Abstract
Current tissue engineering methods lack the ability to properly recreate scaffold-free, cell-dense tissues with physiological structures. Recent studies have shown that the use of nanoscale cues allows for precise control over large-area 2D tissue structures without restricting cell growth or cell density. In this study, we developed a simple and versatile platform combining a thermoresponsive nanofabricated substratum (TNFS) incorporating nanotopographical cues and the gel casting method for the fabrication of scaffold-free 3D tissues. Our TNFS allows for the structural control of aligned cell monolayers which can be spontaneously detached via a change in culture temperature. Utilizing our gel casting method, viable, aligned cell sheets can be transferred without loss of anisotropy or stacked with control over individual layer orientations. Transferred cell sheets and individual cell layers within multilayered tissues robustly retain structural anisotropy, allowing for the fabrication of scaffold-free, 3D tissues with hierarchical control of overall tissue structure.
Keywords: tissue engineering, thermoresponsive, three-dimensional
Tissue engineering seeks to fabricate physiological tissues by utilizing a combination of cells, biomaterials, and engineering methods. However, one of the challenges inherent in tissue engineering is replicating the complex, three-dimensional (3D) structures of most tissues which are critical to their function.1−3 The importance of replicating physiological tissue structure can be seen in the structure–function relationships of many tissues, such as in the orthogonal layering of sheets of connective tissues in the annulus fibrosus of intervertebral discs which allow for their ability to withstand forces applied in multiple directions.4,5 Other examples of tissue-level structure–function relationships include the complex 3D structure of both the sinoatrial and atrioventricular nodes and connected Purkinje fiber network, which are responsible for the appropriate depolarization sequence of the heart,6 and the pennation or angling of muscle fibers in most muscle tissues to the line of action, which allows for a greater degree of control through mechanically diverse functions.7,8 Tissue structure is also often altered in disease states, such as the disarray of cardiac muscle in hypertrophic cardiomyopathy9 and the thinning of bone trabeculae in osteoporosis,10 which can contribute to a deterioration of tissue function.
Despite significant efforts on studying physiologically relevant tissue structures and functions, however, only recently have technological advancements allowed for the engineering of more complex tissue structures. Microfabrication techniques have been used to engineer tissues with defined microscale structures, such as strips, squares, and wells, and have been successful in studies to understand single-cell- or tissue-like structure–function relationships.11,12 Other methods such as directionally defined mechanical strain,13−15 magnetic fields,16,17 and electrical stimulation18,19 have also allowed for the simple alignment or orientation of tissues. Additionally, 3D scaffolds, such as self-assembling peptides,20 are commonly employed to organize cells on biomaterials that can more accurately replicate tissues.21 Finally, methods utilizing thermoresponsive polymers, such as poly(N-isopropylacrylamide) (PNIPAM), termed cell sheet engineering, have been able to fabricate 3D, cell-dense tissues without the use of a supporting scaffolds.22−25 All these methods have started to bridge the gap between the simple plating and culture of cells on flat culture surfaces and the complex tissue structures which are actually found in the human body. However, although promising, most of these techniques are ultimately still constrained to unidirectional, aligned tissue structures which are unable to vary cell alignment through tissue depth.14,18,23 Techniques which have been able to fabricate tissues with controllable 3D structures thus often use scaffolds, which inherently reduce cell–cell contacts and sacrifice the cell-dense nature of physiological tissues.26,27
Subcellular nanoscale cues are a promising alternative for engineering cell and tissue structures which do not restrict the growth of cells. For instance, the addition of nanoscale cues, such as protein-based nanoparticles,28 nanorods,29 and nanotubes,30 have been explored to align both individual cells as well as 2D tissues. Additionally, nanotopographically defined matrix guidance cues have been explored as an alternative method to structurally organize monolayers of tissues.31 Topographical cues at the nanoscale are subcellular and can replicate the well-defined architectures of the local cellular microenvironment, such as mimicking structures of the extracellular matrix (ECM).22 The use of scalable soft nanofabrication techniques thus can allow for the multiscale analysis of complex cell–matrix interactions and their role in tissue engineering. Further, the use of nanotopography has shown more precise control of self-assembled monolayer tissues over conventional alignment techniques and has also shown differential effects on morphological characteristics of cells.31 We previously explored the effects of nanoscale surface topography on single-cell- and tissue-level morphology and function with precise control over cell and tissue structure, such as guided migration in response to topographical gradient32,33 or self-assembly of anisotropic cardiac monolayers.34 The use of these techniques allows for the study and characterization of specific monolayer tissue structures at a larger scale while utilizing a platform which can mimic the local cellular microenvironment.
In this study, we report the development of a thermoresponsive nanofabricated substratum (TNFS) which can be used to structurally organize cell monolayers. By reducing culture temperature, the monolayers detach spontaneously as intact cell sheets. Using a developed gel casting method, the cell sheets can be transferred to a flat surface and will maintain structural organization long-term. The sheets can also be stacked with controlled orientations using our developed method to engineer 3D scaffold-free tissues. These multilayered tissues robustly maintain both individual layer structure as well as the specific angle at which the layers are stacked, providing layer-by-layer control over the fabricated tissue structure. The developed TNFS and gel casting method can be used as a platform to reproducibly engineer more complex tissue structures which can then be used in tissue engineering applications, drug screening, and disease modeling.
Results and Discussion
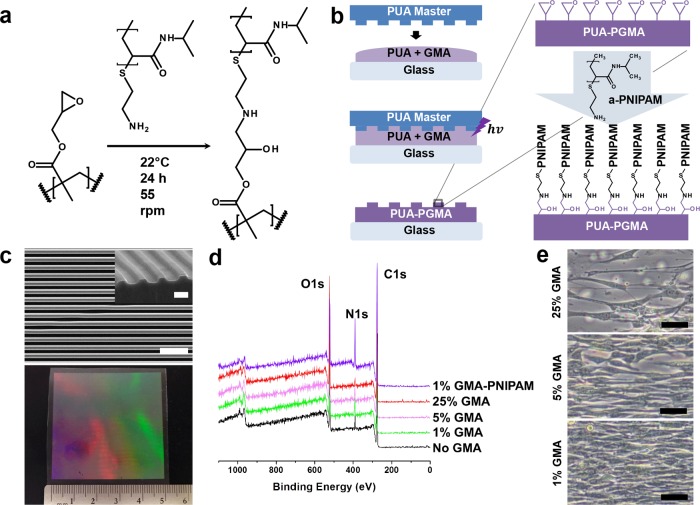
To develop a thermoresponsive nanofabricated substratum, we sought to functionalize substrata engineered with our well-established capillary force lithography (CFL)-based nanofabrication techniques32,33,35 with a thermoresponsive release layer while preserving pattern fidelity. To do this, we utilized epoxy amine chemistry (Figure 1a) to covalently bind amine-terminated poly(N-isopropylacrylamide) to surface epoxy groups present in our modified polymer substratum fabricated with CFL (Figure 1b). Our copolymer consisting of glycidyl methacrylate (GMA) and polyurethane acrylate (PUA) allowed for up to 75 wt %/vol GMA incorporation while still achieving photopolymerization and successful nanofabrication of topographical cues. Nanotopographical cues consisting of 800 nm wide and 500 nm deep parallel ridges and grooves were fabricated. Scanning electron microscopy (SEM) analysis confirmed nanopattern fidelity of a TNFS consisting of 1% GMA (Figure 1c, top). The UV-assisted CFL technique is scalable and can produce a TNFS of large-area size (up to 25 cm2) which is useful for tissue-level fabrication and analysis (Figure 1c, bottom). X-ray photoelectron spectroscopy analysis showed an increase in nitrogen surface compositions of the TNFS reacted with amine-terminated PNIPAM, confirming PNIPAM functionalization of the TNFS (Figure 1d and Table S1 in Supporting Information).
