Abstract
Stem-cell-based therapies have attracted considerable interest in regenerative medicine and oncological research. However, a major limitation of systemic delivery of stem cells is the low homing efficiency to the target site. Here, we report a serendipitous finding that various iron-based magnetic nanoparticles (MNPs) actively augment chemokine receptor CXCR4 expression of bone-marrow-derived mesenchymal stem cells (MSCs). On the basis of this observation, we designed an iron-based nanocluster that can effectively label MSCs, improve cell homing efficiency, and track the fate of the cells in vivo. Using this nanocluster, the labeled MSCs were accurately monitored by magnetic resonance imaging and improved the homing to both traumatic brain injury and glioblastoma models as compared to unlabeled MSCs. Our findings provide a simple and safe method for imaging and targeted delivery of stem cells and extend the potential applications of iron-based MNPs in regenerative medicine and oncology.
Keywords: mesenchymal stem cell, homing, iron oxide nanoparticle, magnetic resonance imaging, CXCR4/SDF-1α
Cell-based therapies are a major focus of regenerative medicine and cancer.1,2 However, a significant barrier to the effective implementation of cell therapy is the inability to deliver sufficient number of cells to the tissue of interest. Culture-expanded mesenchymal stem cells (MSCs) are emerging therapeutics currently under investigation in more than 200 ongoing clinical trials to treat a variety of inflammatory, ischemic, and neurologic diseases.3 The use of MSCs for cell therapy relies on the capacity of these cells to home and long-term engraft into the appropriate target tissue.4,5 Although the exact mechanism by which MSCs cross the endothelial cell layer and are recruited to target tissues remains unresolved, growing evidence suggests that chemokines and their receptors are involved as important factors to control cell migration. For example, interaction between chemokine stromal-derived factor 1α (SDF-1α) and its receptor C-X-C chemokine receptor type 4 (CXCR4)6−8 is of pivotal importance in this process. Various CXCR4-improving methods have been developed including genetic modification of MSCs to overexpress CXCR49 and hypoxic preconditioning of MSCs10,11 to induce CXCR4 expression, typically with less than 1% of MSCs reaching the target tissue.12
The ability to track stem cells noninvasively in vivo is of critical importance to follow the homing ability of stem cells and to monitor the therapeutic efficacy of stem cells after systemic delivery. Magnetic resonance imaging (MRI) is a key tool for cell-tracking studies. Various magnetic nanoparticles (MNPs), such as iron oxide (IO) MNPs, have been extensively used for tracking stem cells because of their superior signal-to-noise ratio, high spatial resolution, and safety of the T2*-weighted imaging.13,14 However, the inherent properties of MSCs after labeling with these MNPs have not been fully explored, despite growing evidence demonstrating that cellular responses to IO MNPs were indeed observed.15,16
Currently, the use of virus-mediated delivery is the most common strategy for modifying stem cells. However, a number of harmful effects limit its clinical applicability, including potential cell toxicity, mutagenesis, and the induction of an immune response. As such, an alternative nonviral delivery method is highly desirable. In this study, we made a serendipitous discovery that various iron-based MNPs themselves actively increase CXCR4 expression of mouse bone-marrow-derived MSCs, which offers a simple and safe method to potentially target MSCs to solid tumor and acute injury sites that secret SDF-1α. The potential molecular mechanism of CXCR4 induction by iron species was also explored. Furthermore, the advancement of MRI nanoprobes has significantly contributed to the tracking of stem cell homing to targeted sites; however, three major limitations have continued to prove daunting including nanoprobe sensitivity, cell labeling efficiency, and stem cell homing efficiency. To address these limitations, based on the newly found properties of MNPs induced to MSCs, we designed an iron-based nanocluster by combining zinc-doped iron oxide (Zn0.4Fe2.6O4) MNPs with hyaluronic acid–cholanic acid (HA–CA) amphiphilic polymer for effective implementation of MRI-guided delivery of cells to the target tissues. We demonstrate that this platform enhances the ability of MSCs to home in both traumatic brain injury (TBI) and glioblastoma models without apparent cytotoxicity and phenotype change of MSCs.
Results
Iron Oxide MNPs Actively Increase CXCR4 Expression of Bone-Marrow-Derived MSCs
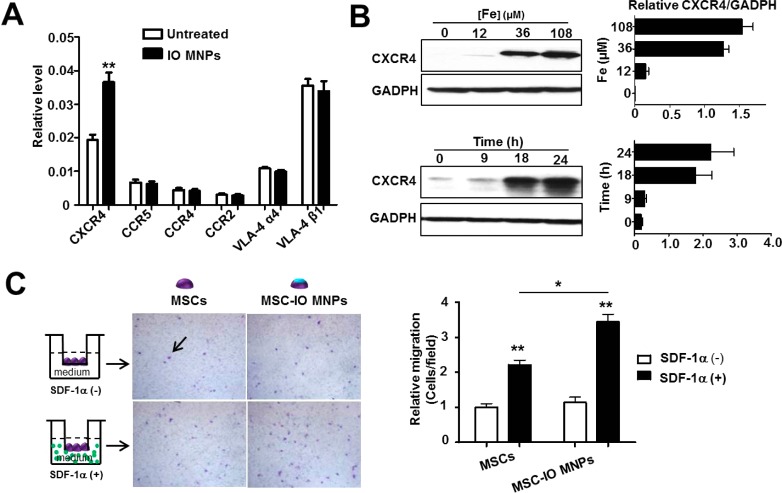
A critical property of MSCs for cell therapy is their intrinsic homing capacity. Several cell surface receptors such as VLA-4,17 as well as certain chemokine receptors including CXCR4 and CCR1-5,18 have been reported to mediate MSC homing. Thus, we sought to determine whether IO MNPs could actively regulate the expression of these MSC receptors. We first treated mouse bone-marrow-derived MSCs with different concentrations of IO MNPs for 2 h, and subsequently, the cells were continued to culture for different periods of time by replacing fresh medium, which mimics the practical application of MSC labeling. As shown in Figure 1A, we examined cell surface receptor expression by real-time PCR and found that CXCR4 mRNA level was significantly increased after IO MNP treatment (**p < 0.01), while other receptors were virtually unaltered. In addition, the CXCR4 protein level was also significantly induced in a concentration- and time-dependent manner (Figure 1B). CXCR4 is generally regarded as a membrane receptor, which is used to enhance homing engraftment of MSCs through increased cell invasion in response to SDF-1α, the ligand of CXCR4. To further explore the distribution of increased CXCR4 in MSCs, different subcellular protein fractions, including those from the cytoplasm, cell surface membrane, and nucleus, were stepwise isolated and CXCR4 expression was analyzed by Western blot. Supporting Information Figure S1 shows that IO MNP-labeled MSCs had increased CXCR4 expression on both cell surface membrane and cytoplasm. These results suggest the potential of improved homing of MSCs to inflammatory sites after labeling.
Figure 1.
Iron oxide magnetic nanoparticles (IO MNPs) increase CXCR4 expression in mesenchymal stem cells (MSCs). (A) Real-time PCR analysis of cell surface receptor expression of IO MNP-treated MSCs. MSCs were incubated with IO MNPs for 2 h. After changing to fresh medium, the cells were continued to culture for 22 h. (B) Western blot analysis of MSC CXCR4 expression after treatment with different concentrations of IO MNPs at the indicated culture time after labeling. The results of Western blotting were quantified by measuring the band density and then normalizing it to GADPH. (C) Effects of IO MNP labeling on MSC migration in the presence and absence of SDF-1α. Representative images of migrated MSCs (arrow) through 12 μm pore sized membrane. (Left) After particle labeling, the same number of MSCs were plated in the upper chambers at a density of 2 × 104 cell/well. Once the cells were in culture for an additional 22 h, the cells in the upper surface of the membrane were removed, while the cells migrated to the lower membrane surface were fixed and stained with crystal violet. (Right) Quantitative analysis of the migration of MSCs untreated and treated with IO MNPs in the presence and absence of SDF-1α. Values represent mean ± SD (*p < 0.05, **p < 0.01).
