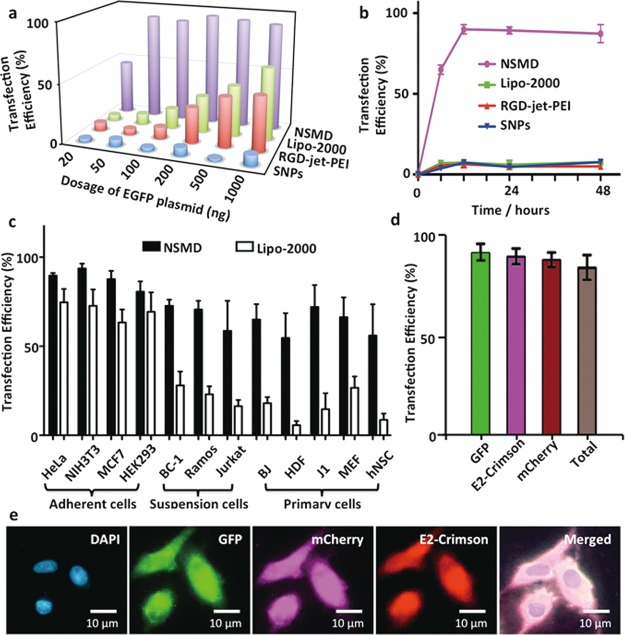
To examine the potency and performance of the NSMD platform for in vitro gene delivery, dose-dependent transfection studies were first conducted on Ad-SiNWS in the presence of pEGFP⊂SNPs34,35 (20–1000 ng of plasmid in 1.0 mL of Dulbecco’s modified Eagle’s medium (DMEM) medium/chamber). Ad-SiNWS (1 × 2 cm2) were placed in two-well chamber slides (Lab-Tek), where 5 × 104 U87 glioblastoma cells were introduced into each chamber. Three control studies were performed in parallel, two of which used commercially available transfection agents (i.e., Lipo-2000 and RGD-jet-PEI), and the third contained pEGFP⊂SNPs in the absence of Ad-SiNWS. The results (Figure 3a) indicated that the NSMD platform exhibited superb transfection performance (up to 95% transfection efficiency) even at a low dosage of EGFP plasmid (e.g., 50 ng/mL), significantly outperforming the two commercial agents. Subsequently, we conducted time-dependent transfection studies using pEGFP⊂SNPs (50 ng plasmid/mL), resulting in an optimal transfection efficiency (Figure 3b, >10-fold improvement than the controls) that was achieved 12 h postadministration of pEGFP⊂SNPs. It is noteworthy that the U87 cells transfected by the NSMD platform expressed GFP signals an average of 4.3 to 7.2 times stronger (Figure S4, Supporting Information) than those observed for U87 cells treated by Lipo-2000, RGD-jet-PEI, and pEGFP⊂SNPs alone. To test the general applicability of the NSMD platform, an array of mammalian cells (Figure 3c) was introduced onto the Ad-SiNWS in the presence of pEGFP⊂SNPs (50 ng plasmid/mL). The NSMD platform exhibited 70–98% transfection efficiencies among these mammalian cells, which is significantly higher than those observed for a Lipo-2000-based delivery system. Generally speaking, the majority of artificial gene delivery systems work well in adherent cell lines, yet challenges remain to reproduce the same performance for suspension and primary cells. As shown in Figure 3c, the NSMD platform exhibits similar performance for transfecting adherent cell lines, suspension cells, and a wide range of primary cells. To explore the feasibility of recycling Ad-SiNWS for multiple rounds of delivery, we repeatedly used the same Ad-SiNWS in more than 10 cycles of transfection studies without observing compromised delivery performance (Figure S5 in the Supporting Information). Finally, we examined continuous and multiround delivery of three different plasmids into the same batch of cells, which were immobilized on the same Ad-SiNWS. Three different DNA⊂SNPs (specifically encoding EGFP, mCherry, and E2-Crimson; 50 ng plasmid/mL) were introduced stepwise into culture medium at 12, 24, and 36 h post cell settlement onto Ad-SiNWS. Each new batch of DNA⊂SNPs competed with the remaining DNA⊂SNPs on the Ad-SiNWS via a dynamic assembly/exchange process, allowing the successive delivery of three genes. Individual genes were effectively delivered into and then expressed by the target cells (U87) without compromising sequential transfection performances. As shown in Figures 3d,e and S6, the U87 cell appears green, cherry, and red, which correspond to the highly efficient (>80%) expression of EGFP, mCherry, and E2-Crimson, respectively.
Figure 3.