Figure 1.
Fabrication and characterization of the thermoresponsive nanofabricated substratum. (a) Epoxy amine addition reaction equation and experimental conditions. (b) Schematic of capillary force lithography to create the nanofabricated substratum and subsequent functionalization of the substratum with amine-terminated PNIPAM (a-PNIPAM). (c) SEM image of PNIPAM-functionalized polymeric nanofabricated substratum (top) and photo of a large-area, scalable TNFS (bottom). Scale bar, 5 μm; 1 μm (inset). (d) XPS analysis of the TNFS surface chemical composition with varying GMA prepolymer concentration before PNIPAM reaction and after 1% GMA TNFS PNIPAM reaction. (e) DIC image of C2C12 cells cultured on 25, 5, and 1% GMA TNFS after PNIPAM reaction for 24 h at 37 °C. Cells demonstrated varying degrees of confluent monolayer formation. Scale bar, 50 μm.
In order to determine the biocompatibility and thermoresponsive characteristics of the TNFS, C2C12 mouse myoblasts were used as the cell model. As varying PNIPAM grafting densities has been previously shown to affect cell adhesion and sheet formation,36 we fabricated the TNFS using varying GMA percentages (1–75%) to modulate the density of available epoxy groups and then reacted the fabricated substrata with amine-terminated PNIPAM solution for 24 h. C2C12 cells were seeded at a high density (1 × 106 cells/cm2) and allowed to attach for 24 h at 37 °C on the TNFS with varying GMA percentages. Cells displayed increased attachment and monolayer formation with decreasing GMA percentage, indicating that high-density grafted PNIPAM inhibits cell adhesion (Figure 1e and Figure S1 in Supporting Information). Specifically, the TNFS consisting of 1% GMA (1% GMA TNFS) as well as a 1% GMA unpatterned, thermoresponsive substratum (control) formed confluent monolayers within 24 h after seeding. Cells on the 1% GMA TNFS also aligned to the direction of the nanopatterns, indicating sensitivity to nanoscale topography (Figure 2a). Cells attached to the 1% GMA TNFS within 2 h of seeding and continued to spread to form a confluent monolayer within 24 h. The self-organization of cells and monolayers to the TNFS cues happened quickly—within 2 h of seeding, allowing for the rapid formation of an aligned monolayer. Additionally, these cell monolayers are very well aligned to the nanotopographical cues even under varying culture conditions such as cell seeding density or culture time, removing variability in the degree of alignment of the forming monolayer. Other studies have shown that topographical guidance cues to align cells even in the presence of conflicting cues such as mechanical strain37 or electrical stimulation,38 suggesting these topographical cues provide a high degree of control over structural organization.
Figure 2.
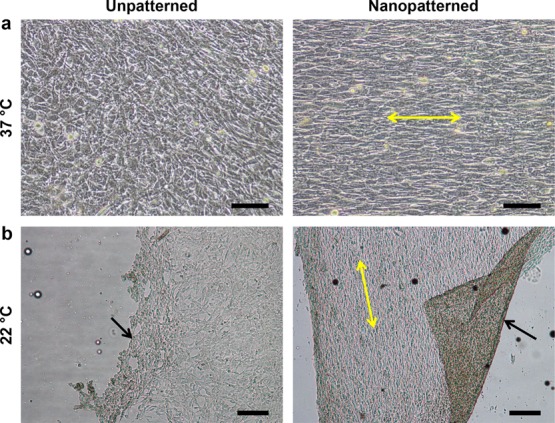
TNFS aligns cell monolayers and spontaneously detaches cell sheets in response to temperature change. (a) DIC image of C2C12 myoblasts cultured on the TNFS and control at 37 °C 24 h after seeding. The TNFS aligns the cell monolayer. Scale bar, 100 μm. (b) Phase contrast image of C2C12 monolayers after incubation at 22 °C for 30 min undergoing spontaneous detachment on both the TNFS and control. Black arrows indicate detaching edge of cell sheet. Yellow arrows indicate TNFS nanopattern direction. Scale bar, 200 μm.
To assess thermoresponsive, spontaneous detachment of cell sheets, 1% GMA TNFS and controls were seeded with cells, removed from the culture incubator (37 °C), and incubated in DPBS for 30 min at room temperature (22 °C). Within 20 min, the edges of the cell sheets on both TNFS and controls began to detach and retract inward, followed by the spontaneous release of the whole sheet (Figure 2b and movie S1 in Supporting Information). During detachment from the 1% GMA TNFS, cells contracted along the longitudinal axis and lost anisotropic morphology, with the entire cell sheet shrinking and losing alignment (Figure S2 and movie S2 in Supporting Information). The detached cell sheet could be transferred to a new surface via forceps or pipets and allowed to reattach. However, the sheet continued to contract and became extremely cell-dense tissues with no discernible anisotropy or organization. Based on the ability to form confluent monolayers rapidly, as well as detach a cell sheet spontaneously, the 1% GMA TNFS was used for all subsequent experiments and will be referred to simply as the TNFS.