We next investigated whether the IO MNP-labeled MSCs had improved homing capacity in vitro. The labeled and unlabeled MSCs were plated in the chambers following the scheme shown in Figure 1C. After incubation for 22 h, the migrated MSCs were stained by crystal violet. In the presence of SDF-1α, the unlabeled MSCs migrated more easily to the lower membrane surface and showed a 2.21 ± 0.53-fold increase in migrated cell number than that in the absence of SDF-1α (*p < 0.01), demonstrating SDF-1α-tropic migration of MSCs. It is worth noting that IO MNP-labeled MSCs showed a 3.44 ± 0.87-fold increase in migrated cell number over the unlabeled ones. The results suggest that IO MNP-treated MSCs have significantly enhanced SDF-1α tropism than the untreated MSCs (*p < 0.05).
Mechanism of Enhanced CXCR4 Expression of MSCs by Iron-Based MNPs
Recent studies have shown that activation of the chemokine receptor CXCR4 by exogenous stimulus in cells is HIF-1α-dependent.19,20 To explore the mechanism by which IO MNPs induce MSC CXCR4 expression, we first examined HIF-1α expression level in MSCs after IO MNP labeling. As shown in Supporting Information Figure S2A, HIF-1α protein level in MSCs was obviously elevated with the increase of particle concentration, demonstrating a similar pattern with CXCR4 expression. To further verify this observation, we knocked down HIF-1α by transducing MSCs with HIF-1α shRNA lentivirus, which significantly blocked IO NP-induced CXCR4 expression (Supporting Information Figure S3), illustrating that HIF-1α mediated the induction of CXCR4 expression by IO NPs. It has been reported that iron overload regulates the changes of HIF-1α expression.21 As such, we next sought to elucidate whether iron level in cells changes signal transduction. After labeling with IO MNPs, increased iron content in MSCs was confirmed by inductively coupled plasma mass spectrometry analysis. In addition, the level of ferritin protein, an iron storage marker, was also significantly improved after treatment (Supporting Information Figure S2B, *p < 0.05). These results imply that iron ions were released from particles after cellular uptake, which in turn enhances HIF-1α and CXCR4 expression (Supporting Information Figure S2C).
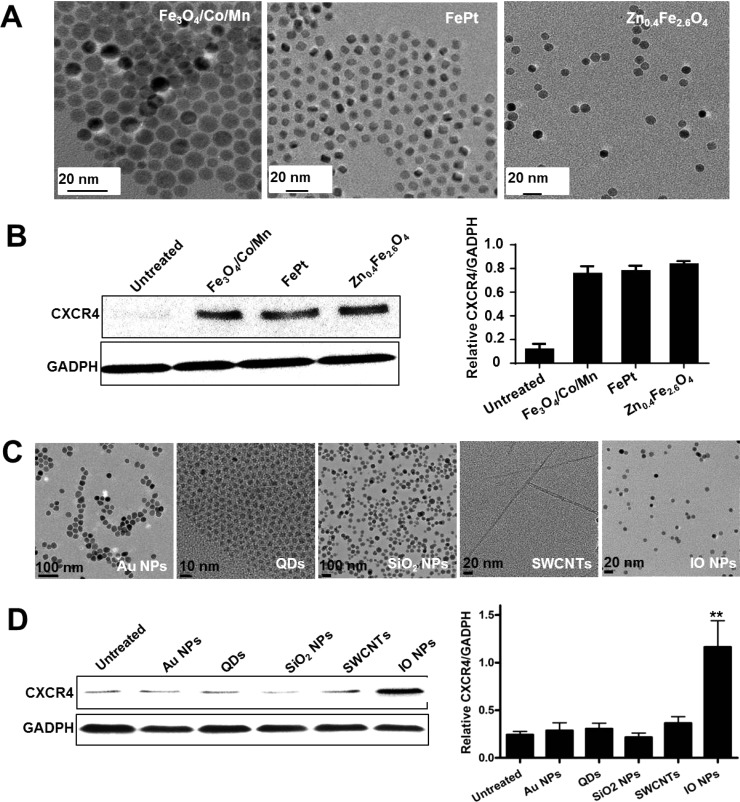
To further identify the iron-dependent mechanism, several other iron-based MNPs, such as Fe3O4/Co/Mn, FePt, and Zn0.4Fe2.6O4 (Figure 2A), with similar sizes and the same coating (PEGylation), were chosen to treat MSCs at the same iron concentration (108 μM). Consistent with IO MNPs, the other three iron-based MNPs also significantly induced CXCR4 expression of MSCs (Figure 2B). Furthermore, the specificity of iron-based NP-regulated CXCR4 expression was confirmed by treating MSCs with other nanomaterials, such as gold NPs, CdSe/ZnS quantum dots (QDs), mesoporous silica NPs (SiO2 NPs), and single-walled carbon nanotubes (SWNTs). No significant effect of these on iron-containing nanomaterials on CXCR4 expression on MSCs was observed (Figure 2C,D). These results verify that the increase of CXCR4 expression is iron-dependent, and that it is a general trend that various iron-based MNPs improve MSC CXCR4 expression.
Figure 2.
Effect of various NPs on CXCR4 expression. (A) TEM images of various iron-based MNPs, such as Fe3O4/Co/Mn, FePt, and Zn0.4Fe2.6O4, with similar sizes and the same coating (PEGylation). (B) After the MSCs were treated with various iron-based MNPs, the CXCR4 expression of cells was analyzed by Western blotting, which was quantified by measuring the band density and then normalized to GADPH. (C) TEM images of gold NPs, QDs, SiO2, SWCNTs, and IO MNPs. (D) Western blot analysis of CXCR4 expression after NP treatment. The bar graphs present the relative values from at least three independent experiments (**p < 0.01).
Improved Labeling Efficiency and Homing Capacity of MSCs by Designing an Iron-Based Nanocluster
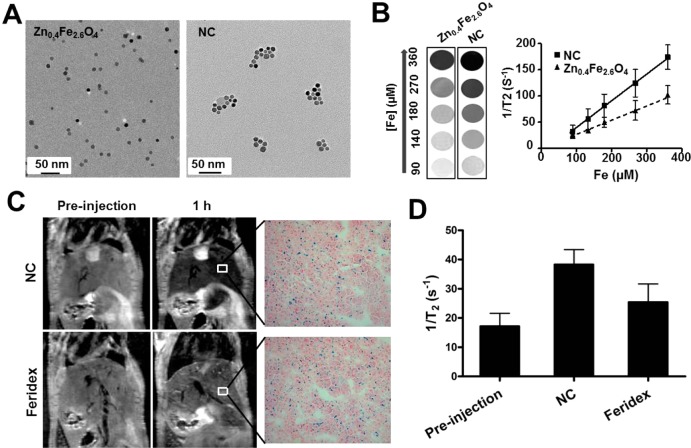
On the basis of the new properties of iron-based MNPs, which can induce CXCR4 expression, we designed an iron-based nanocluster (NC) to challenge the current limitations of tracking MSCs,22 including the suboptimal sensitivity of nanoprobes, relatively poor cell labeling, and unsatisfactory stem cell homing efficiency. To address the sensitivity of nanoprobes, we chose Zn-doped IO MNPs (Zn0.4Fe2.6O4) because of their much higher saturation magnetization value than crystalline IO MNPs and Feridex MNPs.23 Moreover, in our previous report,24 we also found the clustering structure of MNPs has much higher spin–spin (T2) relaxivity than single MNPs. To improve the MSC labeling efficiency, we chose hyaluronic acid (HA)-based polymers to modify the MNPs to improve the interaction with a specific cell surface receptor CD44, a critical surface marker of MSCs.25 Water-soluble HA–Zn0.4Fe2.6O4 nanoformula was first prepared by mixing surfactant-coated Zn0.4Fe2.6O4 with hyaluronic acid–cholanic acid amphiphilic polymer.26 As expected, Figure 3A shows that the Zn0.4Fe2.6O4 particles were uniform and monodispersed, which self-assemble into nanoclusters after HA–CA coating. The hydrodynamic diameter of HA–Zn0.4Fe2.6O4 NC was about 212.3 ± 5.7 nm, which is consistent with a previous report of other HA-based nanoparticles.26 The T2 relaxation times of as-synthesized HA–Zn0.4Fe2.6O4 NCs and the single Zn0.4Fe2.6O4 MNPs were measured at a 7 T MRI scanner with the same concentration and MRI sequence. As expected, HA–Zn0.4Fe2.6O4 NC showed significantly stronger r2 relaxivity than the Zn0.4Fe2.6O4 MNPs (Figure 3B). Next, in order to explore the in vivo T2 sensitivity, the NC was also compared to commercial Feridex MNPs for liver imaging. As shown in Figure 3C, after intravenous injection, both HA–Zn0.4Fe2.6O4 NC and Feridex MNPs showed rapid accumulation in the liver. The similar uptake of HA–Zn0.4Fe2.6O4 NC and Feridex MNPs in liver was found, but HA–Zn0.4Fe2.6O4 NC showed much better liver contrast than Feridex MNPs by quantitative analysis of 1/T2 values of HA–Zn0.4Fe2.6O4 NC and Feridex in liver (Figure 3D).