(a) Dose-dependent transfection studies of NSMD platform and three control studies (i.e., Lipo-2000, RGD-jet-PEI, and pEGFP⊂SNPs in the absence of Ad-SiNWS) with U87 cells were performed in parallel. (b) Time-dependent transfection studies with U87 cells in the presence of pEGFP⊂SNPs (50 ng plasmid/mL); maximum transfection was achieved 12 h postadministration of pEGFP⊂SNPs. (c) A diversity of mammalian cells, including adherent cells (i.e., HeLa, NIH3T3, MCF7, and HEK293), suspension cells (i.e., BC-1, Ramos, and Jurkat), and primary cells (i.e., BJ, HDF, J1, MEF, and hNSC), were introduced onto the NSMD platform in the presence of pEGFP⊂SNPs (50 ng plasmid/mL). NSMD exhibited 70–98% transfection efficiency among different mammalian cells, significantly higher than those observed for a Lipo-2000-based delivery system at similar experimental conditions. (d) A single batch of U87 cells on Ad-SiNWS were sequentially treated by three different SNP vectors with encapsulated DNA plasmids (specifically encoding EGFP, mCherry, and E2-Crimson: 50 ng plasmid/mL) at 12, 24, and 36 h post cell settlement. (e) Micrographs show that each individual gene can be effectively delivered into and then expressed by the cells with superb transfection performance.
To examine how individual features of Ad-SiNWS contribute to the transfection performance of the NSMD platform, three control studies were conducted using flat Si chips (Siflat, Figure 4a), Ad-coated Si (Ad-Siflat, Figure 4b) chips, and SiNWS without Ad coating (SiNW, Figure 4c). These experiments were then examined with Ad-SiNWS (Ad-SiNW, Figure 4d) in the presence of pGFP-loaded SNPs (50 ng plasmid/mL). The fluorescence microscopy images of U87 cells, shown in Figure 4, suggest that both the Ad recognition motif and SiNWS play indispensable roles in achieving enhanced delivery performance. The results of time-dependent Cy5-labeled DNA⊂SNPs taken up by U87 cells on these different silicon substrates also confirmed this conclusion (Figures S7, S8, and S9, Supporting Information). Two possible mechanisms (penetration35,36vs nonpenetration46) have been proposed in the current literature about how nanostructured substrates mediate the delivery of the predeposited payloads. According to our current experimental data, neither one can be ruled out. To examine the toxicity of the NSMD platform, the MTT assay was conducted and indicated that the NSMD-based gene delivery platform exhibited negligible disruption to cell viability (see Figure S10 in the Supporting Information).
Figure 4.
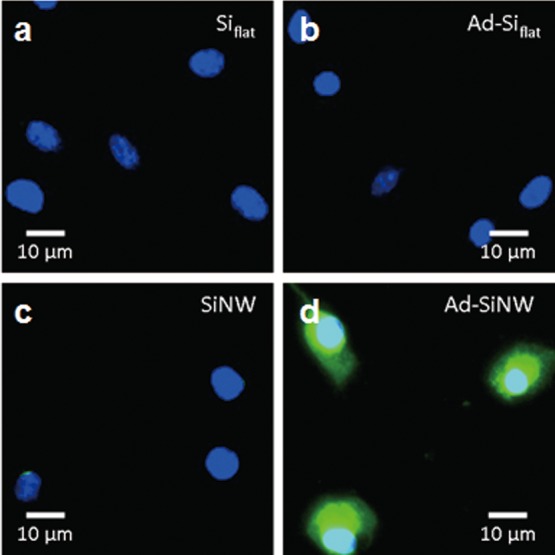
Side-by-side comparison of the transfection studies observed for four categories of substrates including (a) Siflat, (b) Ad-Siflat, (c) SiNWS without Ad coating, and (d) Ad-SiNWS. The nucleus of U87 cells is dyed with DAPI (blue), and expressed GFP is recognized as the green color. As shown in the micrograph images, both the Ad recognition motif and SiNWS play indispensable roles in achieving the enhanced transfection performance of the NSMD platform.
To further test the application potential and performance of the NSMD platform for in vivo gene delivery, the Ad-SiNWS were subcutaneously transplanted into one side of the mouse body (opposite side with no Ad-SiNWS transplanted was used as control) followed by local injection of pGL3 luciferase reporter vectors-encapsulated SNPs (pGL3⊂SNPs, 200 ng plasmid/site) 2 weeks later (to allow in vivo integration). The expressed luciferase was detected at designated times using optical imaging, and the results presented in Figures 5 and S11 show that no significant bioluminescence signals were found at the site without Ad-SiNW (control) or the site with Ad-Siflat, while the Ad-SiNWS sites displayed high bioluminescence intensity levels after 1 day of the pGL3⊂SNP administration. The signals reached their peak by the second day, then steadily declined and eventually reached a natural background level by the ninth day. These results suggest that the subcutaneously transplanted Ad-SiNWS also have the potential to improve the in vivo transfection efficiency of exogenous genes encapsulated in SNPs.
Figure 5.