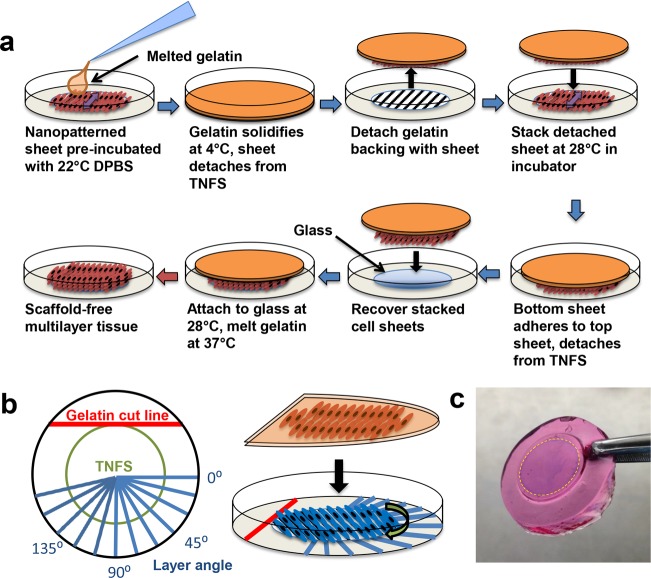
To transfer and manipulate anisotropic cell sheets without loss of structural organization, a previously published cell sheet transfer method was investigated.22 Using this plunger-like manipulator method, a fully intact cell sheet was only able to be transferred from the unpatterned thermoresponsive controls. The cell sheet from the TNFS only partially detached, allowing for transfer of a partial cell sheet or cell sheet fragments. Further, the transferred cell sheet from the TNFS demonstrated some loss of anisotropy immediately after transfer and subsequently lost all anisotropy after 5 days of culture on the flat surface of a standard tissue culture dish (Figure S3a in Supporting Information). We hypothesized that the lack of adhesion strength to the gelatin-coated manipulator from the TNFS may result in partial or difficult cell sheet detachment as well as the lack of morphological maintenance. As a result, a new method was developed to transfer nanopatterned cell sheets, termed the gel casting method (Figure 3a), in which we aimed to encase the cell sheet in the gelatin in order to increase cell sheet–gelatin adhesion strength and preserve morphology. Briefly, melted gelatin is added to the TNFS which is incubated at room temperature and then cooled to solidify and encase the detaching cell sheet prior to full sheet detachment. These sheets can then be stacked onto another TNFS to form multilayer tissues or transferred to a new surface. Using a developed orientation key (Figure 3b), the relative layer angle between the top and subsequent layers can also be controlled during the stacking process. The use of gelatin also allows for the easy manipulation of delicate cell sheets using forceps (Figure 3c). Using the gel casting method, cell sheets from both the TNFS and controls were able to be recovered intact and transferred to glass coverslips. After transfer, cell sheets remained firmly attached to the glass and could be cultured as normal. The transferred nanopatterned cell sheet retained anisotropic cytoskeletal structure immediately (24 h) after transfer as well as up to 7 days post-transfer (Figure 4a and Figure S3b in Supporting Information). Gel casted transferred cell sheets also maintained near 100% viability (Figure S4 in Supporting Information) 24 h after transfer and cell growth continued outward from the edges of the cell sheets, albeit in a random growth pattern for the transferred nanopatterned cell sheet. Immunofluorescent staining of cellular cytoskeletal proteins also indicated that cells within the transferred nanopatterned cell sheet demonstrated a well-aligned cellular cytoskeleton in addition to cell morphology (Figure 4b). Automated image analysis was used to quantify alignment of transferred cell sheets from both TNFS and controls 24 h and 7 days after transfer to a flat glass coverslip. The transferred nanopatterned cell sheet demonstrated significant structural alignment immediately after transfer, which was maintained overall tissue alignment long-term (7 days) comparable to the transferred, unpatterned cell sheet (Figure 4c). Previous studies have shown an increase in ECM production when cells are cultured on topographical surfaces39 as well as structural organization of secreted ECM (such as aligned fibers) which match the surface topography.40−42 Additionally, during cell sheet detachment from PNIPAM, deposited ECM proteins are also detached intact from the surface.43−45 We hypothesize that our nanopatterned surface allows for the structural organization of deposited ECM, which is thus detached and transferred with the gel cast method, allowing for maintenance of cell sheet structure.
Figure 3.
Gel casting method to preserve cell sheet anisotropy and control relative layer angle during stacking. (a) Schematic showing the gel casting method process to stack and transfer nanopatterned cell sheets. (b) Schematic of the orientation key (left) used to control relative layer angle during nanopatterned sheet stacking (right). (c) Image of gel cast sheet after gelatin solidification. Sheet was fixed and stained with trypan blue while casted to enhance visibility.
Figure 4.
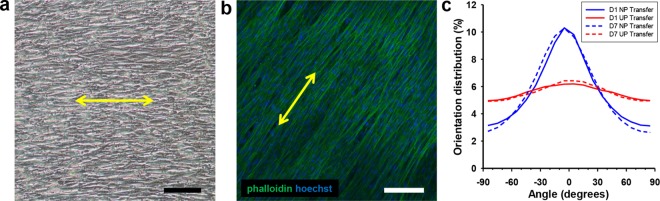
Transferred nanopatterned cell sheets are viable and maintain alignment. (a) DIC image of a transferred nanopatterned cell sheet 7 days after transfer onto glass. Scale bar, 100 μm. (b) Confocal image of phalloidin (green) and Hoechst (blue) staining showing maintenance of aligned cytoskeletal structure of nanopatterned cell sheets. Scale bar, 100 μm. (c) Quantitative analysis of cytoskeletal alignment of transferred nanopatterned (NP) cell sheets and unpatterned (UP) controls 1 day and 7 days after transfer. Yellow arrows indicate direction of transferred cell sheet alignment.
To determine whether the gel casting method could be used to successfully fabricate 3D tissues, nanopatterned cell sheets were stained with fluorescent live cell red and green membrane dyes to discern individual layers when stacked. The gel casting method was then used sequentially to transfer a green-dyed nanopatterned cell sheet onto a red-dyed nanopatterned cell sheet and then the bilayer tissue onto a glass coverslip. After 3 days of culture after stacking and transfer, the transferred bilayer tissue was fixed and imaged. The gel casting method was able to successfully fabricate the bilayer tissue which maintained individual layer morphologies even after transfer (Figure 5a,c) with distinct red and green layers, indicating no layer mixing or migration of cells between layers (Figure 5b). The average overall thickness of the bilayer tissue was 33.2 μm, with both layers having the same thickness.
Figure 5.
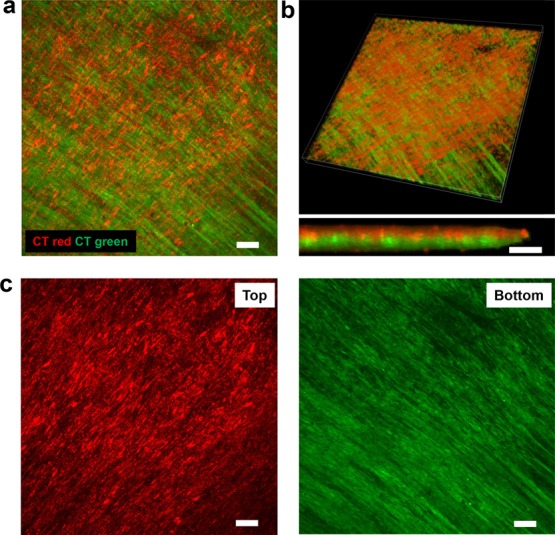
Individual nanopatterned cell sheets can be stacked together using the gel casting method. (a) Max projection confocal image of a z-stack showing two membrane-stained nanopatterned cell sheets stacked together to form a bilayer tissue. Top layer stained with red membrane dye, bottom layer stained with green membrane dye. Bilayer tissue was transferred to a glass coverslip and cultured 3 days before fixation and imaging. Scale bar, 100 μm. (b) Three-dimensional tissue rendering utilizing confocal z-stack images of membrane-stained cell sheets stacked into bilayer tissue, with individual layer colors discernible when magnified (bottom). Scale bar, 40 μm. (c) Confocal image of top and bottom sheets. Scale bar, 100 μm.