Figure 3.
Nanocluster improves T2 contrast effect. (A) TEM images of discrete Zn0.4Fe2.6O4 MNPs and HA–Zn0.4Fe2.6O4 NC. (B) T2*-weighted phantom images of Zn0.4Fe2.6O4 MNPs and HA–Zn0.4Fe2.6O4 NC at different iron concentrations. (C) Comparison of 1/T2 (r2) values of MNPs in the liver of mice using MR imaging. After injection of the same dose of MNPs (0.56 mg/kg iron), liver uptake of MNPs was evaluated by Prussian blue staining. (D) Quantitative analysis of 1/T2 values of HA–Zn0.4Fe2.6O4 NC and Feridex in liver (n = 3).
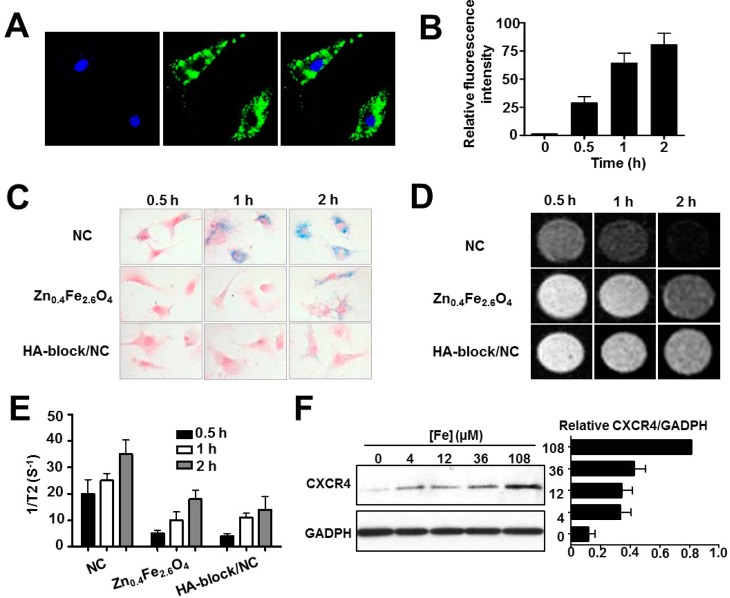
To assess cellular uptake of HA–Zn0.4Fe2.6O4 NC, HA-based polymer was conjugated with FITC prior to the fabrication of NCs. The resulting FITC-labeled HA–Zn0.4Fe2.6O4 (FITC-NCs) MNPs were incubated with MSCs for 2 h at an Fe concentration of 108 μM, and the cell images were then acquired by confocal microscopy. As shown in Figure 4A, bright fluorescence signals of the particles (green) were mainly distributed in the cytoplasm of MSCs. The intracellular fate of the NC was further explored by colocalization studies and transmission electron microscopy (TEM) images (Supporting Information Figure S4A,B), with the majority of the particles accumulated in the endosomes and lysosomes. Furthermore, FACS analysis was used to quantify the signal intensity of the particles taken up by MSCs. Compared with MSCs without adding NC, a gradual enhancement in signal was found with increased incubation time, displaying a 23 ± 8-, 51.3 ± 12.4-, and 64.3 ± 14.3-fold increase by incubation for 0.5, 1, and 2 h, respectively (Figure 4B). To further understand the intracelluar fate of particles, the localization of particles was studied at 24 h after labeling. TEM image illustrated that a portion of the particles escaped from the lysosome and merged into the cytosol of MSCs (Supporting Information Figure 4C).
Figure 4.
Enhanced cellular uptake and CXCR4 expression in HA–Zn0.4Fe2.6O4 NC-labeled MSCs. (A) Confocal images of HA–Zn0.4Fe2.6O4 NC cellular uptake after 2 h incubation. (B) FACS analysis of HA–Zn0.4Fe2.6O4 NC cellular uptake. The MSCs were incubated with FITC-NC for the indicated time periods. (C) Cellular uptake was evaluated by Prussian blue staining. (D,E) T2*-weighted images and quantitative analysis of cellular uptake at the same cell concentration. (F) Western blot analysis of CXCR4 expression in HA–Zn0.4Fe2.6O4 NC-labeled MSCs.
Next, we investigated whether HA coating improved cellular uptake of NCs. Cellular uptake of HA–Zn0.4Fe2.6O4 NC, PEGylated Zn0.4Fe2.6O4 MNPs, and NC plus free HA polymer to block the target (HA-block/NC) was measured by Prussian blue staining (Figure 4C). After incubation with MSCs for 0.5 h, uptake of NC was observed, whereas no significant uptake was found for PEGylated Zn0.4Fe2.6O4 MNPs or HA-block/NC treatment. The NC uptake was obviously increased along with time, showing that the internalization of MNPs was time-dependent. In contrast, HA-block/NC showed very few blue granules in the cytoplasm of MSCs after the same incubation time. Improved cellular uptake after HA coating was also confirmed by MRI study. Quantitative analysis of the relaxation time of the same number of cells (1 × 106/mL) showed consistent results with Prussian blue staining (Figure 4D,E). Collectively, these results indicate that the increased labeling efficiency of the NC is specifically mediated by HA–CD44 binding.
In addition, we determined whether the HA–Zn0.4Fe2.6O4 NC increased CXCR4 expression. The MSCs were treated with different concentrations of NC for 2 h and further cultured for an additional 22 h by replacing the medium. After cell collection, CXCR4 protein expression was analyzed by Western blot. As expected, the CXCR4 level in MSCs was enhanced with the increasing iron concentration (Figure 4F), which corroborates the results of other iron-based MNPs. On the basis of the induction of CXCR4 by the NC, we next tested whether NC labeling could improve the migration capacity of MSCs. In accordance with the IO MNP-labeled MSCs, transwell assay results showed that more HA–Zn0.4Fe2.6O4 NC-labeled MSCs migrated to the lower membrane than unlabeled MSCs in the presence of SDF-1α (Supporting Information Figure S5, *p < 0.05). These results imply that the HA–Zn0.4Fe2.6O4 NC formula might overcome the limitations of tracking targeted delivery of stem cells.