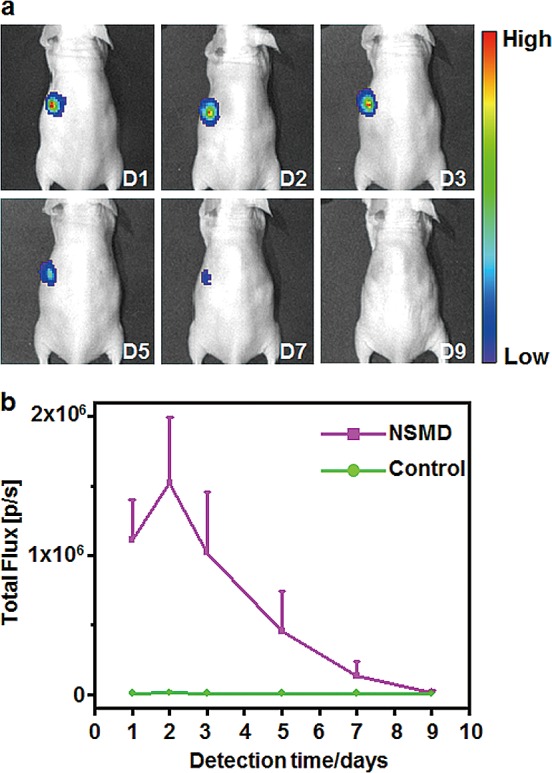
(a) NSMD approach for in vivo delivery of luciferase-encoded plasmid. Optical imaging of luciferase expression was performed in a mouse treated with the in vivo NSMD platform in the presence of luciferase reporter plasmid-encapsulated SNPs (pGL3⊂SNPs; 200 ng/site) at designated time points. The in vivo NSMD study exhibited high expression of pGL3, and a maximum luciferase signal was achieved 48 h postadministration of pGL3⊂SNPs. (b) Continuous monitoring of the luciferase expression in in vivo NSMD-treated mice along with control studies (w/o Ad-SiNWS). The luciferase levels in the NSMD study were significantly higher than those of the control group and peaked at 48 h postadministration of pGL3⊂SNPs.
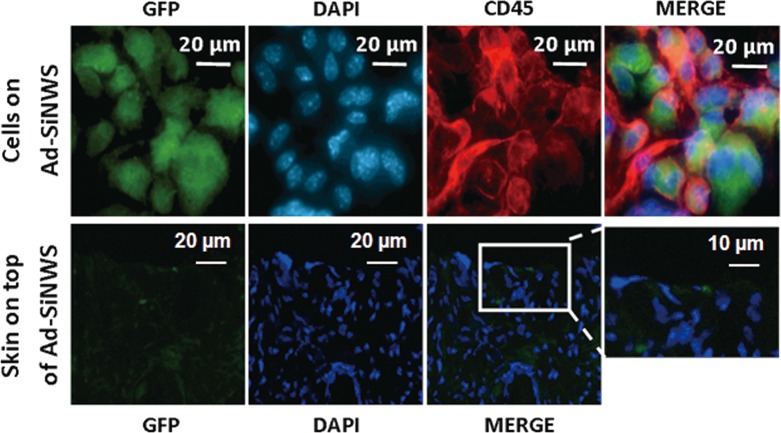
In order to clarify the details of in vivo Ad-SiNWS-mediated DNA⊂SNPs transfection, another group of mice treated with Ad-SiNWS and pEGFP⊂SNPs as mentioned above was sacrificed 2 days after pEGFP⊂SNP administration. Subsequently, the Ad-SiNWS and the skin on top of the Ad-SiNWS were collected for immunostaining and microscopy assay. The fluorescence microscopy images showed that a large number of cells, mostly composed of CD45-positive cells,51 assembled onto the Ad-SiNWS (Figure 6). This may be due to the Ad-SiNWS, a foreign substance, activating the host’s immune system and then recruiting the immune cells onto their surfaces. When administered, the pEGFP⊂SNPs dynamically assembled and locally enriched onto the Ad-SiNWS, while being protected from lymphatic return.52 As we predicted, the cells recruited on Ad-SiNWS were efficiently transfected with pEGFP and showed high GFP fluorescence signals (Figure 6). Meanwhile, no significant fluorescence was detected on the skin tissue above the Ad-SiNWS, suggesting that the skin cells on top of Ad-SiNWS were not transfected with pEGFP in the process of in vivo NSMD of pEGFP (Figure 6). Transfecting immune cells with the NSMD platform might open up additional research opportunities in vaccination and immunotherapy in the future.
Figure 6.
Ex vivo fluorescence imaging of cells located on the Ad-SiNWS and skin tissue on top of the Ad-SiNWS after in vivo NSMD of pEGFP. The micrographs indicated that the immune cells (CD45 positive) were recruited onto the Ad-SiNWS and were transfected with pEGFP⊂SNPs. The cells on the skin tissue section, however, showed no significant fluorescence signals, suggesting that the skin cells on top of Ad-SiNWS were not transfected with pEGFP in the process of the in vivo NSMD study.
Conclusions
To summarize, we have developed an Ad-SiNWS and DNA⊂SNPs based nanosubstrate-mediated delivery platform capable of introducing exogenous genes into an array of mammalian cells with profound performance including high transduction efficiency, low toxicity, wide cell-type adaptability, capability of sequential delivery that sustains a steady supply of genes, and in vivo availability. We believe that this platform will provide a superior system for in vitro and in vivo gene delivery and can be further used for gene therapy applications such as DNA vaccination, cell reprogramming, RNA interference, and many more.
