Abstract
Engineering spatial patterning in mammalian cells, employing entirely genetically encoded components, requires solving several problems. These include how to code secreted activator or inhibitor molecules and how to send concentration-dependent signals to neighboring cells, to control gene expression. The Madin–Darby Canine Kidney (MDCK) cell line is a potential engineering scaffold as it forms hollow spheres (cysts) in 3D culture and tubulates in response to extracellular hepatocyte growth factor (HGF). We first aimed to graft a synthetic patterning system onto single developing MDCK cysts. We therefore developed a new localized transfection method to engineer distinct sender and receiver regions. A stable reporter line enabled reversible EGFP activation by HGF and modulation by a secreted repressor (a truncated HGF variant, NK4). By expanding the scale to wide fields of cysts, we generated morphogen diffusion gradients, controlling reporter gene expression. Together, these components provide a toolkit for engineering cell–cell communication networks in 3D cell culture.
Keywords: morphogen, synthetic patterning, MDCK, HGF, NK4
A main aim of synthetic biology is to design and build biological systems to perform desired functions in a predictable manner.1−3 Within this framework, one area of study is spatial pattern formation: several artificial gene networks have been inspired by biological patterning systems. For example, synthetic stripe patterns have been engineered in transcription-translation reactions in vitro,4 over bacterial cell lawns,5 and over fields of mammalian cells.6
Many of these studies have explored a central paradigm in embryonic developmental patterning: the French Flag model of stripe formation.7 In the model, different genes are expressed in a concentration-dependent manner from a spatial chemical gradient formed by a diffusing signal or morphogen. Our ability to engineer spatial morphogen gradients—and ultimately artificial patterning systems—is only now developing. In this study, we wished to add to the synthetic biology toolbox for building such systems.
Bacteria are the most commonly used chassis for building and studying artificial gene networks because of their robustness8 and general ease-of-use. For example, a synthetic bacterial system was recently developed that employed quorum-sensing machinery to make fluorescent stripe or band patterns, as a function of gradients of diffusible molecules.5 The system used genetically encoded enzymes to produce acyl homoserine lactone molecules (AHLs) that were secreted from sender cells, resulting in concentration-dependent gene expression in nearby receiver cells. Reflecting their importance, several groups have focused on engineering similar cell–cell communication modules5,9−13 and band-forming systems,14 primarily in bacteria but also in yeast.15 Indeed, engineering patterning in 2-dimensional (2D) bacterial systems is developing rapidly; recent examples include highly intricate fractal patterns that emerge from the effects of physical interactions.16,17 Despite these advances, there is a need to develop more patterning and cell–cell communication tools in mammalian systems.
Mammalian models to make patterning systems have been explored far less than their microbial counterparts. Indeed, as a whole, eukaryotic synthetic biology18,19 has fewer developed examples of network engineering. These include the rational design of memory in eukaryotic cells,20 a tunable oscillator,21 a nitric oxide sender–receiver system,22 and a recent mammalian band-forming network.6 The latter study was the first example of a synthetic mammalian morphogen gradient readout system and deployed chemical gradients of tetracycline, which were detected by engineered gene networks in Chinese Hamster Ovary cells, so as to generate different GFP outputs at low, middle, or high tetracycline concentrations. This resulted in single GFP stripes over 2D fields of cells.
In this study, we wished to develop similar mammalian gradient readout systems but using only genetically encoded components that could be delivered entirely as DNA. The main reason was to enable the engineering of stripe-forming networks that require both positive and negative feedback; this cannot be achieved if the diffusing morphogen is chemically synthesized, as with a tetracycline morphogen.6 Certain gradient-readout networks23 (French-flag type) and all reaction-diffusion patterning systems (Turing- or Gierer–Meinhardt-type)24,25 require feedback connections. Our long-term goal is to make stand-alone genetic programs, such as French-flag or Turing systems, that will execute and form patterns when engineered into cells.
When considering this type of engineering problem, we quickly realized that we lack a lot of tools to engineer many types of patterning gene network. For instance, the 3-node activator-inhibitor networks for gradient-detection band-forming systems have been explored exhaustively in a computational atlas,23 but we lacked the tools to make even the simplest activator-inhibitor diffusion systems in mammalian cells. We therefore decided to expand the repertoire of parts for such engineering and, in doing so, we chose a new cell chassis. We selected a cell line, which already had elements of a ’developmental’ program: the Madin–Darby canine kidney (MDCK) cell line spontaneously forms hollow multicellular spheres in 3D collagen culture. We reasoned that we might be able to to graft spatial patterning networks on top of the MDCK cyst scaffold, to generate synthetic morphogen gradients, incorporating diffusing activation and inhibition interactions. Conveniently, MDCK cells are known to activate certain genes in response to an extracellular protein: hepatocyte growth factor (HGF), which could be used as a diffusing extracellular activator. A trunctated form of HGF (NK4) represses HGF-signaling26 and could be used as a diffusing inhibitor in our system.
A short-term aim was simply to explore whether we could make sender–receiver gene expression regions on single cysts. The idea was to show whether one region of a cyst could express and secrete a functional, diffusible activator or inhibitor molecule (HGF or NK4) and thus control gene expression of a fluorescent reporter on a distal region of the same cyst. We envisaged this kind of system to be a prototype for a synthetic developmental pattern on a single cyst, where more complicated patterns might eventually be built-up, layer by layer.
A longer-term aim was to develop tools for spatial patterning network engineering, including the eventual design of artificial French-flag or Turing patterns, as mentioned above. In this study, we therefore developed a toolkit of components and methods to deliver them locally in 3D culture, to make functional sender–receiver interactions, for both activator and inhibitor signaling.
In 2D culture, MDCK cells form polarized epithelial monolayers, with apical, basal, and lateral cell membrane surfaces. By contrast, when single MDCK cells are seeded in a collagen type I matrix, each cell grows to form a polarized spherical monolayer enclosing a fluid-filled lumen.27 After 6 days’ growth, the resulting cysts are hollow spheres of ∼200 cells (∼50 μm diameter)(Figure 1a).
Figure 1.