Cytotoxicity of Nanoclusters on MSCs
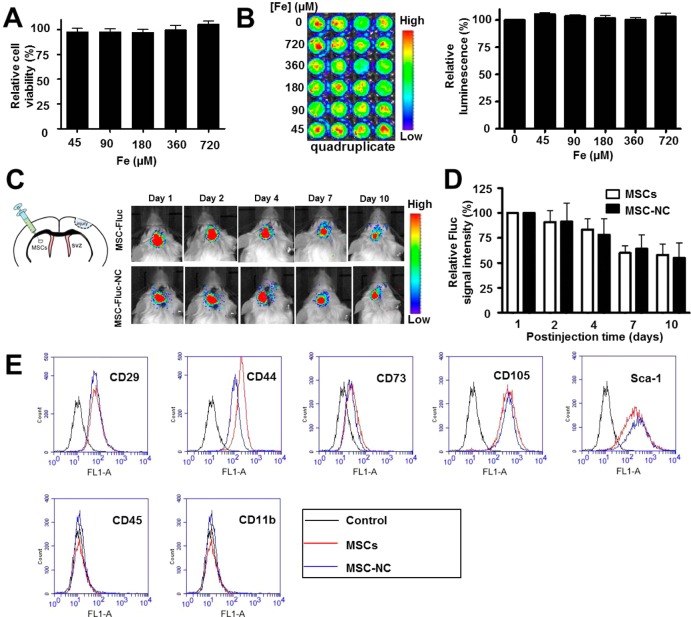
To understand the cytotoxicity of HA–Zn0.4Fe2.6O4 NC labeling on the properties of MSCs, cell viability was evaluated using the standard 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) assay, and the results demonstrated no obvious change in cell proliferation after labeling (Figure 5A and Supporting Information Figure S6). We next imaged luciferase expression of MSCs to confirm the non-cytotoxicity of particles. The MSCs expressing firefly luciferase (MSC-Fluc) were isolated from the luciferase transgenic mice following the same procedure as MSC isolation. The MSC-Fluc cells were treated with different concentrations of HA–Zn0.4Fe2.6O4 NC formula for 48 h, and subsequently, the bioluminescence images were acquired. Quantitative analysis of luciferase expression showed negligible effect of particle labeling on cell proliferation after NC labeling (Figure 5B). To further test the effect of NC on MSC activity, we transplanted 5 × 104 MSC-Fluc cells in a mouse traumatic brain injury model by intraparenchymal injection. The luciferase activities of unlabeled and labeled MSCs were acquired and analyzed by bioluminescence imaging at different time points after cell injection; again no difference in viability was found between the NC-labeled and unlabeled MSCs (Figure 5C,D).
Figure 5.
Effects of HA–Zn0.4Fe2.6O4 NC on MSC properties. (A,B) In vitro cytotoxicity was investigated by (A) MTT assay and (B) firefly luciferase (Fluc) bioluminescence imaging after 48 h treatment with different concentrations of particles. (C) Luciferase activity of unlabeled and labeled MSC-Fluc was acquired and (D) analyzed by a Xenogen IVIS100 imaging system. The MSC-Fluc cells were untreated and treated with HA–Zn0.4Fe2.6O4 NC for 2 h, and subsequently, 5 × 104 cells were injected into brain parenchyma in a traumatic brain injury model. (E) FACS analyses of the surface markers of labeled and unlabeled MSCs after 24 h treatment.
In light of the increased CXCR4 expression on the MSC membrane surface after NC labeling, we next studied whether the cell surface markers of MSCs were also affected. MSCs were treated with NC for 2 h, after which cells were continued to culture for 22 h. Phenotypic analyses of surface markers of both NC-labeled and unlabeled MSCs showed positive staining for CD29, CD44, CD73, CD105, and Sca-1 and negative staining for CD45 and CD11b (Figure 5E). Moreover, long-term effect of NC labeling on MSC surface markers was performed by further culturing MSCs for 10 days. Similar surface marker staining pattern was observed as shown in Supporting Information Figure S7. These results further confirm no obvious effect of NC on the properties of MSCs after labeling.
In Vivo MRI-Guided Therapy in a Traumatic Brain Injury Model
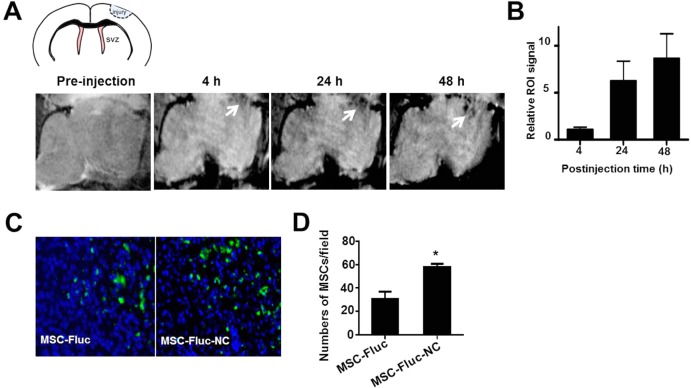
Traumatic brain injury model that highly secretes chemokine SDF-1α is a representative exogenous brain disease in patients.27 Despite considerable advances in therapy, the cure of TBI remains a challenge due to several complicating factors, such as blood-brain barrier, drug resistance, and limited self-renewal capacity of the brain. MSCs have been shown to migrate toward TBI in vivo.28 Direct targeting of TBI by engineered MSCs could be a promising therapeutic approach. To verify whether NC-labeled MSCs can improve migration capacity of MSCs toward TBI, we first assessed the targeted delivery of MSC-NC cells by MRI. MR images of mouse brains with TBI (Figure 6A) were performed pre- and postinjection of 1 × 106 NC-labeled MSCs (n = 4/group). T2* signal at the region of TBI was increased over time after MSC–NC injection, and well-defined hypo-intensities (black) at the injury site were observed at 24 h postinjection. Region of interest analysis showed substantial decrease in gray values for TBI at 24 and 48 h time points than that of preinjection (Figure 6B). The migration capacity of NC-labeled MSCs toward TBI was also approved by Prussian blue staining (Supporting Information Figure S8). In addition, the improved homing capacity of MSC-NC was confirmed by finding more NC-labeled MSC-Fluc than unlabeled MSC-Fluc in the TBI region (Figure 6C, *p < 0.05) via immuno-histostaining of frozen slices at 48 h after intravenous injection of the cells. Furthermore, we showed more tumor homing of NC-labeled MSCs than the unlabeled MSCs in an orthotopic U87MG glioblastoma model (Supporting Information Figure S9), suggesting the potential use of the NC formula in other disease types including cancer.
Figure 6.
In vivo imaging of targeted delivery of MSCs to TBI site after intravenous injection. (A) In vivo MR images of the delivery of HA–Zn0.4Fe2.6O4 NC-labeled MSCs to the injury site (arrow). MSCs (1 × 106) were incubated with and without particles for 2 h, then collected and injected intravenously to TBI mice. MR images were acquired on a 7 T small animal MRI scanner. (B) Region of interest analysis of the TBI site pre- and postinjection of delivery of NC-labeled MSCs. (C) Immunofluorescence staining of frozen brain tissue slices for Fluc at 48 h after intravenous injection of 1 × 106 MSC-Fluc cells. Representative images of MSC-Fluc and MSC-Fluc-NC in injury sites. (D) Quantitative analysis of Fluc-positive MSCs by randomly selected fields in (C) (*p < 0.05).
In view of the substantial homing effect of MSC-NC observed, we next set to explore whether the injection of MSCs could improve TBI recovery. As shown in Figure 7A,B, compared with TBI alone, MSC-treated mice sacrificed at day 10 demonstrated less scarring and decreased contusion volume upon post-mortem examination of brain tissues. It is worth noting that hematoxylin and eosin (H&E) staining of tissue slides revealed significantly enhanced gliosis and increased cellular density with MSC-NC compared with MSC alone treatment following TBI (*p < 0.05).
Figure 7.
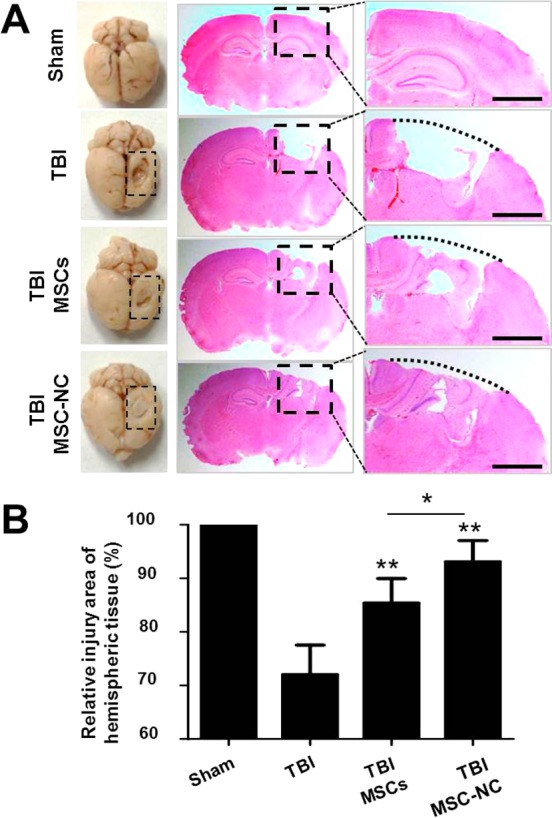
MSCs improve TBI repair. (A) Injury recovery (dashed line box) of brain tissues was observed by H&E staining after tail vein injection of 1 × 106 unlabeled and NC-labeled MSCs. The whole hemispheric tissue was shown in the middle lane, and the injury site was enlarged in the right lane. (B) Injured area was quantified in brain tissues of mice treated with unlabeled and labeled MSCs and compared with TBI alone (*p < 0.05, **p < 0.01).