Methods Section
Materials
Reagents and solvents were purchased from Sigma-Aldrich (St. Louis, MO, USA) and were used as received without further purification unless otherwise noted. Silicon wafers ((100) oriented, p-type, resistivity of ca. 10–20 Ωcm) were obtained from Silicon Quest Int. Two-well Lab-Tek chamber slides were purchased from Thermo Fisher Scientific. Branched polyethylenimine (MW = 10 kDa) was purchased from Polysciences Inc. (Washington, PA, USA). Polymers contain primary, secondary, and tertiary amine groups in approximately a 25/50/25 ratio. First-generation polyamidoamine dendrimer (PAMAM) with 1,4-diaminobutane core and amine terminals in a 20 wt % methanol solution was purchased from Dendritic Nanotechnologies, Inc. (Mount Pleasant, MI, USA). 1-Adamantane hydrochloride and β-cyclodextrin were purchased from TCI America (San Francisco, CA, USA). Phosphate-buffered saline (PBS, 1×, pH 7.2 ± 0.05) was used for sample preparation. 6-Monotosyl-β-cyclodextrin (6-OTs-β-CD) was prepared following the literature-recommended method.53 Octa-Ad-grafted polyamidoamine dendrimer, CD-grafted branched polyethylenimine, and Ad-grafted poly(ethylene glycol) were prepared by the same methods reported previously.32 Dry CH2Cl2 was obtained by refluxing over CaH2 and freshly distilled before use. NIH 3T3 (mouse embryonic fibroblast cell line), U87 (human brain glioblastoma cell line), HeLa (human cervix epithelial carcinoma cells), MCF7 (human breast adenocarcinoma cells), HEK293 (human embryonic kidney cells), BC-1 (lymphoma cells), Ramos (human Burkitt’s lymphoma cells), and Jurkat (human T cell lymphoblast-like cells) were purchased from American Type Culture Collection (ATCC). The Dulbecco’s modified Eagle’s medium, Earl’s modified Eagle’s medium growth medium, and penicillin/streptomycin were obtained from Invitrogen (Carlsbad, CA, USA). Fetal bovine serum and EGFP-encoded plasmid (pMAX EGFP, 4.3 kb) were obtained from Lonza Walkerrsville Inc. (Walkerrsville, MD, USA). mCherry-encoded plasmid (4.7 kb) and E2-Crimson-encoded plasmid (4.6 kb) were purchased from Clontech Laboratories Inc. (Mountain View, CA, USA). Cy5TM monofunctional dye (Cy5-NHS) was purchased from GE Healthcare. pGL3-control plasmid encoding firefly luciferase, d-luciferin, and CellTitra Blue cell viability kit were purchased from Promega Corporation (Madison, WI, USA). BALB/c nude mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). Purified mouse anti-CD45 was obtained from BD Biosciences (San Jose, CA, USA). Alexa Fluor 647 donkey anti-mouse IgG was purchased from Life Technologies (Carlsbad, CA, USA).
Preparation of Adamantane-Grafted Silicon Nanowire Substrates (Ad-SiNWS)
We fabricated SiNWS via a wet chemical etching process. First, the surface of the silicon substrate was made hydrophilic according to the following procedure: the silicon wafer was ultrasonicated in acetone and ethanol at room temperature for 10 and 5 min, respectively, to remove contamination from organic grease. Then, the degreased silicon substrate was heated in boiling piranha solution (4:1 (v/v) H2SO4/H2O2) and RCA solution (1:1:5 (v/v/v) NH3/H2O2/H2O) each for 1 h. Subsequently, the silicon substrate was rinsed several times with deionized water. Then, the clean silicon substrate was used in a wet chemical etching process. An etching mixture consisting of deionized water, 4.6 M HF, and 0.2 M silver nitrate was used at room temperature. The etching duration was dependent upon the required length of the nanowires. After etching, the substrate was immersed in boiling aqua regia (3:1 (v/v) HCl/HNO3) for 15 min to remove the silver film. Finally, the substrate was rinsed with DI water and dried under nitrogen and was then ready for surface modification. The surface modifications of the SiNWS and Siflat were processed with 4% (v/v) 3-aminopropyl trimethoxysilane in ethanol at room temperature for 45 min. Then, the Siflat and SiNWS were treated with the 1-adamantane isocyanate (1.0 mM) in DMSO for 30 min. The modified Siflat and SiNWS were then washed with DMSO twice to remove excess 1-adamantane isocyanate. The substrates were rinsed with DI water three times and stored at 4–8 °C before cell seeding.
Treatment of Cells with DNA⊂SNPs Using NSMD Platform
Synthesis of Plasmid DNA-Encapsulated Supramolecular Nanoparticles (DNA⊂SNPs)
A self-assembly procedure was employed to achieve the plasmid DNA-encapsulated supramolecular nanoparticles (DNA⊂SNPs). A 2.0 μL amount of DMSO solution containing Ad-PAMAM (3.96 μg) was added into a 600 μL PBS mixture with DNA, Ad-PEG (10.56 μg), and CD-PEI (9.0 μg) under vigorous stirring. The mixture was kept at 4 °C for 30 min, yielding the pEGFP⊂SNPs with a size of 106 ± 5 nm.