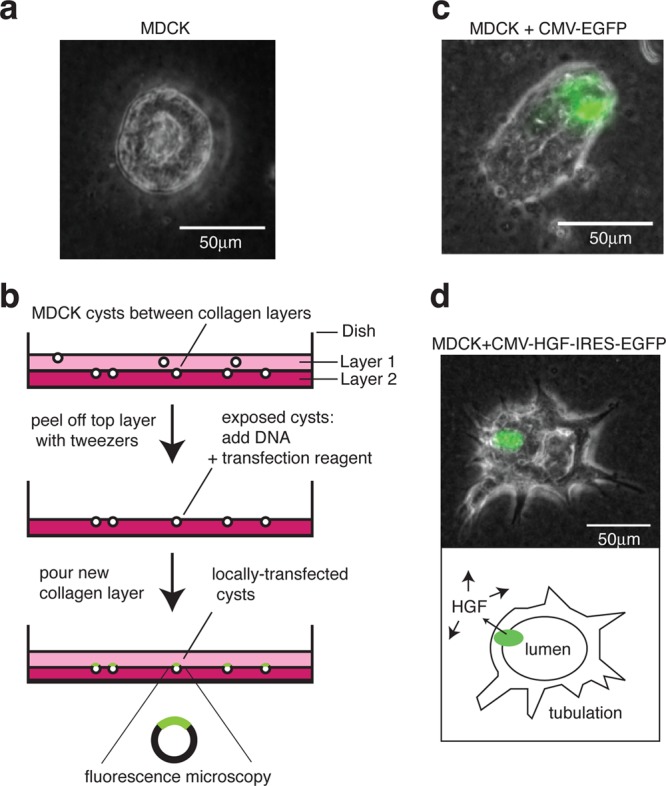
Localized transfection method for 3D MDCK culture. (a) MDCK cells seeded into collagen grow into hollow spherical cysts (∼200 cells) over a period of 6 days. (b) Schematic view showing two collagen layers in a 35 mm diameter dish. A second layer, comprising collagen mixed with the seeding cells, allows cysts to develop at the interface between the layers. The top layer is peeled away to allow transfection of one face of the cysts. A new collagen layer is then poured on top. (c) A typical MDCK cyst, locally transfected with EGFP under a constitutive CMV promoter. (d) Local transfection with HGF under a CMV promoter, followed by an internal ribosome entry site (IRES) and the EGFP gene. A schematic view (below) shows the transfected region (green) secreting functional HGF, which diffuses around the cyst to induce tubulation. Phase contrast light microscopy images are superimposed with the GFP fluorescence channel. Scale bars: 50 μm.
Hepatocyte growth factor (HGF) binds the extracellular c-Met receptor tyrosine kinase on cysts,28 ultimately inducing branching hollow tubules, and differential gene regulation.29−31 This led us to hypothesize that HGF could be used as a synthetic morphogen signaling molecule.
As our aim was to use DNA-encoded components, we first tested HGF gene expression by transient transfection. Whereas we could get ∼60% DNA transfection efficiency with lipofectamine in standard 2D MDCK culture and <1% with PCR-coated beads32 (Supporting Information Figure S1), we were initially unable to see transfection in collagen 3D culture. To overcome this issue, we developed a new localized transfection method for MDCK cysts, by growing the cysts in between two layers of collagen (Figure 1b). The top layer was peeled away with fine tweezers to reveal accessible portions of the cysts for transfection. Consequently, a CMV-EGFP reporter could be delivered to one face of a cyst with high efficiency: 72 ± 7% s.d. of cysts were locally transfected (142 cysts total; 3 independent experiments)(Figure 1c). Within these transfected cysts, as determined by confocal microscopy, the transfection efficiency was estimated to be 22 ± 11% s.d. (73 cysts; 3 independent experiments).
A bicistronic construct expressing both HGF and EGFP could also be locally transfected and resulted in cyst tubulation (Figure 1d). This demonstrated that HGF DNA could be locally delivered, expressed as protein, and secreted in a functional form, at levels sufficient to exert a morphogenic effect on nearby receiver cells.
To build a synthetic gene network where cells would secrete HGF to change gene expression in neighboring cells, it was first necessary to engineer an HGF-dependent reporter. HGF activates c-Met signaling pathways, altering the expression of many genes, including matrix metalloproteinases (MMPs). For example, HGF activates the MMP-1 promoter.33 Based on this, we designed an MMP-1 promoter construct driving a destabilized d2EGFP34 (Supporting Information S1).
We generated stable cell lines with MMP-1-d2EGFP and examined the response of cysts to HGF in 3D cell culture. Qualitatively, the fluorescence increased markedly over 16 h of HGF-exposure (Figure 2a–h; Supporting Information Movie S1). Not all cells in this isogenic cell line turned green in response to uniform HGF, because of biological variability in individual cell responses within cysts. We observed this variability throughout our experiments, and the effect is not to be confused with the localized transfections, where only some cells contain the gene constructs (e.g., Figure 1). Tubulation can also be seen in Figure 2; although this phenotypic output is the natural result of HGF signaling, it was not the focus of our study per se. Rather, we concentrated on fluorescent outputs which are more easily quantified. In fact, to reduce potential interference with downstream engineered networks, it is possible that the tubulation response could be eliminated with shRNAs.35
Figure 2.