Discussion
Acute TBI breaks the impermeability of blood-brain barrier, which triggers immune response and release of inflammatory cytokines.29 Despite the multipotency and self-renewing characteristics of MSCs, it is still a subject of debate whether the regenerative potential of TBI tissue exerted by these cells is due to transdifferentiation and substitution of dead cells or the secretion of a soluble factor which stimulates local progenitor cells to survive, proliferate, and differentiate.30,31 Accumulating evidence suggests that the efficiency of MSCs homing to injured tissues is an important parameter influencing the outcome of MSC therapy by immunoregulating the local environment of injury sites.32 Although the precise molecular mechanisms by which MSCs migrate and home into sites of injury have not yet been fully understood, high expression of CXCR4 on MSCs using genetic modification, specific cytokines, or hypoxic preconditioning can promote MSCs to home to damaged tissues following systemic administration.
This study represents the first evidence that iron-based MNPs can actively increase the expression of chemokine receptor CXCR4 in bone-marrow-derived MSCs without the need for genetic modification and result in improved homing of MSCs to the injury sites and tumor. The new property that MNPs enhance CXCR4 expression of stem cells undoubtedly expands the current bioapplications of MNPs beyond conventional DNA, protein and cell separation, magnetic biosensors, MRI, tissue engineering, and hyperthermia, etc. Unlike the complex process and potential safety concerns of genetic modification, this simple method was implemented by various iron-based MNPs with different iron states (Figure 2). In fact, iron is vital for almost all organisms.33 It serves as a cofactor for many proteins and enzymes necessary for oxygen and energy metabolism, as well as for several other essential processes. Regulation of iron uptake, storage, intracellular trafficking, and utilization is involved in the cellular iron metabolism process.34 Most cells in the body acquire iron from transferrin (Tf). Iron loaded in Tf binds to Tf receptors on the cell membrane and subsequently enters endosomes via a clathrin-dependent endocytosis mechanism. Similarly, IO MNPs are degraded by endosome to release iron ions after cellular uptake (Supporting Information Figures S4 and S10) and further regulate molecular expression in MSCs. Previous studies demonstrated that SDF-1α and CXCR4 are expressed in complementary patterns35 which guide primordial stem cells to sites of rapid vascular expansion. Induction of SDF-1α/CXCR4 expression via HIF-1α can directly guide regenerative progenitor cells to areas of injury.36,37 In this study, we also found increased CXCR4 after iron-based MNP labeling was regulated by HIF-1α. Furthermore, CXCR4 expression by IO MNPs can be induced in a time- and concentration-dependent manner. These results imply that CXCR4 regulation might be a complex process involving varied mechanisms in different cellular environments. The detailed mechanism will be further explored in future studies.
The ability to track stem cells noninvasively in vivo is of critical importance to follow the homing ability of stem cells and to monitor the therapeutic efficacy of stem cells after systemic delivery. The current stem-cell-tracking methods are challenged by limited sensitivity, low cell labeling efficiency, and poor stem cell homing efficiency. To address these, we designed iron-based NCs by combining the superiority of zinc-doped IO (Zn0.4Fe2.6O4) MNPs and the new findings that chemokine receptor CXCR4 expression was increased after IO MNP labeling. Our design of HA–Zn0.4Fe2.6O4 NC has several advantages. First, previous reports have shown that zinc-doped iron oxide MNPs have superior T2 MR imaging contrast effect than those of undoped Fe3O4 and Feridex.23,38 Second, HA coating is introduced to improve the interaction with a specific cell surface receptor CD44, a critical surface marker of MSCs.39 Iron content in the Zn0.4Fe2.6O4 increases CXCR4 expression in MSCs, which is used to enhance the homing of MSCs to the target site. As expected, T2-weighted contrast effect of HA–Zn0.4Fe2.6O4 NC is not only better than undoped Fe3O4 and Feridex but also Zn0.4Fe2.6O4 (Figure 3). One possible reason is that the r2 value of MNPs is strongly related to interparticle distance.24,40 The clustering of Zn0.4Fe2.6O4 MNPs in the HA–CA nanoparticle leads to much higher spin–spin (T2) relaxation times than single MNPs. Furthermore, specific binding of HA to CD44 overexpressed on the surface of MSCs leads to highly effective cellular uptake of the HA–Zn0.4Fe2.6O4 NC. More importantly, MSCs can be induced to express high level of the chemokine receptor CXCR4 in the presence of NC for improved TBI tropism and therapeutic effect, suggesting the potential application of this new finding.
Conclusions
In this work, we demonstrate for the first time that iron-based MNPs not only can label MSCs and track the fate of cells by MRI but also mediate MSC homing to the target site. This MR imaging-guided targeted delivery of magnetic nanoparticle-labeled stem cell approach opens up a wide range of new applications in regenerative medicine and oncological research and provides a new insight into the interface between nanotechnology and medicine. The findings that iron intake by mesenchymal stem cells from the carefully designed zinc-doped IO MNPs may be rapidly translatable into the clinic to improve cell therapy efficacy without genetic modifications.
Methods
Preparation of Mouse MSCs and MSC-Fluc
Isolation and culture of MSCs and MSC-Fluc were performed by following our previously reported procedures.41,42 Briefly, balb/c mice and FVB mice transfected luciferase were sacrificed by cervical dislocation. The marrow was harvested by inserting a syringe needle (27-gauge) into one end of the bone and flushing with PBS. The bone marrow cells were filtered through a 70 mm nylon mesh filter. After collection by centrifugation at 2000 rpm, cells were dispersed into MesenCult MSC basal medium (Mouse) (STEMCELL Technology), a standardized basal medium for the in vitro culture of mouse MSCs or MSC-Fluc. Then, cells were cultured at 37 °C in a humidified atmosphere containing 95% air and 5% CO2. The adhered cells were split when they reached 80–90% confluence. The MSC surface marker expression profile was confirmed by FACS prior to the use of MSCs and MSC-Fluc.
CXCR4 Expression of MSCs
The MSCs were treated without and with the indicated concentration of particles for 2 h and, subsequently, washed twice by PBS. Fresh medium was added and cultured for different time points. After collection of cells, CXCR4 expression of MSCs was performed by FACS and Western blot.
To analyze CXCR4 expression in different subcellular protein fractions, MSCs were incubated without or with IO MNPs (108 μM) for 2 h. The cells were cultured for 22 h following medium being replaced. After collection of cells, the proteins expressed in cytoplasm, membranem and nucleus were stepwise separated by a subcellular protein fractionation kit (Thermo Scientific).
HIF-1α Knockdown
The following plasmids were used: nonspecific or mouse HIF-1α shRNA for gene knockdown (Open Biosystems). Lentiviruses for knockdown were prepared using the respective plasmids according to manufacturer’s protocol (Sigma-Aldrich). Cells were incubated with virus (MOI ∼ 1) for 24 h followed by NP treatment. Western blotting was performed for detecting expression of HIF-1α.