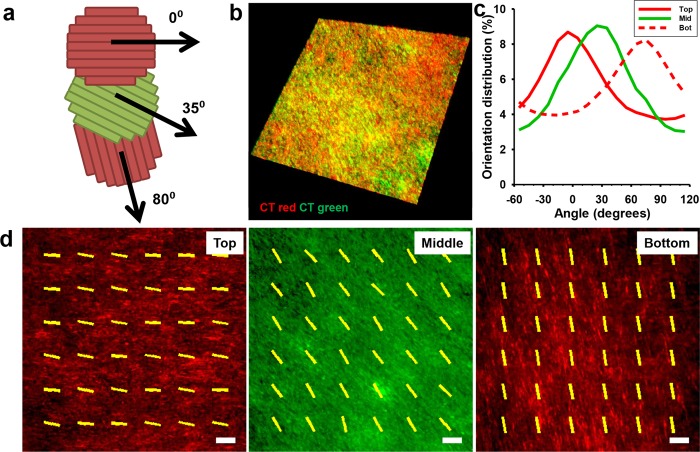
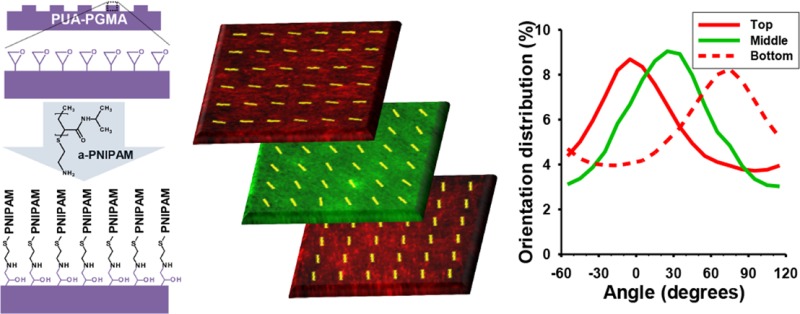
Next, using the gel casting method and orientation key, a 3D tissue with specific layer orientations and specific interlayer angles was fabricated. To confirm this, stained red and green nanopatterned cell sheets were detached and stacked sequentially with specific interlayer angles (Figure 6a) to form a trilayer tissue and transferred to a flat glass coverslip and cultured for 3 days. The fabricated tissue demonstrated 3D, multicolored structures when analyzed using confocal microscopy (Figure 6b). Individual layers were easily discernible (Figure 6d) and did not mix. Further, the red/green signal from individual cell sheets was analyzed using a modified image analysis technique to quantitatively evaluate alignment of individual layers within the tissue. The individual layers maintained alignment as well as the specific layer angle at which they were stacked, indicating no reorganization both within the sheets and between layers (Figure 6c and movie S3). The overall thickness of the layered tissue was 48.9 μm, with an average layer thickness of 16.3 μm, similar to the results from the bilayer stacking experiments. The tissue maintained this distinct structural organization 3 days after transfer, and no retraction or detachment of either individual sheets or the whole tissue was noted during this time. In contrast, unpatterned controls were also stacked into trilayer tissues using the gel casting method but did not maintain discrete individual layers (movie S4), indicating that the gel casting method does not directly prevent interlayer migration.
Figure 6.
Stacked nanopatterned cell sheets maintain individual layer alignment and demonstrate precise control over 3D structure. (a) Schematic of the trilayer myoblast tissue with individual layer orientations. (b) Three-dimensional tissue rendering utilizing confocal z-stack images of membrane-stained cell sheets stacked into trilayer tissue. Top and bottom layers were stained with red membrane dye; middle layer was stained with green membrane dye. Trilayer tissue was transferred to a glass coverslip and cultured 3 days before fixation and imaging. (c) Quantitative analysis of cell sheet alignment of individual layers demonstrates retained alignment 3 days after stacking as well as maintained interlayer angles. (d) Confocal images of the individual layers with representative alignment vectors generated via the image analysis program overlaid. Scale bar, 100 μm.
While other groups have fabricated aligned tissues using cell sheet engineering23 and have demonstrated multilayered tissues with specifically arranged layers,24 to our knowledge, this is the first demonstration of three-layered tissues with clear anisotropy, defined individual layers, and controllable interlayer angles. Interestingly, we observed the maintenance of both layer anisotropy and stacked layer angle of tissues utilizing myoblasts. It has recently been reported using human myoblasts that aligned cell sheets reorganize themselves to match the alignment of the top sheet, generating a completely aligned 3D muscle tissue.23 This rearrangement was observed within 24 h of stacking and included migration between individual sheets, resulting in a tissue of intermixed layers. In contrast to these results, our method allows the maintenance of individual layer orientations as well as prevention of intermixing of individual sheets for 3 days. Myoblasts are well-known as migratory cells due to their migration to sites of injury for muscle repair, and recent studies have also shown that myoblasts can migrate vertically through multilayered tissue constructs.46,47 This cell fluidity has been hypothesized to contribute to the loss of individual layer orientations in multilayered myoblast tissues constructs. However, we did not observe this phenomenon in our experiments utilizing a mouse myoblast line and our nanotopography-based platform. Our recent study showed an upregulation of myogenic markers of myoblasts when cultured on the nanopatterned substratum and an increase in dystrophin expression when nanopatterned monolayered muscle patches were implanted in an in vivo mouse muscular dystrophy model, relative to unpatterned controls.48 Other studies have reported differential response of myoblasts to nanotopography versus micropatterning.49 It is possible that the utilization of nanotopographical cues to structurally organize monolayers may evoke a different biological response within the chosen cell model when compared to methods such as micropatterning, which could affect migratory behavior or protein expression. Specifically, the organization of the deposited ECM which is subsequently transferred during the stacking process may provide structural cues which serve to both allow for sheet-to-sheet attachment as well as maintain individual sheet orientation and prevent dissolution of sheets or migration of cells between sheets. Our results also suggest that the structural maintenance in 3D tissues is not a result of the developed gel casting transfer method, as unpatterned controls stacked into 3D tissues did not demonstrate discrete individual layers (movies S3 and S4). With such results, we believe this platform can allow for the facile, reproducible fabrication of well-defined 3D tissue structures which may not be achievable via other means.