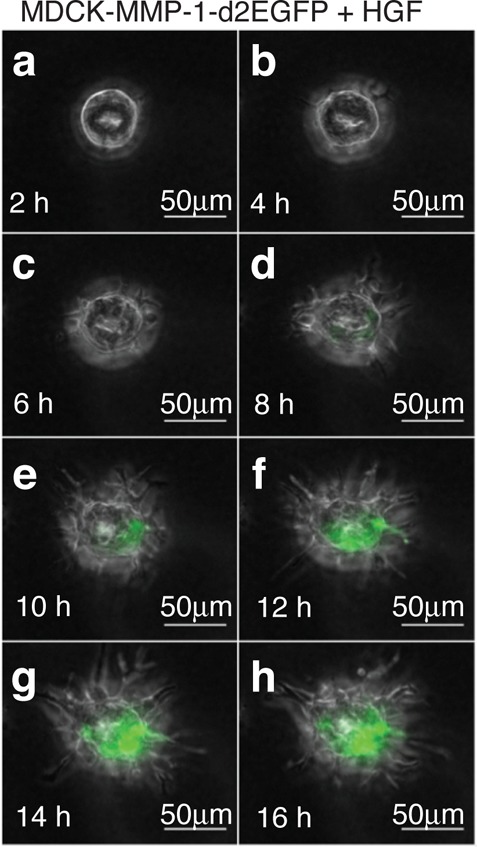
Engineering an HGF-responsive reporter cell line. HGF induces green fluorescence and tubulation in MDCK-MMP-1-d2EGFP cysts. (a–h) A cyst was treated with 10 ng/mL HGF for 16 h. Images were taken every two hours after adding HGF. Note the low basal fluorescence in MDCK cysts in 3D culture (panel a). A typical cyst is shown and can also be seen in timelapse (Supporting Information Movie S1). Scale bars, 50 μm.
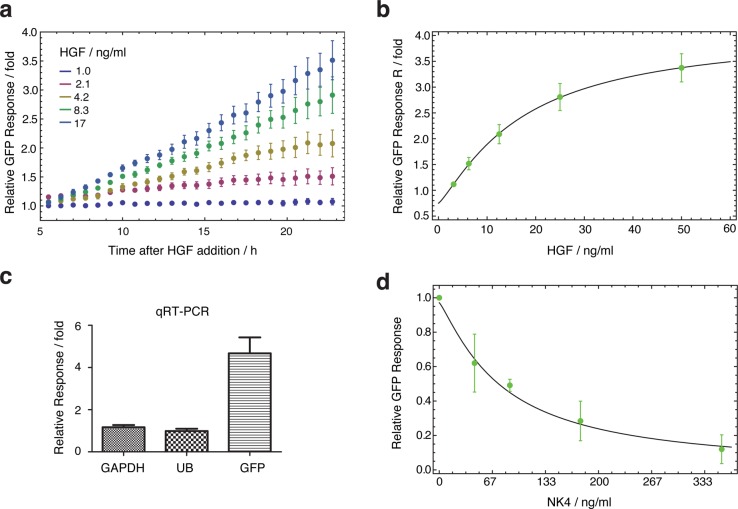
To quantify HGF induction, we analyzed timelapse microscopy images of cysts in collagen. We found that the fold-induction of GFP with different HGF concentrations was highly reproducible (Figure 3a). Thus, we were able to fit an HGF-GFP dose-reponse curve (Figure 3b). HGF had an effective concentration for 50% GFP activation (EC50) of ∼17 ng/mL and a maximum response of ∼4-fold (Supporting Information S1). HGF-GFP induction was also verified at the RNA level with quantitative real time PCR (qRT-PCR): 17 ng/mL HGF (50 ng in a 3 mL well) induced GFP about 4-fold (Figure 3c). Above this HGF concentration, the cysts tend to dissociate (scatter) into single cells, and so, we avoided such conditions in subsequent experiments.
Figure 3.
HGF and NK4 dose–responses in the MDCK:MMP-1-d2EGFP stable cell line. (a) HGF response over 24 h for 5 HGF concentrations. The relative GFP response is calculated by normalizing to the fluorescence from 0 ng/mL HGF samples (all data: mean of 4 experiments). (b) Maximum responses reached in 24 h as function of HGF concentration. These allow an estimation of maximum EGFP response (Rmax = ∼4-fold), the effective concentration for 50% induction (EC50 = 17 ng/mL), and a Hill coefficient for activation (n = 1.2). (c) qRT-PCR of EGFP RNA with 17 ng/mL HGF (mean of 3 experiments; comparison to two housekeeping genes, GAPDH and UB). (d) NK4 repression of 17 ng/mL HGF. Maximal GFP responses (relative to 0 ng/mL HGF samples) over a 24 h period are plotted as a function of NK4 concentration. The NK4 concentration for 50% inhibition of an Rmax GFP signal can also be calculated (IC50 = 38 ng/mL). For all calculations see Supporting Information S1.
It should be noted that we also tested the cell line in 2D culture and found a slightly lower maximum induction up to ∼1.7-fold (Supporting Information Figure S2). Since MDCK cells in 2D culture tend to be highly motile and give lower signal to background, we focused on the 3D system. Overall, having established the conditions for reporter activation, the next step was to see if we could control the system with a repressor.
NK4 is a synthetic HGF fragment, composed of the N-terminal 447 amino acids, including four kringle domains.26 NK4 binds to c-Met but does not induce receptor dimerization and activation. In fact, NK4 inhibits the mitogenic, motogenic, and morphogenic activities of HGF and is the most complete HGF antagonist described.26 Thus, NK4 was the ideal candidate for adding a repressor function to our HGF-induced MMP-1-d2EGFP cyst system.
To quantitate NK4 repression, we used the same microscopy approach as for assaying HGF induction. We tested increasing concentrations of NK4, versus the reporter cell line with 17 ng/mL HGF (Figure 3d). We were thus able to estimate the effective NK4 concentration for 50% inhibition (IC50): 38 ng/mL (Supporting Information S1). Furthermore, 350 ng/mL of NK4 repressed the system fully, so that GFP was undetectable above background (Supporting Information Figure S3). NK4 is therefore a suitable repressor for our synthetic system.
To build dynamic systems that respond to changing conditions, it is necessary that reporter induction be reversible. In principle, system reversibility could be influenced by several processes including the degradation of HGF inducer, the shut-down of signaling to the pMMP-1 promoter, or the degradation of expressed GFP. We therefore designed experiments to test whether the system was reversible at any of these levels.