Preparation of Various NPs and Zn0.4Fe2.6O4–HA Nanoclusters
Iron oxide Fe3O4 (IO),43 Fe3O4/Co/Mn,44 FePt45 and Zn0.4Fe2.6O446 MNPs, SiO2 NPs,47 and SWNTs48 were synthesized based on the previous reports. PEGylated iron-based MNPs were prepared following a previous procedure.43 Gold NPs and CdSe/ZnS QDs were obtained from Nanocs Inc. and Ocean NanoTech, LLC, respectively.
To obtain water-soluble HA–Zn0.4Fe2.6O4 nanoclusters, HA-based polymer was first synthesized by following a previously reported procedure.49 Then, 3 mg of HA-based polymer or HA–FITC-based polymer was solubilized into 1.5 mL of distilled water, and subsequently, 0.3 mL (5.2 mg/mL) of Zn0.4Fe2.6O4 MNPs in toluene was added. The solutions were dispersed via probe sonication using a VCX-750 ultrasonic processor (Sonics & Materials, Newtown, CT). The probe was driven at 80% of the instruments’ maximum amplitude in an ice bath. The particles were well-dispersed in solution around 5 min sonication. After sonication, the solution was heated to 50 °C in an opened vial for overnight. The particles were redispersed into 1 mL of distilled water and then purified by a high strength magnet. Finally, the NC was separated by a disposable PD-10 desalting column (GE Healthcare). The NC was observed by transmission electron microscopy. The iron concentration was quantified by an inductively coupled plasma mass spectrometry.
Cellular Uptake of NCs
MSCs were plated 24 h before the start of the experiment in chamber slides at a density of 5 × 103 cell/cm2. The cells were incubated with 10 μg/mL NCs and PEGylated Zn0.4Fe2.6O4 for the indicated time periods. For the fluorescence imaging of NCs, the cells were incubated with 10 μg/mL NCs and then the cells were fixed with Z-fix solution (Anatech, Battle Creek, MI) for 15 min. The cells were incubated with 0.1% Triton X-100 in PBS at room temperature for 5 min and subsequently incubated with Alexa Fluor 568 phalloidin (Invitrogen) for staining F-actin for 20 min, followed by 1.5 μg/mL DAPI staining at room temperature. The slides were observed, and the images were acquired with an Olympus confocal microscope (Olympus FV10i).
To demonstrate HA-dependent cellular uptake of particles, the control experiment was also performed by adding 10-fold excess of HA-based polymer prior to incubation with NCs. After the cells were washed three times with PBS buffer, the cells were stained with Prussian blue solution containing 20% (v/v) hydrochloric acid and 10% (v/v) potassium ferrocyanide solution or MRI phantom observation.
In Vitro Chemotaxis Assay
Cell chemotaxis assay was performed using a modified protocol from the manufacturer (12 mm pore size, Corning Incorporated). After incubation with and without particles for 2 h, MSCs were trypsinized and resuspended in the medium. The cells were plated in the upper chambers at the density of 1 × 104 cell/well. The lower chamber containing nonserum medium was added without and with 0.1 μg/mL SDF-1α. After incubation at 37 °C overnight, cells remaining at the upper surface of the membrane were removed using a swab, whereas the cells that migrated to the lower membrane surface were fixed with Z-fix solution and stained with 2% crystal violet. The number of cells migrating through the filter was counted and plotted as the number of migrating cells per optic field (×10).
Orthotopic Glioblastoma Model and Traumatic Brain Injury Model
The procedures for developing an orthotopic glioblastoma model and TBI model were performed according to a protocol approved by the National Institutes of Health Clinical Center Animal Care and Use Committee (NIH CC/ACUC). For the orthotopic glioblastoma model, briefly, male athymic nude mice (4–6 weeks) were injected intracranially with 1 × 105 U87MG cells in the right frontal lobe at coordinates 1.5 mm lateral from the bregma, 0.5 mm anterior, and 2.5 mm intraparenchymal. Tumor cells were allowed to engraft for 3 weeks. Successful tumor model was confirmed by MRI prior to the injection of MSCs.
TBI model was induced in male balb/c mice (6–8 weeks) by a controlled cortical impact model (CCI). Briefly, mice were anesthetized by isoflurane (3% for induction and 1.5% for maintenance) and then placed in a stereotaxic device. After drilling a hole (4 mm × 4 mm) on the right area of the skull, an impact tip with a diameter of 2 mm was used to contuse the brain tissue. The tip penetration depth was 1.5 mm and velocity was 5 m/s. A warming pad was used to keep body temperatures around 37 °C during the surgery. Intraparenchymal injection of MSCs was performed 24 h after TBI. Briefly, mice were placed in a stereotaxic device, and a small hole (0.2 mm) was drilled on the left area of skull (1.0 mm caudal to the bregma and −1.3 mm lateral to the midline at a depth of 3.0 mm). Then 5 × 104 MSCs in 6 μL of PBS were injected carefully into brain parenchyma with a Hamilton syringe within 10 min.
In Vivo Imaging
The luciferase activity of MSC-Fluc was visualized and quantified by a Xenogen IVIS-100 system (Caliper Life Sciences, Hopkinton, MA, USA). In brief, mice were anesthetized with 2% isoflurane in O2 and received i.p. injection of d-luciferin solution in PBS at a dose of 150 mg/kg. Serial images were acquired between 5 and 20 min after d-luciferin administration, and the bioluminescence signal intensity was quantified.
VMRI studies were conducted in a 7 T horizontal bore small animal MRI scanner (Bruker Biospin). All mice were anesthetized with 1–2% isoflurane mixed with pure oxygen via a nose cone and were placed in a stretched supine position with a respiratory sensor. Axial and coronal two-dimensional (2D) fast spin–echo sequence images were first acquired to ensure the imaging position of the implanted tumor. The following parameters were adopted in data acquisition: (1) T1-weighted multislice gradient-echo images: TR/TE = 250/4.5 ms, matrix = 256 × 256, FA = 30, 9 contiguous slices; (2) T2-weighted multislice spin–echo images: TR/TE = 2000/48 ms, matrix = 256 × 256, 9 contiguous slices; (3) T2*-weighted images: TR/TE = 1500/4 ms, matrix = 256 × 256, FA = 30, 9 contiguous slices; (4) T2-map: TR = 2000/48 ms, matrix = 256 × 256, 1 slice, T2 relaxation measurements with TE of 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 110, 120, 130, 140, 150, and 160 ms. For each data set, one slice with comparable locations within the tumor was selected to determine signal intensities. Signal intensities were measured in defined regions of interest with ImageJ (National Institutes of Health).
The 3–7 generations of MSCs were prepared for mouse imaging after particle labeling. Briefly, MSCs were incubated with particles (108 μM) for 2 h. After the cells were washed twice, MSCs were trypsinized and resuspended in the PBS. Mice were anesthetized with isoflurane, and subsequently, 1 × 106 MSCs were injected by tail vein injection at a speed of 0.1 mL/min. The mice were imaged by MRI at the indicated time points.
Frozen glioblastoma and TBI tissue slices (5 μm) were fixed with cold acetone for 20 min and dried in the air for 30 min at room temperature. After blocking with 1% BSA for 30 min, the sections were incubated with anti-luciferase primary antibody (Abcam) for 2 h at room temperature and then visualized with Alexa Fluor 488-conjugated secondary antibodies (1:200). After being washed with PBS, the whole slides were mounted with DAPI-containing mounting medium. Fluorescence images were acquired with an epifluorescence microscope (Olympus, X81).
Data and Statistical Analysis
All graphs were constructed and statistical analysis performed using Graphpad Prism software v.5.00 (GraphPad Software). A one-way ANOVA with a posthoc Tukey test was used to identify significant differences among treatment groups. Significance was set at p < 0.05 unless otherwise stated.
Acknowledgments
This work was supported, in part, by the Intramural Research Program (IRP) of the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), and by the Department of Defense (DOD) Center for Neuroscience and Regenerative Medicine (CNRM).
Author Contributions
⊥ X.H. and F.Z. contributed equally to this work.