Conclusions
In this study, we have developed a simple, yet versatile platform for the fabrication of scaffold-free 3D tissues using our newly developed TNFS and gel casting method. This platform enables the reproducible and robust fabrication of scaffold-free 3D tissues with controllable architecture via nanoscale control of cell and tissue structure. Although other groups have demonstrated the fabrication of 3D tissues using scaffolds with controllable 3D architectures,21,50 micropatterned molds, or scaffolds functionalized with thermoresponsive polymers,23,24,51 the use of nanotopographical cues allows for the structural organization of cells and cell monolayers at a subcellular level. Nanotopographical cues do not prevent or restrict cell growth and thereby reduce variabilities such as the degree of alignment due to pattern size or cell seeding density. Additionally, the fabrication method of our TNFS is highly reproducible, cost-effective, and scalable, allowing for a variety of applications in cell biology and tissue engineering. The developed substratum incorporated our CFL-based nanofabrication method which allows us to fabricate a variety of nanotopographical cues in a large-area format. Epoxy-based chemistry enables simple surface immobilization of a thermoresponsive polymer. The TNFS had variable cell attachment based on GMA percentage incorporation and thus PNIPAM grafting density, with the 1% GMA TNFS allowing for both rapid monolayer formation as well as spontaneous, thermoresponsive detachment of intact cell sheets. Next, the gel casting method was used to transfer aligned sheets to a flat surface, and the transferred cell sheets maintain long-term structural organization without presentation of additional nanotopographical cues. The platform also allowed for the stacking of aligned sheets at specific angles between layers. The multilayered tissues retained specific layer organization, layer integrity, and overall tissue architecture upon transfer to a flat surface for 3 days, demonstrating the feasibility of our platform in the creation of complex 3D tissues with highly controllable architecture. Interestingly, when stacked into multilayered tissues, individual sheets also retain their specific alignment and cells from individual sheets do not mix or migrate. This allows for the fabrication of 3D tissues with clear layer structures which can have precisely controlled structures, allowing for the engineering of complex and physiological tissues. Our developed platform could be readily expanded to the engineering of other complex, defined 3D tissues of interest, and used in quantitative studies of structure–function relationship.
Experimental Section
Fabrication of a Poly(urethane acrylate)–Poly(glycidyl methacrylate) (PUA-PGMA) Nanopatterned Substratum
A UV-curable poly(urethane acrylate) (PUA, Minutatek, Korea) mold was fabricated using capillary force lithography as published previously32,33,35 and was used as the nanopatterned template for the PUA-PGMA substratum (Figure 1a). To allow for epoxy functionalization of the fabricated nanopatterned substratum, 1% GMA weight/volume monomer (Sigma-Aldrich) was added to the liquid PUA precursor (Norland Optical Adhesive), sonicated for 1 h, and then hand mixed for 10 min. Solution was then degassed under a vacuum for 1 h to remove air bubbles. A glass coverslip (Ø18 mm, Fisher) was cleaned using isopropyl alcohol and brush coated with an adhesion promoter (Glass Primer, Minuta Tech) to allow for attachment of the polymer to the glass surface and air-dried. Twenty microliters of PUA-PGMA prepolymer was added to the coverslip and covered with the PUA template consisting of 800 nm wide and 500 nm deep parallel grooves and ridges. The PUA-PGMA prepolymer was spontaneously drawn into the nanofeatures of the PUA template via capillary force. The template–prepolymer–glass was cured under 365 nm UV light to initiate photopolymerization for 5 min. After polymerization, the PUA template was peeled off from the PUA-PGMA substratum using forceps and the substratum was UV-cured overnight to finalize polymerization.
PNIPAM Grafting to the PUA-PGMA Substratum
Amine groups react spontaneously with epoxy groups in an addition reaction to form a hydroxyl group and a secondary amine (Figure 1a). Amine-terminated poly(N-isopropylacrylamide) (Mn = 2500, Sigma-Aldrich) was dissolved in deionized (DI) water at room temperature at a concentration of 1 g/30 mL. The PNIPAM solution was reacted with the PUA-PGMA substratum in a shaker at room temperature for 24 h at 55 rpm to allow for thermoresponsive functionalization. The TNFS was then washed three times with DI water and sterilized with 294 nm UV light overnight prior to use.
Characterization of the TNFS
Scanning electron microscopy (SEM, FEI Sirion) was used to confirm nanotopographical fidelity. X-ray photoelectron spectroscopy (XPS, Surface Science Instruments S-probe spectrometer) was utilized to determine surface composition of the TNFS.
Cell Culture and Seeding on the TNFS
C2C12 mouse myoblasts were cultured in Dulbecco’s minimal essential media (DMEM, Gibco) supplemented with 20% fetal bovine serum (FBS, Sigma), 1% penicillin–streptomycin (Sigma), and 1% amphotericin-B (Sigma) in an incubator at 37 °C, 5% CO2. Cells were split at 80% confluency to prevent spontaneous differentiation. To seed cells onto the TNFS, cells were split and seeded at a density of 1 × 106 cells/cm2 to form a confluent monolayer within 24 h of culture and cultured normally with a daily media change. Cells were imaged with a bright-field microscope (Nikon TS100) during culture. To visualize individual layers, cells were labeled with either 2 μM CellTracker CMFDA Green (Invitrogen) or 2 μM CellTracker Red CMTPX (Invitrogen) for 30 min before seeding onto the TNFS and then washed gently two times with warmed media after attachment.
Gel Casting Transfer and Stacking of Nanopatterned Cell Sheets
To prevent loss of anisotropy after sheet detachment, a gel casting method was developed to preserve cell morphology. A 7.5% (wt/v) gelatin (Sigma) in media solution was made, which forms a solid at room temperature but melts at 37 °C. To detach the cell sheet, the TNFS was removed from the incubator and placed in a tissue culture hood at 22 °C; the medium was aspirated and room temperature (22 °C) DPBS (Gibco) was added to the TNFS and incubated for 25 min to initiate cell sheet detachment. After room temperature incubation, the DPBS was aspirated and melted gelatin solution (37 °C) was then added to the TNFS, and the TNFS and gelatin were then incubated at 4 °C for 5 min to allow the gelatin to solidify rapidly. The solidified gelatin (gel cast) was then removed from the TNFS surface with forceps with the cell sheet adhered to the bottom surface and transferred to a new surface. The cell sheet was allowed to adhere to new surfaces in a 28 °C, 5% CO2 incubator for 1 h, and then the gel cast was melted by transferring the sheet to the 37 °C, 5% CO2 incubator for 1 h. After the gel cast was melted, the transferred cell sheet was washed gently three times with warm media and cultured as normal. To create multilayered tissues, during the 4 °C incubation of the top sheet, the next layered sheet would first be removed from the 37 °C incubator, placed in the tissue culture hood, and the media removed. The gel cast from the first cell sheet (with the first sheet still adhered) would then be carefully placed on top of the second cell sheet (still attached to the TNFS) with forceps, and the gel cast, cell sheets, and the TNFS would then be transferred to the 28 °C incubator and allowed to attach for 30 min (Figure 3a). After the 30 min incubation, the gel cast, cell sheets, and the TNFS would then be incubated at 4 °C for 5 min to firm the gel cast for handling, at which point a subsequent cell sheet could be prepared for stacking or the multilayered tissue could be transferred to a new surface (Figure 3a).