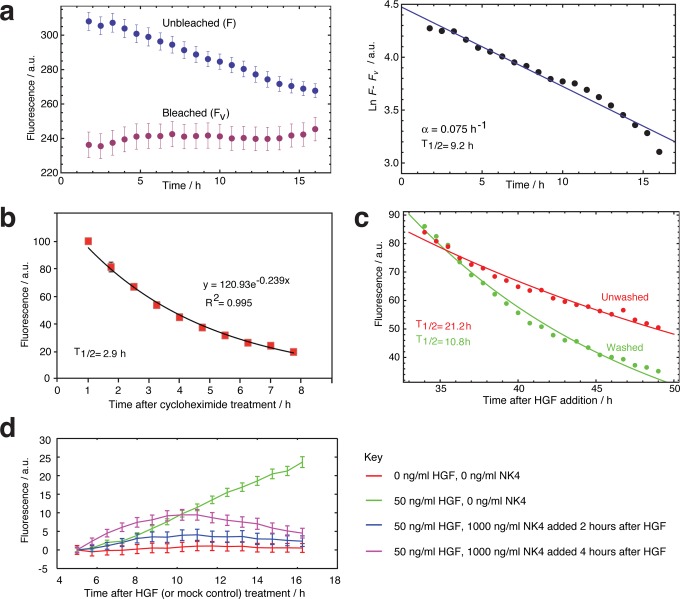
First, we measured the half-life of the destabilized d2EGFP34 in MDCK cysts using a bleach-chase approach36 (Figure 4a). The GFP half-life (T1/2 = 9.2 h) is longer than the two hours reported in other cells but does confirm protein turnover. We compared this result to measuring GFP half-life using a standard cycloheximide treatment.37 We thus obtained T1/2 = 2.9 h (Figure 4b), which is closer to the 2 h half-life reported in the literature.34 Cycloheximide inhibits protein biosynthesis and is toxic to the cell, compared with the less invasive bleach-chase method. The two methods may therefore represent upper and lower bounds for d2EGFP protein degradation under varying conditions of cell metabolism. It may be possible that cycloheximide stimulates protein decay machinery as a response to the shock of ribosome inhibition, decreasing apparent half-lives. Notably, the bleach-chase method is said to compare well to the gold standard of radioactive pulse chase to measure half-lives,37 and both methods are less invasive than cycloheximide treatment. Further studies to elucidate differences between these methods would be desirable in the future.
Figure 4.
Reversibility of HGF induction of MDCK:MMP-1-d2EGFP reporter cysts. (a) Measuring the half-life of d2EGFP in cysts with bleach-chase.36 Fluorescence after bleaching (red) converges to the nonbleached profile (blue) with exponential dynamics, revealing the protein removal rate by calculating the slope of a linear fit between the difference of bleached and unbleached samples, on a semilog plot (below). Data are from 68 cysts (unbleached) and 43 cysts (bleached). α = 0.075 h–1 ± 0.003; R2 = 0.97; T1/2 = 9.24 h. (b) d2EGFP half-life measurement after inducing cysts with 8.3 ng/mL HGF for 15 h and then blocking protein synthesis with 6.6 μg/mL cycloheximide. T1/2 = 2.9 h (mean of 3 experiments). (c) Reversibility of induction after adding 8.3 ng/mL HGF, from T = 0–24 h, and either washing (green) or leaving unwashed (red). Quantification is from microscopy between T = 30–50 h (mean of 3 experiments). (d) NK4 represses HGF-induced fluorescence even when NK4 is added 2 or 4 h after HGF (mean of 3 experiments). All error bars: 1 s.e.m.
We next tested the effect of adding and washing away HGF. Washed samples (Figure 4c) should comprise the decay of cell signaling and d2EGFP expression, after HGF removal (apparent T1/2 = 10.8 h). Since this HGF removal result corresponds closely to the measured upper bound of the half-life of d2EGFP (bleach-chase), we can conclude that signaling switches-off rapidly once HGF is gone and that GFP protein degradation is the main limiting process in system reversibility.
We further tested whether HGF itself degraded in long-term incubation (Figure 4c). The Unwashed sample should comprise the intrinsic decay of HGF, cell signaling and d2EGFP expression (apparent T1/2 = 21.2 h). Since cell signaling and d2EGFP degrade much faster, the majority of this slow decline is likely to be from HGF degradation. Overall, signal switch-off is a combination of HGF half-life, pathway signaling decay, and GFP half-life; these factors will all have to be considered when modeling outputs in downstream network engineering studies.
Finally, we tested whether NK4 could repress activation even after HGF was allowed to activate the system for a defined period, such as 2 or 4 h. The result was that NK4 could indeed block activation even after a sustained exposure to HGF (Figure 4d). In summary, the reporter system is reversible at various levels, which is useful for building dynamic synthetic networks.
Having established that the MDCK:MMP-1-d2EGFP cell line formed cysts that could respond reversibly to recombinant HGF, we next combined this with the localized transfection technique (Figure 1b) to see whether locally delivered DNA (encoding HGF) could send signals to neighboring cells, within the same cyst. We therefore constructed a bicistronic CMV-promoter expression construct, with red fluorescent reporters for marking transfection (mCherry or dsRed, as indicated, depending on the FACS or microscopy applications). The HGF coding sequence was expressed via an internal ribosome entry site (IRES). The pCMV-mCherry-IRES-HGF plasmid could be locally transfected to form red patches on single cysts (Figure 5). These mCherry-expressing cells also secreted functional HGF, as determined by the nearby induction of green fluorescence (via MMP-1-d2EGFP) and tubulation (Figure 5h,j; Supporting Information Movie S2, S3). Secreted HGF could also be detected in the cell culture medium by ELISA, up to ∼0.5 ng/mL (Supporting Information Figure S4).
Figure 5.