Supporting Information Available
Supplementary figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Mooney D. J.; Vandenburgh H. Cell Delivery Mechanisms for Tissue Repair. Cell Stem Cell 2008, 2, 205–213. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Jahagirdar B. N.; Reinhardt R. L.; Schwartz R. E.; Keene C. D.; Ortiz-Gonzalez X. R.; Reyes M.; Lenvik T.; Lund T.; Blackstad M.; et al. Pluripotency of Mesenchymal Stem Cells Derived from Adult Marrow. Nature 2002, 418, 41–49. [DOI] [PubMed] [Google Scholar]
- Voswinkel J.; Francois S.; Simon J. M.; Benderitter M.; Gorin N. C.; Mohty M.; Fouillard L.; Chapel A. Use of Mesenchymal Stem Cells (MSC) in Chronic Inflammatory Fistulizing and Fibrotic Diseases: A Comprehensive Review. Clin. Rev. Allergy Immunol. 2013, 45, 180–192. [DOI] [PubMed] [Google Scholar]
- Karp J. M.; Leng Teo G. S. Mesenchymal Stem Cell Homing: The Devil Is in the Details. Cell Stem Cell 2009, 4, 206–216. [DOI] [PubMed] [Google Scholar]
- Sarkar D.; Spencer J. A.; Phillips J. A.; Zhao W.; Schafer S.; Spelke D. P.; Mortensen L. J.; Ruiz J. P.; Vemula P. K.; Sridharan R.; et al. Engineered Cell Homing. Blood 2011, 118, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled A.; Petit I.; Kollet O.; Magid M.; Ponomaryov T.; Byk T.; Nagler A.; Ben-Hur H.; Many A.; Shultz L.; et al. Dependence of Human Stem Cell Engraftment and Repopulation of Nod/Scid Mice on CXCR4. Science 1999, 283, 845–848. [DOI] [PubMed] [Google Scholar]
- Dong F.; Harvey J.; Finan A.; Weber K.; Agarwal U.; Penn M. S. Myocardial CXCR4 Expression Is Required for Mesenchymal Stem Cell Mediated Repair Following Acute Myocardial Infarction. Circulation 2012, 126, 314–324. [DOI] [PubMed] [Google Scholar]
- Cheng Z.; Ou L.; Zhou X.; Li F.; Jia X.; Zhang Y.; Liu X.; Li Y.; Ward C. A.; Melo L. G.; et al. Targeted Migration of Mesenchymal Stem Cells Modified with CXCR4 Gene to Infarcted Myocardium Improves Cardiac Performance. Mol. Ther. 2008, 16, 571–579. [DOI] [PubMed] [Google Scholar]
- Zhang D.; Fan G. C.; Zhou X.; Zhao T.; Pasha Z.; Xu M.; Zhu Y.; Ashraf M.; Wang Y. Over-Expression of CXCR4 on Mesenchymal Stem Cells Augments Myoangiogenesis in the Infarcted Myocardium. J. Mol. Cell Cardiol. 2008, 44, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azab A. K.; Hu J.; Quang P.; Azab F.; Pitsillides C.; Awwad R.; Thompson B.; Maiso P.; Sun J. D.; Hart C. P.; et al. Hypoxia Promotes Dissemination of Multiple Myeloma through Acquisition of Epithelial to Mesenchymal Transition-like Features. Blood 2012, 119, 5782–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.; Lu C.; Liu H.; Rao S.; Cai J.; Liu S.; Kriegel A. J.; Greene A. S.; Liang M.; Ding X. Hypoxic Preconditioning with Cobalt of Bone Marrow Mesenchymal Stem Cells Improves Cell Migration and Enhances Therapy for Treatment of Ischemic Acute Kidney Injury. PLoS One 2013, 8, 62703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash I. M.; Chouraqui P.; Baron J.; Feinberg M. S.; Etzion S.; Tessone A.; Miller L.; Guetta E.; Zipori D.; Kedes L. H.; et al. Systemic Delivery of Bone Marrow-Derived Mesenchymal Stem Cells to the Infarcted Myocardium: Feasibility, Cell Migration, and Body Distribution. Circulation 2003, 108, 863–868. [DOI] [PubMed] [Google Scholar]
- Hinds K. A.; Hill J. M.; Shapiro E. M.; Laukkanen M. O.; Silva A. C.; Combs C. A.; Varney T. R.; Balaban R. S.; Koretsky A. P.; Dunbar C. E. Highly Efficient Endosomal Labeling of Progenitor and Stem Cells with Large Magnetic Particles Allows Magnetic Resonance Imaging of Single Cells. Blood 2003, 102, 867–872. [DOI] [PubMed] [Google Scholar]
- Loebinger M. R.; Kyrtatos P. G.; Turmaine M.; Price A. N.; Pankhurst Q.; Lythgoe M. F.; Janes S. M. Magnetic Resonance Imaging of Mesenchymal Stem Cells Homing to Pulmonary Metastases Using Biocompatible Magnetic Nanoparticles. Cancer Res. 2009, 69, 8862–8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T. H.; Hsiao J. K.; Hsu S. C.; Yao M.; Chen Y. C.; Wang S. W.; Kuo M. Y.; Yang C. S.; Huang D. M. Iron Oxide Nanoparticle-Induced Epidermal Growth Factor Receptor Expression in Human Stem Cells for Tumor Therapy. ACS Nano 2011, 5, 9807–9816. [DOI] [PubMed] [Google Scholar]
- Kim H. S.; Oh S. Y.; Joo H. J.; Son K. R.; Song I. C.; Moon W. K. The Effects of Clinically Used MRI Contrast Agents on the Biological Properties of Human Mesenchymal Stem Cells. NMR Biomed. 2010, 23, 514–522. [DOI] [PubMed] [Google Scholar]
- Ruster B.; Gottig S.; Ludwig R. J.; Bistrian R.; Muller S.; Seifried E.; Gille J.; Henschler R. Mesenchymal Stem Cells Display Coordinated Rolling and Adhesion Behavior on Endothelial Cells. Blood 2006, 108, 3938–3944. [DOI] [PubMed] [Google Scholar]
- Ponte A. L.; Marais E.; Gallay N.; Langonne A.; Delorme B.; Herault O.; Charbord P.; Domenech J. The In Vitro Migration Capacity of Human Bone Marrow Mesenchymal Stem Cells: Comparison of Chemokine and Growth Factor Chemotactic Activities. Stem Cells 2007, 25, 1737–1745. [DOI] [PubMed] [Google Scholar]
- Staller P.; Sulitkova J.; Lisztwan J.; Moch H.; Oakeley E. J.; Krek W. Chemokine Receptor CXCR4 Downregulated by von Hippel–Lindau Tumour Suppressor pVHL. Nature 2003, 425, 307–311. [DOI] [PubMed] [Google Scholar]
- Ceradini D. J.; Kulkarni A. R.; Callaghan M. J.; Tepper O. M.; Bastidas N.; Kleinman M. E.; Capla J. M.; Galiano R. D.; Levine J. P.; Gurtner G. C. Progenitor Cell Trafficking Is Regulated by Hypoxic Gradients through HIF-1 Induction of SDF-1. Nat. Med. 2004, 10, 858–864. [DOI] [PubMed] [Google Scholar]
- Andrews M.; Arredondo M. Hepatic and Adipocyte Cells Respond Differentially to Iron Overload, Hypoxic and Inflammatory Challenge. Biometals 2012, 25, 749–759. [DOI] [PubMed] [Google Scholar]
- Kircher M. F.; Gambhir S. S.; Grimm J. Noninvasive Cell-Tracking Methods. Nat. Rev. Clin. Oncol. 2011, 8, 677–688. [DOI] [PubMed] [Google Scholar]
- Cho M. H.; Lee E. J.; Son M.; Lee J. H.; Yoo D.; Kim J. W.; Park S. W.; Shin J. S.; Cheon J. A Magnetic Switch for the Control of Cell Death Signalling in In Vitro and In Vivo Systems. Nat. Mater. 2012, 11, 1038–1043. [DOI] [PubMed] [Google Scholar]