Use of Orientation Key To Control Stacked Layer angle
To create tissues with specific layer orientation, an orientation key was fabricated out of transparent polystyrene. After gel casting, the gel cast and the TNFS could be visualized under a microscope at a 20× objective, and the nanopatterns were aligned to the 0° position of the orientation key. The gel cast was then cut parallel to the 0° position as a reference. During stacking, the detaching layer was then placed upon the orientation key and oriented to the desired angle relative to the casted layer and confirmed under the microscope. The gelatin cut line of the casted layer was then oriented to the key, and the casted layer was then stacked and allowed to attach as normal (Figure 3b).
Assessment of Transferred Cell Sheet Viability
A live/dead fluorescent staining kit (Invitrogen) utilizing calcein AM as a live cell reporting dye and ethidium homodimer-1 as a dead cell reporting dye was used according to manufacturer’s instructions. Briefly, the kit reagents were added to sterile 1× DPBS to create a 1 mM calcein AM and 4 mM ethidium homodimer-1 solution. Cells were washed gently with warm 1× DPBS, and then 200 μL of calcein AM/ethidium homodimer-1 solution was added to the cells and incubated protected from light for 30 min at 37 °C, 5% CO2. After incubation, cells were gently rinsed with 1× DPBS and then imaged with a confocal microscope (Nikon A1R).
Immunofluorescent Staining and Imaging
Cells were washed with phosphate buffered saline (PBS, Sigma) and fixed in 4% paraformaldehyde (Sigma) for 15 min at room temperature (22 °C). Fixed cells were then washed with PBS and permeabilized and blocked with a solution of 5% bovine serum albumin (Sigma) and 0.25% Triton X-100 (Sigma) in PBS for 1 h at room temperature, then washed with PBS. For F-actin staining, cells were then incubated in AlexaFluor488-conjugated phalloidin (Invitrogen) at a dilution of 1:200 in 1% BSA in PBS for 1 h at 37 °C. All samples were then stained with a Hoechst stain (Sigma) at a dilution of 1:1000, washed with PBS once, then treated with Vectashield (Vector Laboratories), mounted on coverslips, and imaged using a confocal microscope.
Quantitative Analysis of Cell Alignment
To assess alignment, immunofluorescent images of phalloidin or CellTracker-stained cells were taken at three representative fields at 10× magnification and analyzed using a modified, previously published MATLAB script utilizing pixel gradient analysis.52 Briefly, the images were passed through a Gaussian low-pass filter and Sobel horizontal edge-emphasized filter (predefined MATLAB Image Analysis toolbox functions) to generate a 2D convolution. The Sobel filter was then transposed to extract the vertical edge, and the horizontal and vertical edges were combined to calculate the gradient magnitude of each pixel in the image. The image was then thresholded to determine the borders of the areas of interest, and the orientation of the gradient was calculated via respect to the x-axis (0°). A representative image of the filtering, vector generation, and histogram creation process is demonstrated in Figure S5 (in Supporting Information). The orientation gradient data obtained from the pixel gradient analysis MATLAB script was then used to generate representative plots of overall cell alignment using a second custom MATLAB script. Each image was segmented into a grid of user-designated size. The gradient orientation angle for each pixel in a given grid square was then shifted in order to take a circular average with respect to the angle present with the highest incidence within that grid square’s orientation angle data. This circular average is representative of the mean orientation of each pixel gradient within a given grid square.
Acknowledgments
D.-H.K. thanks the Department of Bioengineering at the University of Washington for the new faculty startup fund. This work was also supported by a Muscular Dystrophy Association (MDA) Research Grant (MDA 255907) (D.-H.K.), an American Heart Association (AHA) Scientist Development Grant (13SDG14560076) (D.-H.K.), the Wallace H. Coulter Foundation Translational Research Partnership Award (D.-H.K.), a NIH Bioengineering Cardiovascular Training Grant Fellowship (T32EB001650) (A.J.), NIH R01 HL084642, P01 HL094374, P01 GM81619, and U01 HL100405 (C.E.M.). Authors thanks Dr. Sung Gap Im for discussions on epoxy amine polymer chemistry, and Dr. Eung-Sam Kim for critical reading of this manuscript.
Supporting Information Available
TNFS XPS table, cell response to varying TNFS GMA compositions, loss of cell morphology after detachment, comparisons of cell sheet transfer methods, transferred cell sheet viability, quantitative alignment computation schematic, and movies of cell sheet detachment and stained trilayer nanopatterned tissues and unpatterned controls. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Pampaloni F.; Reynaud E. G.; Stelzer E. H. K. The Third Dimension Bridges the Gap between Cell Culture and Live Tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [DOI] [PubMed] [Google Scholar]
- Griffith L. G.; Swartz M. A. Capturing Complex 3d Tissue Physiology in Vitro. Nat. Rev. Mol. Cell Biol. 2006, 7, 211–224. [DOI] [PubMed] [Google Scholar]
- Gauvin R.; Khademhosseini A. Microscale Technologies and Modular Approaches for Tissue Engineering: Moving toward the Fabrication of Complex Functional Structures. ACS Nano 2011, 5, 4258–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin H. L.; Elliott D. M. Quantifying the Contributions of Structure to Annulus Fibrosus Mechanical Function Using a Nonlinear, Anisotropic, Hyperelastic Model. J. Orthop. Res. 2007, 25, 508–516. [DOI] [PubMed] [Google Scholar]
- Lundon K. Structure and Function of the Lumbar Intervertebral Disk in Health, Aging, and Pathologic Conditions. J. Orthop. Sports Phys. Ther. 2001, 31, 291–306. [DOI] [PubMed] [Google Scholar]