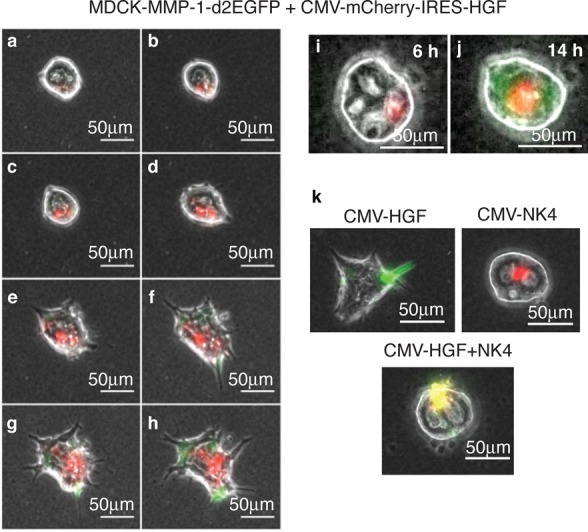
Localized sender–receiver patterns in single MDCK cysts. The MDCK-MMP-1-d2EGFP reporter cyst is locally transfected with pCMV-mCherry-IRES-HGF (red region). The red region secretes HGF which diffuses extracellularly to induce distal GFP expression and tubule formation. (a–h) Images taken from 6 h after transfection, at 2 h intervals. A typical cyst is shown and can also be seen in timelapse (Supporting Information Movie S2). (i, j) An independent example of the same setup at 6 h (i) and 14 h (j); this cyst goes on to tubulate after 14 h (Supporting Information Movie S3). (k) Locally transfecting MDCK wt cysts with CMV-EGFP-IRES-HGF activator (green) or mCherry-IRES-NK4 (red), or both together (yellow). Images taken after 24 h. Cotransfecting NK4 blocks HGF-induced tubulation. Scale bars, 50 μm.
The induction of green cells around the red source region was a first step toward our aim of a synthetic ‘developmental’ program. The next step was to see if we could control this process with a repressor. We therefore transiently transfected HGF and NK4 constructs together and achieved a repression of HGF signaling (Figure 5k). Repression was stable over 24 h, although we did not perform any observations after this time point. Overall, this shows that secreted NK4 can be produced at sufficient concentrations to block secreted HGF.
Having explored HGF and NK4 activities on single cysts, we moved to larger fields containing hundreds of cysts, on 35 mm culture plates. Our motivation was to set up a system employing larger spatial scales for diffusion-gradient based patterning. The fields were made with the same 2-layer method as before (Figure 1b), but 3 mm diameter glass beads or 2 mm diameter microcapillaries were used to mold or gouge out ‘wells’ in the collagen. The aim was to visualize the diffusion of activator, in the presence or absence of inhibitor, by adding sender cells to the wells.
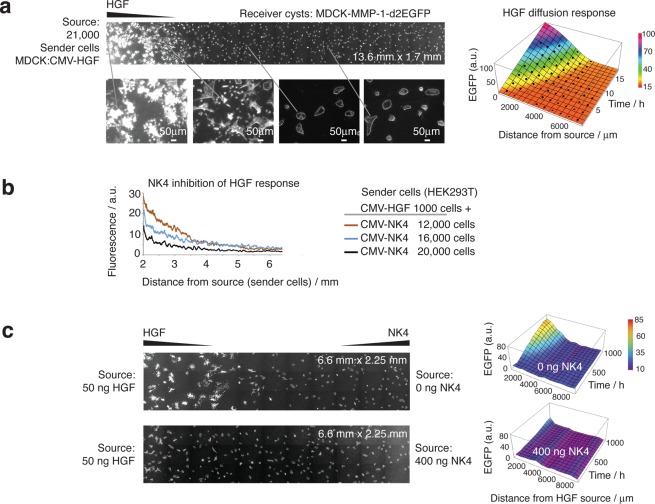
To visualize cell fields, we used an automated wide-field inverted fluorescence microscopy system. Using low magnification settings, we took multiple overlapping regions of the collagen gel culture (e.g., 750 × 600 μm), using the custom software and scripting tools of the microscope. Phase contrast microscopy allowed us to focus automatically on the cyst plane. The images were then aligned and stitched together to make a large image (e.g., 13.6 mm ×1.7 mm; Figure 6a).
Figure 6.
Sender cell regions produce gradients of HGF-induced and NK4-repressed fluorescence in collagen matrices containing MDCK-MMP-1-d2EGFP receiver cysts. (a) A widefield view of the receiver region next to a well (not shown) containing 21 000 MDCK sender cells (transfected with CMV-HGF-IRES-dsRED and sorted by FACS for dsRED). The composite image is stitched from multiple 752 × 599 mm images and shows the fluorescence and tubulation of the cysts at time point 16.5 h, with close-up views in the insets below. Timelapse data were thus collected and used to obtain the diffusion gradient response profile over time (right). Average intensity values at particular spatial points (black dots) were used to fit the diffusion model shown by the color axis surface profile (see Supporting Information for diffusion constant calculations). (b) Quantification of NK4 repression of HGF-induced GFP gradients from mixtures of transfected HEK293T sender cells added to single sender wells (HEK293T were used because of high transfection efficiency). Sender cells expressed CMV-HGF-IRES-DsRed plus CMV-NK4-CBD-IRES-DsRed. Fluorescence images were quantified with ImageJ (Radial profile plugin). (c) Interaction of an HGF gradient with an NK4 gradient. Sender wells were created approximately 9 mm apart and the indicated amounts of purified HGF or NK4 were added at opposite ends, allowing diffusion over a field of reporter cysts, while taking timelapse GFP fluorescent images (images shown: 18.5 h). The images were quantitated (right; see Supporting Information) to obtain response profiles over time, showing that NK4 restricts the HGF-induced fluorescent gradient.
Using a region of HGF-secreting MDCK sender cells resulted in gradients of GFP expression over a field of receiver cysts (Figure 6a). Tubulation could be seen to occur along the diffusion gradient (Figure 6a; insets). We quantified and visualized the gradients of reporter gene output using timelapse series of data (Figure 6a; right; Supporting Information S1). Thus, we calculated an effective diffusion constant for HGF by fitting a diffusion model onto the observed gradients (Supporting Information S1; Figure S5). The value we obtained of 3.1 × 10–3 mm2/min is similar to another published estimate of HGF diffusion in collagen.38
Next, we looked at the effects of NK4 repression on the HGF signaling gradient, by producing different amounts of HGF and NK4 from the same sender well (Figure 6b). HEK293T sender cells were transfected with plasmids expressing CMV-HGF-IRES-DsRed plus CMV-NK4-CBD-IRES-DsRed (HEK293T were used here rather than MDCK because of their high transfection efficiency). We found that NK4 repression modulated the observed gradients, according to the dose of repressor cells (Figure 6b).