Experimental Procedure of Cell Treatment with DNA⊂SNPs Using NSMD Platform
A total of 5 × 104 target cells were introduced into each well of a two-well chamber slide (Lab-Tek), in which a 1 × 2 cm2 Ad-SiNWS was placed in the bottom of the chamber. After the cells fully attached onto Ad-SiNWS (for 12 h), the chambers were washed with PBS and refilled with fresh cell culture medium (DMEM). Then, different quantities of 100 nm DNA⊂SNPs (containing 20 ng to 1 μg of plasmid DNA) were introduced into individual chambers and co-incubated with attached cells for the designated time. After the chamber was washed with PBS, the cells were immediately fixed with 2% PFA and stained with DAPI. Transfection performances of individual conditions were quantified using microscopy-based image cytometry.
Characterization Methods and Settings
Dynamic Light Scattering
DLS experiments were performed with a Zetasizer Nano instrument (Malvern Instruments Ltd., United Kingdom) equipped with a 10 mW helium–neon laser (λ = 632.8 nm) and a thermoelectric temperature controller. Measurements were taken at a 90° scattering angle. The hydrodynamic size of the DNA⊂SNPs was measured by using DLS. The sizes and the standard derivations were obtained by averaging the values of three or more measurements.
Transmission Electron Microscopy
The morphology and sizes of Ad-SiNWS and DNA⊂SNP-enriched Ad-SiNWS were examined using a transmission electron microscope. The studies were carried out on a Philips CM 120 electron microscope, operating at an acceleration voltage of 120 kV. The TEM samples were prepared by drop-coating 2 μL of sample suspension solutions onto carbon-coated copper grids. Excess amounts of solution were removed by filter papers after 45 s. Subsequently, the samples were negatively stained with 2% uranyl acetate for 45 s before TEM studies.
Fluorescent Microscopy, Imaging Processing, and Data Analysis
The Ad-SiNWS chip was mounted onto a Nikon TE2000S inverted fluorescent microscope with a CCD camera (QImaging, Retiga 4000R), X-Cite 120 Mercury lamp, automatic stage, and filters for five fluorescent channels (W1: 325–375 nm, W2: 465–495 nm, W3: 570–590 nm, W4: 590–650 nm, and W5: 650–900 nm). Following image acquisition, MetaMorph (Molecular Devices, version 7.5.6.0) was used to quantify the cells’ EGFP expression and their mCherry and Cy5 fluorescence intensity. The multiwavelength cell scoring module of the Metamorph software allows for image analysis. A cell-counting application in the module allowed us to calculate the total number of cells. In order to determine the gene transfection efficiency, the EGFP-expressed cells were counted with the same exposure time (0.2 s) by the MetaMorph program, which distinguishes the transfected cells from the nontransfected cells. The background of each image is selected with the spot without cells, and then its intensity is compared to the spot with cells. The autofluorescence signals in nontransfected cells are also quantified using the aforementioned protocol. Cells with an EGFP fluorescence intensity 4 times higher than the background are recognized as transfected cells (Figure S1). The gene transfection efficiency was defined as the EGFP-expressed cell number divided by the total cell number.
In Vivo NSMD of Plasmid DNA Encapsulated in Supramolecular Nanoparticles
Six- to 8-week-old female BALB/c nude mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). All animal manipulations were performed with sterile technique and were approved by the University of California at Los Angeles Animal Research Committee. After the mice were anesthetized with 2% isoflurane in a heated (30 °C) induction chamber, the Ad-SiNWS were subcutaneously transplanted into one side of the mouse’s body. The symmetrically opposite side with no Ad-SiNWS transplanted was used as a control. Two weeks after Ad-SiNWS transplantation, pGL3⊂SNPs (200 ng pGL3/site) were administered via local subcutaneous injection. The luciferase expressed in the mice was then measured using in vivo optical imaging at designated time points. After the mice were anesthetized, 2 mg of d-luciferin in 200 μL of PBS solution was administered into the mice by tail vein injection, and then bioluminescence imaging was acquired approximately 10 min after luciferin administration on an IVIS Lumina II optical system (Caliper Life Science, MA, USA).
Ex Vivo Fluorescence Imaging of Cells and Skin Tissues Located on Ad-SiNWS after in Vivo NSMD of pEGFP
Another group of mice, treated the same as described above in the in vivo study, was sacrificed 2 days after pEGFP⊂SNP administration. The Ad-SiNWS and the skin on top of the Ad-SiNWS were subsequently collected for immunostaining and microscopy assay. Cells on Ad-SiNWS were fixed with 4% paraformaldehyde and blocked with 5% BSA before incubation with the mouse anti-CD45 (1:200) at room temperature for 2 h. Subsequently, cells were incubated with Alexa Fluor 647 donkey anti-mouse IgG (1:1000) for 1 h at room temperature. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) for 10 min. Cells were then examined using a TE2000S inverted fluorescent microscope (Nikon, Japan). The GFP fluorescence on skin sections was assayed using fluorescent microscopy after DAPI staining.