- Nikolaidou T.; Aslanidi O. V.; Zhang H.; Efimov I. R. Structure–Function Relationship in the Sinus and Atrioventricular Nodes. Pediatr. Cardiol. 2012, 33, 890–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard P.; Andersen J. L.; Dyhre-Poulsen P.; Leffers A.-M.; Wagner A.; Magnusson S. P.; Halkjær-Kristensen J.; Simonsen E. B. A Mechanism for Increased Contractile Strength of Human Pennate Muscle in Response to Strength Training: Changes in Muscle Architecture. J. Physiol. 2001, 534, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi E.; Brainerd E. L.; Roberts T. J. Variable Gearing in Pennate Muscles. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 1745–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki Y.; Yamauchi Y.; Okabe M.; Terasaki F.; Ishizaka N. Three-Dimensional Architecture of Cardiomyocytes and Connective Tissues in Hypertrophic Cardiomyopathy: A Scanning Electron Microscopic Observation. Circulation 2012, 125, 738–739. [DOI] [PubMed] [Google Scholar]
- Fonseca H.; Moreira-Gonçalves D.; Coriolano H.-J.; Duarte J. Bone Quality: The Determinants of Bone Strength and Fragility. Sports Med. 2014, 44, 37–53. [DOI] [PubMed] [Google Scholar]
- McDevitt T. C.; Angello J. C.; Whitney M. L.; Reinecke H.; Hauschka S. D.; Murry C. E.; Stayton P. S. In Vitro Generation of Differentiated Cardiac Myofibers on Micropatterned Laminin Surfaces. J. Biomed. Mater. Res. 2002, 60, 472–479. [DOI] [PubMed] [Google Scholar]
- Kuo P.-L.; Lee H.; Bray M.-A.; Geisse N. A.; Huang Y.-T.; Adams W. J.; Sheehy S. P.; Parker K. K. Myocyte Shape Regulates Lateral Registry of Sarcomeres and Contractility. Am. J. Pathol. 2012, 181, 2030–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kada K.; Yasui K.; Naruse K.; Kamiya K.; Kodama I.; Toyama J. Orientation Change of Cardiocytes Induced by Cyclic Stretch Stimulation: Time Dependency and Involvement of Protein Kinases. J. Mol. Cell. Cardiol. 1999, 31, 247–259. [DOI] [PubMed] [Google Scholar]
- Neidlinger-Wilke C.; Grood E. S.; Wang J. H.-C.; Brand R. A.; Claes L. Cell Alignment Is Induced by Cyclic Changes in Cell Length: Studies of Cells Grown in Cyclically Stretched Substrates. J. Orthop. Res. 2001, 19, 286–293. [DOI] [PubMed] [Google Scholar]
- Salameh A.; Wustmann A.; Karl S.; Blanke K.; Apel D.; Rojas-Gomez D.; Franke H.; Mohr F. W.; Janousek J.; Dhein S. Cyclic Mechanical Stretch Induces Cardiomyocyte Orientation and Polarization of the Gap Junction Protein Connexin43. Circ. Res. 2010, 106, 1592–602. [DOI] [PubMed] [Google Scholar]
- Sato M.; Ito A.; Akiyama H.; Kawabe Y.; Kamihira M. Effects of B-Cell Lymphoma 2 Gene Transfer to Myoblast Cells on Skeletal Muscle Tissue Formation Using Magnetic Force-Based Tissue Engineering. Tissue Eng., Part A 2012, 19, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletti D.; Teodori L.; Albertini M. C.; Rocchi M.; Pristerà A.; Fini M.; Molinaro M.; Adamo S. Static Magnetic Fields Enhance Skeletal Muscle Differentiation in Vitro by Improving Myoblast Alignment. Cytometry, Part A 2007, 71A, 846–856. [DOI] [PubMed] [Google Scholar]
- Radisic M.; Park H.; Shing H.; Consi T.; Schoen F. J.; Langer R.; Freed L. E.; Vunjak-Novakovic G. Functional Assembly of Engineered Myocardium by Electrical Stimulation of Cardiac Myocytes Cultured on Scaffolds. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 18129–18134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon N.; Cannizzaro C.; Chao P.-h. G.; Maidhof R.; Ting H.; Au H.; Radisic M.; Vunjak-Novakovic G. Electrical Stimulation Systems for Cardiac Tissue Engineering. Nat. Protoc. 2009, 4, 155–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelain F.; Silva D.; Caprini A.; Taraballi F.; Natalello A.; Villa O.; Nam K. T.; Zuckermann R. N.; Doglia S. M.; Vescovi A. Bmhp1-Derived Self-Assembling Peptides: Hierarchically Assembled Structures with Self-Healing Propensity and Potential for Tissue Engineering Applications. ACS Nano 2011, 5, 1845–1859. [DOI] [PubMed] [Google Scholar]
- Caldwell D. J.; Rao R. R.; Stegemann J. P. Assembly of Discrete Collagen–Chitosan Microenvironments into Multiphase Tissue Constructs. Adv. Healthcare Mater. 2013, 2, 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi Y.; Shimizu T.; Sasagawa T.; Sekine H.; Sakaguchi K.; Kikuchi T.; Sekine W.; Sekiya S.; Yamato M.; Umezu M.; Okano T. Fabrication of Functional Three-Dimensional Tissues by Stacking Cell Sheets in Vitro. Nat. Protoc. 2012, 7, 850–858. [DOI] [PubMed] [Google Scholar]
- Takahashi H.; Shimizu T.; Nakayama M.; Yamato M.; Okano T. The Use of Anisotropic Cell Sheets To Control Orientation during the Self-Organization of 3D Muscle Tissue. Biomaterials 2013, 34, 7372–7380. [DOI] [PubMed] [Google Scholar]
- Williams C.; Xie A. W.; Yamato M.; Okano T.; Wong J. Y. Stacking of Aligned Cell Sheets for Layer-by-Layer Control of Complex Tissue Structure. Biomaterials 2011, 32, 5625–5632. [DOI] [PubMed] [Google Scholar]
- Xia Y.; He X.; Cao M.; Chen C.; Xu H.; Pan F.; Lu J. R. Thermoresponsive Microgel Films for Harvesting Cells and Cell Sheets. Biomacromolecules 2013, 14, 3615–3625. [DOI] [PubMed] [Google Scholar]
- Radisic M.; Malda J.; Epping E.; Geng W.; Langer R.; Vunjak-Novakovic G. Oxygen Gradients Correlate with Cell Density and Cell Viability in Engineered Cardiac Tissue. Biotechnol. Bioeng. 2006, 93, 332–343. [DOI] [PubMed] [Google Scholar]
- Vunjak-Novakovic G.; Tandon N.; Godier A.; Maidhof R.; Marsano A.; Martens T. P.; Radisic M. Challenges in Cardiac Tissue Engineering. Tissue Eng., Part B 2010, 16, 169–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatkiewicz W. I.; Seras-Franzoso J.; García-Fruitós E.; Vazquez E.; Ventosa N.; Peebo K.; Ratera I.; Villaverde A.; Veciana J. Two-Dimensional Microscale Engineering of Protein-Based Nanoparticles for Cell Guidance. ACS Nano 2013, 7, 4774–4784. [DOI] [PubMed] [Google Scholar]
- Zan X.; Feng S.; Balizan E.; Lin Y.; Wang Q. Facile Method for Large Scale Alignment of One Dimensional Nanoparticles and Control over Myoblast Orientation and Differentiation. ACS Nano 2013, 7, 8385–8396. [DOI] [PubMed] [Google Scholar]