Finally, we explored whether a gradient of HGF-induced GFP expression in receiver cysts could be modulated by a gradient of NK4 repressor. For this, we set up a collagen matrix with two sender wells, approximately 9 mm apart. Purified HGF or NK4 were added at opposite ends, allowing diffusion over a field of reporter cysts, while taking timelapse GFP fluorescent images (Figure 6c). By quantifying the time series, we observed the evolution of the response (Figure 6c; right). We found that NK4 can both repress and restrict the spread of HGF-induced gradient.
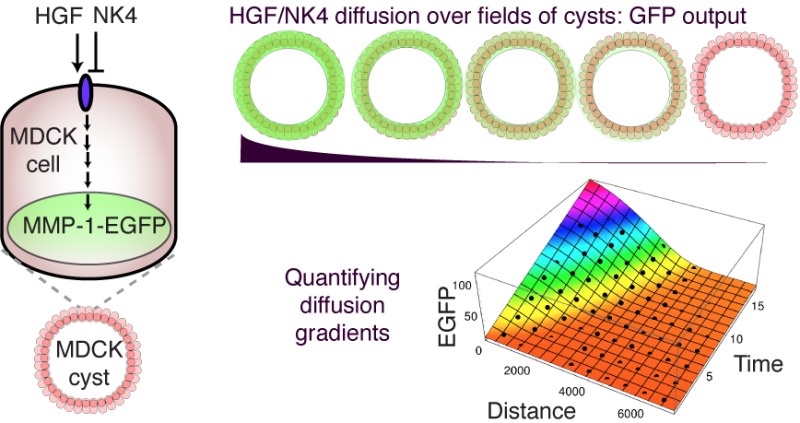
Overall, the system we have developed is reproducibly and reversibly controlled by secreted HGF activator and NK4 repressor, both on the scale of single cysts and in widefield views, suggesting that these components are promising candidates for engineering synthetic patterns.
In this study, we built a set of tools for linking gene expression and secreted extracellular activator or inhibitor molecules, in the MDCK mammalian cell line, to allow the engineering of synthetic patterns.
The MDCK cell line was chosen because it has been reported to respond with changes on gene expression to the diffusing extracellular activator HGF33 and to a related inhibitor, NK4.26 Thus, the cells contained potential elements that could be developed toward engineering spatial patterning networks, such as French-flag-type stripes,5,23 or reaction-diffusion patterns.24,25 In this pilot study, we aimed to test whether sender cells could secrete functional activators, or inhibitors, and thus change reporter gene expression in distal receiver cells.
MDCK cells can be cultured in both 2D and 3D and we found that that the 3D MDCK cyst system was preferable for our purposes: (i) the cells in 3D were not motile as in 2D; (ii) reporter background was lower in 3D (resulting in a greater signal/background ratio); (iii) the collagen culture matrix provided a suitable environment for controlled diffusion of HGF and NK4.
Furthermore, the 3D system allowed us to develop a localized transfection method, employing a dual-layer collagen dish, where cysts growing at the layer interface could be locally transfected in a bull’s eye pattern. This technique enabled us to make a simple patterning program: a circular red mCherry-expressing region on a cyst secreted HGF and, over time, neighboring colorless regions began to turn EGFP green and to tubulate. The motivation was thus to work toward building an artificial developmental program step-by-step. (see Supporting Information Movies S1–S3).
It should be noted that throughout the present study the cysts tubulated in response to HGF. Because there are no known feedbacks between tubulation and HGF signaling, this did not impair our downstream efforts to engineer cell signaling and communication; in fact, the tubulation acted as a convenient additional phenotypic output to indicate the presence of active HGF. However, for some network engineering applications, it could be advantageous to block tubulation, to program cells without any change in physiology. For this purpose, Lipschutz and colleagues have reported that an shRNA targeted to MMP-13 can inhibit tubulogenesis.35 In the future, it would be interesting to see whether an HGF-GFP reporter cell line, stably expressing this shRNA, could function without tubulation.
Moving beyond patterning at the scale of single MDCK cysts, a larger scale spatial configuration was chosen because this was more appropriate for studying morphogen systems, where gradients and stripes are engineered on the scale of fields of cells.5,6 Using the larger-scale setups, we showed that gradients of activation could be modulated by the dosage of activator and inhibitor.
Although it is outside the scope of the current work, dowstream experiments will test the possibility of using these components to build simple wide-field cell–cell communication networks, involving positive and negative feedback, such as the French-flag or Turing systems mentioned above. To enable such dynamic network engineering, we established here that the MMP-1-HGF fluorescence induction process was reversible and could be repressed by the NK4 repressor. Moreover, the components we have developed are modular and work together consistently, in dose-dependent manners. Using these criteria, the toolkit could be further expanded component-by-component in the future, to make more sophisticated gene network programs based on secretion, diffusion, activation and repression. Thus, we hope to contribute to the field of patterning network engineering within mammalian synthetic biology.
Mammalian systems provide a diverse range of cellular machinery for synthetic biology. First, the mammalian single plasma membrane lends itself well to synthetic cell–cell communication applications because it can be easily traversed by fusions with secretory protein signal peptides. Second, a wide range of receptors and secreted signaling factors are available as candidates for re-engineering. Third, eukaryotes often contain many splice variants within the same gene, with different agonist or antagonist function. The NK4 used in this study is a good example of a truncated protein that retains only part of its original function: receptor binding without any activation. Although bacterial protein domains are often modular, the ease with which eukaryotic activators can be converted into inhibitors via differing exon usage, raises interesting questions about their potential to evolve antagonistic or self-repressive networks.39
Although we are far away from building structures or organs from the bottom-up, the stepwise approach reported here reveals that modular network components can indeed be isolated and grafted onto existing systems. This results in reproducible systems behavior, using both intra- and extracellular components. In mammalian synthetic biology, we are still at the beginning of finding out what is possible and what is not. Ultimately, our drive is to understand systems by building them.
Methods
Cell Culture
MDCK cells were cultured in MEM with 10% FBS, 100 IU/ml penicillin, 100 mg/mL streptomycin, and 2 mM l-glutamine at 37 °C, 5% CO2.