Acknowledgments
This research was supported by the National Institutes of Health through two research grants (R21 GM098982 and R21 EB016270 to H.R.T.). J.S.C. acknowledges a postdoctoral fellowship from the Basic Science Research Program with the National Research Foundation in Korea (2013R1A6A3A03024902). A.D.P. was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R01AR064327.
The authors declare no competing financial interest.
Supporting Information Available
Determination of gene-transfection of cells, control experiments of the host guest recognition of adamantane and cyclodextrin, SEM study on the interaction between cells and SINWS, intensities of EGFP transfected by different platforms, transfection performance of NSMD for repeatedly treatment, gene transfection of cell with various types of DNA⊂SNPs, time-dependent transduction of DNA⊂SNPs into cells on different substrates, cell viability assay, and in vivo gene expression test with Ad-modified flat Si chip. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Glover D. J.; Lipps H. J.; Jans D. A. Towards Safe, Non-Viral Therapeutic Gene Expression in Humans. Nat. Rev. Genet. 2005, 6, 299–310. [DOI] [PubMed] [Google Scholar]
- Kim D. H.; Rossi J. J. Strategies for Silencing Human Disease Using RNA Interference. Nat. Rev. Genet. 2007, 8, 173–184. [DOI] [PubMed] [Google Scholar]
- Aoi T.; Yae K.; Nakagawa M.; Ichisaka T.; Okita K.; Takahashi K.; Chiba T.; Yamanaka S. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science 2008, 321, 699–702. [DOI] [PubMed] [Google Scholar]
- Kay M. A.; Glorioso J. C.; Naldini L. Viral Vectors for Gene Therapy: The Art of Turning Infectious Agents into Vehicles of Therapeutics. Nat. Med. 2001, 7, 33–40. [DOI] [PubMed] [Google Scholar]
- Thomas C. E.; Ehrhardt A.; Kay M. A. Progress and Problems with the Use of Viral Vectors for Gene Therapy. Nat. Rev. Genet. 2003, 4, 346–358. [DOI] [PubMed] [Google Scholar]
- Davis M. E.; Chen Z. G.; Shin D. M. Nanoparticle Therapeutics: An Emerging Treatment Modality for Cancer. Nat. Rev. Drug Discovery 2008, 7, 771–782. [DOI] [PubMed] [Google Scholar]
- Niemeyer C. M. Nanoparticles, Proteins, and Nucleic Acids: Biotechnology Meets Materials Science. Angew. Chem., Int. Ed. 2001, 40, 4128–4158. [DOI] [PubMed] [Google Scholar]
- Ferrari M. Cancer Nanotechnology: Opportunities and Challenges. Nat. Rev. Cancer 2005, 5, 161–171. [DOI] [PubMed] [Google Scholar]
- De M.; Ghosh P. S.; Rotello V. M. Applications of Nanoparticles in Biology. Adv. Mater. 2008, 20, 4225–4241. [Google Scholar]
- Nie S.; Xing Y.; Kim G. J.; Simons J. W. Nanotechnology Applications in Cancer. Annu. Rev. Biomed. Eng. 2007, 9, 257–288. [DOI] [PubMed] [Google Scholar]
- Torney F.; Trewyn B. G.; Lin V. S. Y.; Wang K. Mesoporous Silica Nanoparticles Deliver DNA and Chemicals into Plants. Nat. Nanotechnol. 2007, 2, 295–300. [DOI] [PubMed] [Google Scholar]
- Rosi N. L.; Giljohann D. A.; Thaxton C. S.; Lytton-Jean A. K. R.; Han M. S.; Mirkin C. A. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science 2006, 312, 1027–1030. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Cai W.; He L.; Nakayama N.; Chen K.; Sun X.; Chen X.; Dai H. In Vivo Biodistribution and Highly Efficient Tumour Targeting of Carbon Nanotubes in Mice. Nat. Nanotechnol. 2007, 2, 47–52. [DOI] [PubMed] [Google Scholar]
- Tseng Y. C.; Mozumdar S.; Huang L. Lipid-Based Systemic Delivery of siRNA. Adv. Drug Delivery Rev. 2009, 61, 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Wagner E. Bioresponsive Polymers for Non-Viral Gene Delivery. Curr. Opin. Mol. Ther. 2009, 11, 165–178. [PubMed] [Google Scholar]
- Woodrow K. A.; Cu Y.; Booth C. J.; Saucier-Sawyer J. K.; Wood M. J.; Saltzman W. M. Intravaginal Gene Silencing Using Biodegradable Polymer Nanoparticles Densely Loaded with Small-Interfering RNA. Nat. Mater. 2009, 8, 526–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow W. Y.; Yang Y. Y. Functional Polycarbonates and Their Self-Assemblies as Promising Non-Viral Vectors. J. Controlled Release 2009, 139, 40–47. [DOI] [PubMed] [Google Scholar]
- Yu H. J.; Zou Y.; Jiang L.; Yin Q.; He X.; Chen L.; Zhang Z.; Gu W.; Li Y. Induction of Apoptosis in Non-Small Cell Lung Cancer by Downregulation of MDM2 Using pH-Responsive PMPC-b-PDPA/siRNA Complex Nanoparticles. Biomaterials 2013, 34, 2738–2747. [DOI] [PubMed] [Google Scholar]