- Liu G.; Xie J.; Zhang F.; Wang Z.; Luo K.; Zhu L.; Quan Q.; Niu G.; Lee S.; Ai H.; et al. N-Alkyl-PEI-Functionalized Iron Oxide Nanoclusters for Efficient Sirna Delivery. Small 2011, 7, 2742–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toole B. P. Hyaluronan: From Extracellular Glue to Pericellular Cue. Nat. Rev. Cancer 2004, 4, 528–539. [DOI] [PubMed] [Google Scholar]
- Choi K. Y.; Jeon E. J.; Yoon H. Y.; Lee B. S.; Na J. H.; Min K. H.; Kim S. Y.; Myung S. J.; Lee S.; Chen X.; et al. Theranostic Nanoparticles Based on PEGylated Hyaluronic Acid for the Diagnosis, Therapy and Monitoring of Colon Cancer. Biomaterials 2012, 33, 6186–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H.; Natsume A.; Wakabayashi T.; Aoshima C.; Shimato S.; Ito M.; Ishii J.; Maeda Y.; Hara M.; Kim S. U.; et al. Intravenously Transplanted Human Neural Stem Cells Migrate to the Injured Spinal Cord in Adult Mice in an SDF-1- and HGF-Dependent Manner. Neurosci. Lett. 2007, 426, 69–74. [DOI] [PubMed] [Google Scholar]
- Francois S.; Bensidhoum M.; Mouiseddine M.; Mazurier C.; Allenet B.; Semont A.; Frick J.; Sache A.; Bouchet S.; Thierry D.; et al. Local Irradiation Not Only Induces Homing of Human Mesenchymal Stem Cells at Exposed Sites but Promotes Their Widespread Engraftment to Multiple Organs: A Study of Their Quantitative Distribution after Irradiation Damage. Stem Cells 2006, 24, 1020–1029. [DOI] [PubMed] [Google Scholar]
- Ziebell J. M.; Morganti-Kossmann M. C. Involvement of Pro- and Anti-inflammatory Cytokines and Chemokines in the Pathophysiology of Traumatic Brain Injury. Neurotherapeutics 2010, 7, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Mal N.; Kiedrowski M.; Chacko M.; Askari A. T.; Popovic Z. B.; Koc O. N.; Penn M. S. SDF-1 Expression by Mesenchymal Stem Cells Results in Trophic Support of Cardiac Myocytes after Myocardial Infarction. FASEB J. 2007, 21, 3197–3207. [DOI] [PubMed] [Google Scholar]
- Phinney D. G.; Prockop D. J. Concise Review: Mesenchymal Stem/Multipotent Stromal Cells: The State of Transdifferentiation and Modes of Tissue Repair—Current Views. Stem Cells 2007, 25, 2896–2902. [DOI] [PubMed] [Google Scholar]
- Xie J.; Wang W.; Si J. W.; Miao X. Y.; Li J. C.; Wang Y. C.; Wang Z. R.; Ma J.; Zhao X. C.; Li Z.; et al. Notch Signaling Regulates CXCR4 Expression and the Migration of Mesenchymal Stem Cells. Cell Immunol. 2013, 281, 68–75. [DOI] [PubMed] [Google Scholar]
- Fleming R. E.; Ponka P. Iron Overload in Human Disease. N. Engl. J. Med. 2012, 366, 348–359. [DOI] [PubMed] [Google Scholar]
- Pantopoulos K.; Porwal S. K.; Tartakoff A.; Devireddy L. Mechanisms of Mammalian Iron Homeostasis. Biochemistry 2012, 51, 5705–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath K. E.; Koniski A. D.; Maltby K. M.; McGann J. K.; Palis J. Embryonic Expression and Function of the Chemokine SDF-1 and Its Receptor, CXCR4. Dev. Biol. 1999, 213, 442–456. [DOI] [PubMed] [Google Scholar]
- Ceradini D. J.; Kulkarni A. R.; Callaghan M. J.; Tepper O. M.; Bastidas N.; Kleinman M. E.; Capla J. M.; Galiano R. D.; Levine J. P.; Gurtner G. C. Progenitor Cell Trafficking Is Regulated by Hypoxic Gradients through HIF-1 Induction of SDF-1. Nat. Med. 2004, 10, 858–864. [DOI] [PubMed] [Google Scholar]
- Staller P.; Sulitkova J.; Lisztwan J.; Moch H.; Oakeley E. J.; Krek W. Chemokine Receptor CXCR4 Downregulated by von Hippel–Lindau Tumour Suppressor Pvhl. Nature 2003, 425, 307–311. [DOI] [PubMed] [Google Scholar]
- Jang J. T.; Nah H.; Lee J. H.; Moon S. H.; Kim M. G.; Cheon J. Critical Enhancements of MRI Contrast and Hyperthermic Effects by Dopant-Controlled Magnetic Nanoparticles. Angew. Chem., Int. Ed. 2009, 48, 1234–1238. [DOI] [PubMed] [Google Scholar]
- Khan W. S.; Hardingham T. E. The Characterization of Mesenchymal Stem Cells: A Stem Cell Is Not a Stem Cell. J. Stem Cells 2012, 7, 87–95. [PubMed] [Google Scholar]
- Matsumoto Y.; Jasanoff A. T2 Relaxation Induced by Clusters of Superparamagnetic Nanoparticles: Monte Carlo Simulations. Magn. Reson. Imaging 2008, 26, 994–998. [DOI] [PubMed] [Google Scholar]
- Wang H.; Cao F.; De A.; Cao Y.; Contag C.; Gambhir S. S.; Wu J. C.; Chen X. Trafficking Mesenchymal Stem Cell Engraftment and Differentiation in Tumor-Bearing Mice by Bioluminescence Imaging. Stem Cells 2009, 27, 1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.; Zhang F.; Wang H.; Niu G.; Choi K. Y.; Swierczewska M.; Zhang G.; Gao H.; Wang Z.; Zhu L.; et al. Mesenchymal Stem Cell-Based Cell Engineering with Multifunctional Mesoporous Silica Nanoparticles for Tumor Delivery. Biomaterials 2013, 34, 1772–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Huang X.; Zhu L.; Guo N.; Niu G.; Swierczewska M.; Lee S.; Xu H.; Wang A. Y.; Mohamedali K. A.; et al. Noninvasive Monitoring of Orthotopic Glioblastoma Therapy Response Using RGD-Conjugated Iron Oxide Nanoparticles. Biomaterials 2012, 33, 5414–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.; Zeng H.; Robinson D. B.; Raoux S.; Rice P. M.; Wang S. X.; Li G. Monodisperse MFe2O4 (M = Fe, Co, Mn) Nanoparticles. J. Am. Chem. Soc. 2004, 126, 273–279. [DOI] [PubMed] [Google Scholar]
- Chen M.; Liu J. P.; Sun S. One-Step Synthesis of FePt Nanoparticles with Tunable Size. J. Am. Chem. Soc. 2004, 126, 8394–8395. [DOI] [PubMed] [Google Scholar]
- Jang J. T.; Nah H.; Lee J. H.; Moon S. H.; Kim M. G.; Cheon J. Critical Enhancements of MRI Contrast and Hyperthermic Effects by Dopant-Controlled Magnetic Nanoparticles. Angew. Chem., Int. Ed. 2009, 48, 1234–1238. [DOI] [PubMed] [Google Scholar]
- Huang X.; Swierczewska M.; Choi K. Y.; Zhu L.; Bhirde A.; Park J.; Kim K.; Xie J.; Niu G.; Lee K. C.; et al. Multiplex Imaging of an Intracellular Proteolytic Cascade by Using a Broad-Spectrum Nanoquencher. Angew. Chem., Int. Ed. 2012, 51, 1625–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.; Zhang F.; Sun X.; Choi K. Y.; Niu G.; Zhang G.; Guo J.; Lee S.; Chen X. The Genotype-Dependent Influence of Functionalized Multiwalled Carbon Nanotubes on Fetal Development. Biomaterials 2014, 35, 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. Y.; Chung H.; Min K. H.; Yoon H. Y.; Kim K.; Park J. H.; Kwon I. C.; Jeong S. Y. Self-Assembled Hyaluronic Acid Nanoparticles for Active Tumor Targeting. Biomaterials 2010, 31, 106–114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.