For 3D MDCK culture,40 cells were trypsinized and resuspended to 3 × 104 cells/ml in a type I collagen solution containing PureCol (3 mg/mL), Advanced BioMatrix 5005-B, 10× MEM, NaHCO3 (2.35 mg/mL), l-glutamine (29.2 mg/mL), and 1 M HEPES (pH 7.6). Cells in suspension were added to wells containing a cell-free collagen mix (polymerized previously at 37 °C) and were incubated at 37 °C (without CO2) for 20–30 min. Two milliliters of liquid media was added above the collagen layers and replaced every 2–3 days. For the analysis of gene expression diffusion patterns, cysts were seeded as described above, in a 35 mm plate containing a 2 mm diameter glass capillary, which was then removed to make a ‘well’ for seeding sender cells.
Localized Cyst Transfection in Three-Dimensional Collagen Gel Culture
MDCK cysts were grown (as above) in 35 mm plates. After 8 days, the top collagen layer was carefully peeled off with the help of fine tweezers.27 Cysts were transfected with 4.5 μg plasmid DNA and 20 μL Lipofectamine per plate, with incubation at 37 °C for 5 h. The medium was removed and a new collagen layer added. The plate was then incubated in a 37 °C oven (in the absence of CO2) for 20–30 min. Two milliliters of fresh liquid medium was then added for 24 h further culture.
Stable Cell Lines
MDCK cells were transfected with Lipofectamine according to the manufacturer’s instructions. Twenty-four hours post transfection, green FITC+ cells were enriched with a FACSaria cell sorter, and were grown for 1 week in MEM with a supplement of G418 (500 μg/mL; PAA Laboratories P11–012)). The cells were again sorted by FITC signal, into individual wells of a 96-well plate, and were grown to confluency. Cells were treated with HGF (50 ng/mL, 24 h), and candidates with increased fluorescence were further tested for their response to HGF, in 3D culture. The cell line V384A2 had the strongest induction and was used in all subsequent experiments.
Production and Quantification of NK4
HEK 293T cells were transfected with CMV-NK4 using Effectene (Qiagen) according to the manufacturer’s instructions. Twenty-four hours post transfection, the MEM medium was replaced with fresh medium.
After a further 24 h, the supernatant was collected and concentrated using Amicon Ultra Centrifugal Filters (30K molecular weight cut off). NK4 in the concentrated supernatant was quantified with a Quantikine ELISA kit for human HGF (R&D Systems), according to the manufacturer’s instructions.
Microscopy Quantification
All images shown were taken with a Zeiss Live observer microscope system, at 10× (0.3 NA) or 20× (0.5 NA) magnification (indicated), 37 °C and 5% CO2. A 300 W Xenon arc bulb was used for illumination and a 38HE filter set for acquisition. Cysts were imaged every 45 min with an exposure of 5s. The cysts were grown as described above on MatTek 6-well glass bottom dishes (thickness no. 1.0; glass diameter 20 mm). For the differential response experiments, at T = 0 h, medium was replaced with 1 mL of MEM, with the appropriate HGF concentrations, to a total collagen-plus-MEM volume of 3 mL. For the reversibility experiments, 25 ng/mL HGF was added at T = 0 h and washed 2 × 1 h with MEM and 1 × 1 h with PBS at T = 24 h. GFP signals were collected with a 5 s exposure time and 15 ms in the phase contrast channel. Twenty images were taken for each concentration. Images were analyzed with a custom MATLAB script. Regulatory functions were fitted to estimate the 50% effective or inhibitory concentrations of HGF and NK4 (EC50, IC50), maximum fold-responses, and the Hill coefficient (n) (Supporting Information).
qRT-PCR
RNA was isolated with the Qiagen RNA mini Kit. After 20 h of HGF induction, the top collagen layer was removed. RLT buffer was added directly to the lower layer and immediately transferred to a 1.5 mL polyethylene tube and processed according to the manufacturer’s protocol. RNA was reverse transcribed with the SuperScript III first-strand synthesis mix (Invitrogen). Primers for GFP sequence and controls are as follows: GFP Fwd, CCTGAAGTTCATCTGCACCA; Rev, AAGTCGTGCTGCTTCATGTG; canine Glyceraldehyde 3-phosphate dehydrogenase, GAPDH Fwd, AACATCATCCCTGCTTCCAC; Rev, GACCACCTGGTCCTCAGTGT; Ubiquitin-specific Peptidase UB Fwd, CAGCTAGAAGATGGCCGAAC; Rev, ACTTCTTCTTGCGGCAGTTG. The fold change was calculated using the Pfaffl method.41
Wide-Field Fluorescence Microscopy
MDCK cysts were grown as above in 35 mm plates. A 2 mm diameter glass microcapillary was fixed vertically in the 2-layer collagen culture to make a well. After 8 days, the capillary was removed and transiently transfected HEK293 sender cells were injected into the well. After 20 min settling time, the top collagen layer was carefully peeled off with the help of fine tweezers,27 and a new collagen layer added, as above.
Automatic mosaic imaging of large areas (Zeiss Cell Observer HS system: AxioObserver Z1 microscope; AxioCam cooled CCD camera; 10×, 0.3 NA objective): overlapping fields were imaged in fluorescence and phase contrast. The mosaic pattern was generated and acquired using autofocusing of the transmission channel, with custom Zeiss Visual Basic for Applications (VBA) and Commander Module routines for the pattern generation. Large field images were then aligned and stitched using ImageJ functions.
Acknowledgments
We thank James Sharpe, Ben Lehner, and Phil Sanders for critical reading. A.C. was funded by GABBA and the Portuguese Fundação para a Ciência e Tecnologia (FCT), Studentship BD/15897/2005. D.B. and V.R.S. are both funded by La Caixa PhD Fellowships. L.D. is funded by CONICET (Argentina). M.I. is funded by FP7 ERC 201249 ZINC-HUBS, Ministerio de Ciencia e Innovación Grant BFU2010-17953 and the MEC-EMBL agreement.
Supporting Information Available
Figures S1–S5, Movies S1–S3, annotated DNA sequences of the final constructs used in this study and the computational model of diffusion and repression. This information is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Author Contributions
∇ A.C. and D.B.M. contributed equally. A.C., D.B.M. and M.I. designed the experiments. A.C., D.B.M. and V.R.S. performed the experiments. T.Z. supervised microscopy and developed scripts. L.D. performed data analysis and modeling.