- Gabrielson N.; Lu H.; Yin L.; Li D.; Wang F.; Cheng J. Reactive and Bioactive Cationic g-Helical Polypeptide Template for Non-Viral Gene Delivery. Angew. Chem., Int. Ed. 2012, 51, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W. D.; Selim K. M. K.; Lee C. H.; Kang I. K. Bioinspired Application of Dendrimers: From Biomimicry to Biomedical Applications. Prog. Polym. Sci. 2009, 34, 1–23. [Google Scholar]
- Zelikin A. N.; Lynn D. M.; Farhadi J.; Martin I.; Shastri V.; Langer R. Erodible Conducting Polymers for Potential Biomedical Applications. Angew. Chem., Int. Ed. 2002, 41, 141–144. [DOI] [PubMed] [Google Scholar]
- Zhang J. T.; Lynn D. M. Ultrathin Multilayered Films Assembled from “Charge-Shifting” Cationic Polymers: Extended, Long-Term Release of Plasmid DNA from Surfaces. Adv. Mater. 2007, 19, 4218–4223. [Google Scholar]
- Jewell C. M.; Lynn D. M. Surface-Mediated Delivery of DNA: Cationic Polymers Take Charge. Curr. Opin. Colloid Interface Sci. 2008, 13, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell C. M.; Lynn D. M. Multilayered Polyelectrolyte Assemblies as Platforms for the Delivery of DNA and Other Nucleic Acid-Based Therapeutics. Adv. Drug Delivery Rev. 2008, 60, 979–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Zhang J.; Lynn D. M. Ultrathin Multilayered Films that Promote the Release of Two DNA Constructs with Separate and Distinct Release Profiles. Adv. Mater. 2008, 20, 4148–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurer E. M.; Flessner R. M.; Sullivan S. P.; Prausnitz M. R.; Lynn D. M. Layer-by-Layer Assembly of DNA- and Protein-Containing Films on Microneedles for Drug Delivery to the Skin. Biomacromolecules 2010, 11, 3135–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flessner R. M.; Yu Y.; Lynn D. M. Rapid Release of Plasmid DNA from Polyelectrolyte Multilayers: A Weak Poly(acid) Approach. Chem. Commun. 2011, 47, 550–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aytar B. S.; Prausnitz M. R.; Lynn D. M. Rapid Release of Plasmid DNA from Surfaces Coated with Polyelectrolyte Multilayers Promoted by the Application of Electrochemical Potentials. ACS Appl. Mater. Interfaces 2012, 4, 2726–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea L. D.; Smiley E.; Bonadio J.; Mooney D. J. DNA Delivery from Polymer Matrices for Tissue Engineering. Nat. Biotechnol. 1999, 17, 551–554. [DOI] [PubMed] [Google Scholar]
- Shen H.; Tan J.; Saltzman W. M. Surface-Mediated Gene Transfer from Nanocomposites of Controlled Texture. Nat. Mater. 2004, 3, 569–574. [DOI] [PubMed] [Google Scholar]
- Salvay D. M.; Zelivyanskaya M.; Shea L. D. Gene Delivery by Surface Immobilization of Plasmid to Tissue-Engineering Scaffolds. Gene Ther. 2010, 17, 1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Diaz L. G.; Nandivada H.; Ding J.; Nogueira-De-Souza N. C.; Krebsbach P. H.; O’Shea K. S.; Lahann J.; Smith G. D. Synthetic Polymer Coatings for Long-Term Growth of Human Embryonic Stem Cells. Nat. Biotechnol. 2010, 28, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. H. K.; Pun S. H. Substrate-Mediated Nucleic Acid Delivery from Self-Assembled Monolayers. Trends Biotechnol. 2011, 29, 119–126. [DOI] [PubMed] [Google Scholar]
- Wu C. C.; Xu H.; Otto C.; Reinhoudt D. N.; Lammertink R. G. H.; Huskens J.; Subramaniam V.; Velders A. H. Porous Multilayer-Coated AFM Tips for Dip-Pen Nanolithography of Proteins. J. Am. Chem. Soc. 2009, 131, 7526–7527. [DOI] [PubMed] [Google Scholar]
- Kim W.; Ng J. K.; Kunitake M. E.; Conklin B. R.; Yang P. D. Interfacing Silicon Nanowires with Mammalian Cells. J. Am. Chem. Soc. 2007, 129, 7228–7229. [DOI] [PubMed] [Google Scholar]