- Namgung S.; Baik K. Y.; Park J.; Hong S. Controlling the Growth and Differentiation of Human Mesenchymal Stem Cells by the Arrangement of Individual Carbon Nanotubes. ACS Nano 2011, 5, 7383–7390. [DOI] [PubMed] [Google Scholar]
- Kim H. N.; Jiao A.; Hwang N. S.; Kim M. S.; Kang D. H.; Kim D.-H.; Suh K.-Y. Nanotopography-Guided Tissue Engineering and Regenerative Medicine. Adv. Drug Delivery Rev. 2013, 64, 536–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-H.; Seo C.-H.; Han K.; Kwon K. W.; Levchenko A.; Suh K.-Y. Guided Cell Migration on Microtextured Substrates with Variable Local Density and Anisotropy. Adv. Funct. Mater. 2009, 19, 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-H.; Han K.; Gupta K.; Kwon K. W.; Suh K.-Y.; Levchenko A. Mechanosensitivity of Fibroblast Cell Shape and Movement to Anisotropic Substratum Topography Gradients. Biomaterials 2009, 30, 5433–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-H.; Lipke E. A.; Kim P.; Cheong R.; Thompson S.; Delannoy M.; Suh K.-Y.; Tung L.; Levchenko A. Nanoscale Cues Regulate the Structure and Function of Macroscopic Cardiac Tissue Constructs. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.; Kim D. H.; Kim B.; Choi S. K.; Lee S. H.; Khademhosseini A.; Langer R.; Suh K. Y. Fabrication of Nanostructures of Polyethylene Glycol for Applications to Protein Adsorption and Cell Adhesion. Nanotechnology 2005, 16, 2420–2426. [DOI] [PubMed] [Google Scholar]
- Nagase K.; Watanabe M.; Kikuchi A.; Yamato M.; Okano T. Thermo-Responsive Polymer Brushes as Intelligent Biointerfaces: Preparation via ATRP and Characterization. Macromol. Biosci. 2011, 11, 400–409. [DOI] [PubMed] [Google Scholar]
- Mudera V. C.; Pleass R.; Eastwood M.; Tarnuzzer R.; Schultz G.; Khaw P.; McGrouther D. A.; Brown R. A. Molecular Responses of Human Dermal Fibroblasts to Dual Cues: Contact Guidance and Mechanical Load. Cell Motil. Cytoskeleton 2000, 45, 1–9. [DOI] [PubMed] [Google Scholar]
- Au H. T. H.; Cheng I.; Chowdhury M. F.; Radisic M. Interactive Effects of Surface Topography and Pulsatile Electrical Field Stimulation on Orientation and Elongation of Fibroblasts and Cardiomyocytes. Biomaterials 2007, 28, 4277–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaglia L.; Capece G.; Di Spigna G.; Bruno M. P.; Buonocore N.; Postiglione L. Effects of Titanium Surface Topography on Morphology and in Vitro Activity of Human Gingival Fibroblasts. Minerva Stomatol. 2013, 62, 267–280. [PubMed] [Google Scholar]
- Karamichos D.; Funderburgh M. L.; Hutcheon A. E. K.; Zieske J. D.; Du Y.; Wu J.; Funderburgh J. L. A Role for Topographic Cues in the Organization of Collagenous Matrix by Corneal Fibroblasts and Stem Cells. PLoS One 2014, 9, e86260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y.; Chen J.; Zhang T.; Gu D.; Zhang C.; Li Z.; Lin S.; Fu X.; Schultze-Mosgau S. Effect of 3d Microgroove Surface Topography on Plasma and Cellular Fibronectin of Human Gingival Fibroblasts. J. Dent. 2013, 41, 1109–1121. [DOI] [PubMed] [Google Scholar]
- Wu J.; Du Y.; Watkins S. C.; Funderburgh J. L.; Wagner W. R. The Engineering of Organized Human Corneal Tissue through the Spatial Guidance of Corneal Stromal Stem Cells. Biomaterials 2012, 33, 1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canavan H. E.; Cheng X.; Graham D. J.; Ratner B. D.; Castner D. G. Surface Characterization of the Extracellular Matrix Remaining after Cell Detachment from a Thermoresponsive Polymer. Langmuir 2004, 21, 1949–1955. [DOI] [PubMed] [Google Scholar]
- Canavan H. E.; Cheng X.; Graham D. J.; Ratner B. D.; Castner D. G. Cell Sheet Detachment Affects the Extracellular Matrix: A Surface Science Study Comparing Thermal Liftoff, Enzymatic, and Mechanical Methods. J. Biomed. Mater. Res., Part A 2005, 75A, 1–13. [DOI] [PubMed] [Google Scholar]
- Kushida A.; Yamato M.; Konno C.; Kikuchi A.; Sakurai Y.; Okano T. Decrease in Culture Temperature Releases Monolayer Endothelial Cell Sheets Together with Deposited Fibronectin Matrix from Temperature-Responsive Culture Surfaces. J. Biomed. Mater. Res. 1999, 45, 355–362. [DOI] [PubMed] [Google Scholar]
- Goetsch K. P.; Myburgh K. H.; Niesler C. In Vitro Myoblast Motility Models: Investigating Migration Dynamics for the Study of Skeletal Muscle Repair. J. Muscle Res. Cell Motil. 2013, 34, 333–347. [DOI] [PubMed] [Google Scholar]
- Kino-oka M.; Ngo T. X.; Nagamori E.; Takezawa Y.; Miyake Y.; Sawa Y.; Saito A.; Shimizu T.; Okano T.; Taya M. Evaluation of Vertical Cell Fluidity in a Multilayered Sheet of Skeletal Myoblasts. J. Biosci. Bioeng. 2012, 113, 128–131. [DOI] [PubMed] [Google Scholar]
- Yang H.-S.; Ieronimakis N.; Tsui J. H.; Kim H. N.; Suh K.-Y.; Reyes M.; Kim D.-H. Nanopatterned Muscle Cell Patches for Enhanced Myogenesis and Dystrophin Expression in a Mouse Model of Muscular Dystrophy. Biomaterials 2014, 35, 1478–1486. [DOI] [PubMed] [Google Scholar]
- Huang N. F.; Patel S.; Thakar R. G.; Wu J.; Hsiao B. S.; Chu B.; Lee R. J.; Li S. Myotube Assembly on Nanofibrous and Micropatterned Polymers. Nano Lett. 2006, 6, 537–542. [DOI] [PubMed] [Google Scholar]
- Ovsianikov A.; Gruene M.; Pflaum M.; Koch L.; Maiorana F.; Wilhelmi M.; Haverich A.; Chichkov B. Laser Printing of Cells into 3D Scaffolds. Biofabrication 2010, 2, 014104. [DOI] [PubMed] [Google Scholar]
- Hosseini V.; Ahadian S.; Ostrovidov S.; Camci-Unal G.; Chen S.; Kaji H.; Ramalingam M.; Khademhosseini A. Engineered Contractile Skeletal Muscle Tissue on a Microgrooved Methacrylated Gelatin Substrate. Tissue Eng., Part A 2012, 18, 2453–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.; Jönsson H.; Campbell K.; Melke P.; Williams J. W.; Jedynak B.; Stevens A. M.; Groisman A.; Levchenko A. Self-Organization in High-Density Bacterial Colonies: Efficient Crowd Control. PLoS Biol. 2007, 5, e302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.