- Shalek A. K.; Robinson J. T.; Karp E. S.; Lee J. S.; Ahn D.-R.; Yoon M.-H.; Suttona A.; Jorgollic M.; Gertnera R. S.; Gujrala T. S.; et al. Vertical Silicon Nanowires as a Universal Platform for Delivering Biomolecules into Living Cells. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 1870–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Wang S.; Su H.; Chen K. J.; Armijo A. L.; Lin W. Y.; Wang Y.; Sun J.; Kamei K.; Czernin J.; et al. A Supramolecular Approach for Preparation of Size-Controlled Nanoparticles. Angew. Chem. 2009, 48, 4344–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Chen K. J.; Wu T. H.; Wang H.; Lin W. Y.; Ohashi M.; Chiou P. Y.; Tseng H. R. Photothermal Effects of Supramolecularly Assembled Gold Nanoparticles for the Targeted Treatment of Cancer Cells. Angew. Chem., Int. Ed. 2010, 49, 3777–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Liu K.; Chen K.-J.; Lu Y.; Wang S.; Lin W.-Y.; Guo F.; Kamei K.-i.; Chen Y.-C.; Ohash M.; et al. A Rapid Pathway toward a Superb Gene Delivery System: Programming Structural and Functional Diversity into a Supramolecular Nanoparticle Library. ACS Nano 2010, 4, 6235–6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Chen K. J.; Wang S.; Ohashi M.; Kamei K.; Sun J.; Ha J. H.; Liu K.; Tseng H. R. A Small Library of DNA-Encapsulated Supramolecular Nanoparticles for Targeted Gene Delivery. Chem. Commun. 2010, 46, 1851–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Wang H.; Kamei K.; Yan M.; Chen K. J.; Yuan Q.; Shi L.; Lu Y.; Tseng H. R. Delivery of Intact Transcription Factor by Using Self-assembled Supramolecular Nanoparticles. Angew. Chem., Int. Ed. 2011, 50, 3058–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. J.; Tang L.; Garcia M. A.; Wang H.; Lu H.; Lin W. Y.; Hou S.; Yin Q.; Shen C. K.; Cheng J.; et al. The Therapeutic Efficacy of Camptothecin-Encapsulated Supramolecular Nanoparticles. Biomaterials 2012, 33, 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. J.; Wolahan S. M.; Wang H.; Hsu C. H.; Change H.-W.; Durazob A.; Hwang L.-P.; Garcia M. A.; Jiang Z. K.; Wu L.; et al. A Small MRI Contrast Agent Library of Gadolinium(III)-Encapsulated Supramolecular Nanoparticles for Improved Relaxivity and Sensitivity. Biomaterials 2011, 32, 2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K. Q.; Yan Y. J.; Gao S. P.; Zhu J. Synthesis of Large-Area Silicon Nanowire Arrays via Self-Assembling Nanoelectrochemistry. Adv. Mater. 2002, 14, 1164–1167. [Google Scholar]
- Wang S.; Wang H.; Jiao J.; Chen K. J.; Owens G. E.; Kamei K.; Sun J.; Sherman D. J.; Behrenbruch C. P.; Wu H.; et al. Three-Dimensional Nanostructured Substrates toward Efficient Capture of Circulating Tumor Cells. Angew. Chem., Int. Ed. 2009, 48, 8970–8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson L.; Lin Z. C.; Xie C.; Cui Y.; Cui B. Characterization of the Cell-Nanopillar Interface by Transmission Electron Microscopy. Nano Lett. 2012, 12, 5815–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H.; Chen K.-J.; Noh S.-H.; Garcia M. A.; Wang H.; Lin W.-Y.; Jeong H.; Kong B. J.; Stout D. B.; Cheon J.; et al. On-Demand Drug Release System for In Vivo Cancer Treatment through Self-Assembled Magnetic Nanoparticles. Angew. Chem., Int. Ed. 2013, 52, 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Liu K.; Chen K.-J.; Lu Y.; Wang S.; Lin W.-Y.; Guo F.; Kamei K.-i.; Chen Y. C.; Ohashi M.; et al. A Rapid Pathway toward a Superb Gene Delivery System: Programming Structural and Functional Diversity into a Supramolecular Nanoparticle Library. ACS Nano 2010, 4, 6235–6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.; Wang H.; Chen K. J.; Guo F.; Lin W. Y.; Chen Y. C.; Phung D. L.; Tseng H. R.; Shen C. K. F. A Digital Microfluidic Droplet Generator Produces Self-Assembled Supramolecular Nanoparticles for Targeted Cell Imaging. Nanotechnology 2010, 21, 445603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Wang H.; Kamei K.; Yan M.; Chen K. J.; Yuan Q. H.; Shi L. Q.; Lu Y. F.; Tseng H. R. Delivery of Intact Transcription Factor by Using Self-Assembled Supramolecular Nanoparticles. Angew. Chem., Int. Ed. 2011, 50, 3058–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. L. The Leukocyte Common Antigen Family. Annu. Rev. Immunol. 1989, 7, 339–369. [DOI] [PubMed] [Google Scholar]
- McLennan D. N.; Porter C. J. H.; Charman S. A. Subcutaneous Drug Delivery and the Role of the Lymphatics. Drug Discovery Today 2005, 2, 89–96. [DOI] [PubMed] [Google Scholar]
- Petter R. C.; Salek J. S.; Sikorski C. T.; Kumaravel G.; Lin F. T. Cooperative Binding by Aggregated Mono-6-(Alkylamino)-Beta-Cyclodextrins. J. Am. Chem. Soc. 1990, 112, 3860–3868. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.