Supplementary Material
References
- Andrianantoandro E.; Basu S.; Karig D. K.; Weiss R. (2006) Synthetic biology: New engineering rules for an emerging discipline. Mol. Syst. Biol. 2, 2006–0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endy D. (2005) Foundations for engineering biology. Nature 438, 449–453. [DOI] [PubMed] [Google Scholar]
- Drubin D. A.; Way J. C.; Silver P. A. (2007) Designing biological systems. Genes Dev. 21, 242–254. [DOI] [PubMed] [Google Scholar]
- Isalan M.; Lemerle C.; Serrano L. (2005) Engineering gene networks to emulate Drosophila embryonic pattern formation. PLoS Biol. 3, e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S.; Gerchman Y.; Collins C. H.; Arnold F. H.; Weiss R. (2005) A synthetic multicellular system for programmed pattern formation. Nature 434, 1130–1134. [DOI] [PubMed] [Google Scholar]
- Greber D.; Fussenegger M. (2010) An engineered mammalian band-pass network. Nucleic Acids Res. 38, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L. (1968) The French Flag problem: A contribution to the discussion on pattern development and regulation. Towards a Theoretical Biology, Vol. 1, pp 125–133, Edinburgh University Press, Edinburgh. [Google Scholar]
- Isalan M.; Lemerle C.; Michalodimitrakis K.; Horn C.; Beltrao P.; Raineri E.; Garriga-Canut M.; Serrano L. (2008) Evolvability and hierarchy in rewired bacterial gene networks. Nature 452, 840–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino T.; Mondragon-Palomino O.; Tsimring L.; Hasty J. (2010) A synchronized quorum of genetic clocks. Nature 463, 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H.; Kaern M.; Araki M.; Chung K.; Gardner T. S.; Cantor C. R.; Collins J. J. (2004) Programmable cells: Interfacing natural and engineered gene networks. Proc. Natl. Acad. Sci. U.S.A. 101, 8414–8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L.; Cox R. S. 3rd; Weiss R.; Arnold F. H. (2004) Programmed population control by cell–cell communication and regulated killing. Nature 428, 868–871. [DOI] [PubMed] [Google Scholar]
- Bulter T.; Lee S. G.; Wong W. W.; Fung E.; Connor M. R.; Liao J. C. (2004) Design of artificial cell–cell communication using gene and metabolic networks. Proc. Natl. Acad. Sci. U.S.A. 101, 2299–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A. S.; Collins J. J. (2010) Synthetic biology: Applications come of age. Nat. Rev. Genet. 11, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohka T.; Heins R. A.; Phelan R. M.; Greisler J. M.; Townsend C. A.; Ostermeier M. (2009) An externally tunable bacterial band-pass filter. Proc. Natl. Acad. Sci. U.S.A 106, 10135–10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. T.; Weiss R. (2005) Artificial cell–cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat. Biotechnol. 23, 1551–1555. [DOI] [PubMed] [Google Scholar]
- Rudge T. J.; Steiner P. J.; Phillips A.; Haseloff J. (2012) Computational modeling of synthetic microbial biofilms. ACS Synth. Biol. 1, 345–352. [DOI] [PubMed] [Google Scholar]
- Rudge T. J.; Federici F.; Steiner P. J.; Kan A.; Haseloff J. (2013) Cell polarity-driven instability generates self-organized, fractal patterning of cell layers. ACS Synth. Biol. 10.1021/sb400030p. [DOI] [PubMed] [Google Scholar]
- Haynes K. A.; Silver P. A. (2009) Eukaryotic systems broaden the scope of synthetic biology. J. Cell Biol. 187, 589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber D.; Fussenegger M. (2007) Mammalian synthetic biology: Engineering of sophisticated gene networks. J. Biotechnol. 130, 329–345. [DOI] [PubMed] [Google Scholar]
- Ajo-Franklin C. M.; Drubin D. A.; Eskin J. A.; Gee E. P.; Landgraf D.; Phillips I.; Silver P. A. (2007) Rational design of memory in eukaryotic cells. Genes Dev. 21, 2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigges M.; Marquez-Lago T. T.; Stelling J.; Fussenegger M. (2009) A tunable synthetic mammalian oscillator. Nature 457, 309–312. [DOI] [PubMed] [Google Scholar]
- Wang W. D.; Chen Z. T.; Kang B. G.; Li R. (2008) Construction of an artificial intercellular communication network using the nitric oxide signaling elements in mammalian cells. Exp. Cell Res. 314, 699–706. [DOI] [PubMed] [Google Scholar]
- Cotterell J.; Sharpe J. (2010) An atlas of gene regulatory networks reveals multiple three-gene mechanisms for interpreting morphogen gradients. Mol. Syst. Biol. 6, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turing A. M. (1952) The chemical basis of morphogenesis. Philos. Trans. R. Soc., B 237, 37–72. [Google Scholar]
- Gierer A.; Meinhardt H. (1972) A theory of biological pattern formation. Kybernetik 12, 30–39. [DOI] [PubMed] [Google Scholar]
- Date K.; Matsumoto K.; Shimura H.; Tanaka M.; Nakamura T. (1997) HGF/NK4 is a specific antagonist for pleiotrophic actions of hepatocyte growth factor. FEBS Lett. 420, 1–6. [DOI] [PubMed] [Google Scholar]
- McAteer J. A.; Evan A. P.; Gardner K. D. (1987) Morphogenetic clonal growth of kidney epithelial cell line MDCK. Anat. Rec. 217, 229–239. [DOI] [PubMed] [Google Scholar]
- Park M.; Dean M.; Cooper C. S.; Schmidt M.; O’Brien S. J.; Blair D. G.; Vande Woude G. F. (1986) Mechanism of met oncogene activation. Cell 45, 895–904. [DOI] [PubMed] [Google Scholar]
- Montesano R.; Matsumoto K.; Nakamura T.; Orci L. (1991) Identification of a fibroblast-derived epithelial morphogen as hepatocyte growth factor. Cell 67, 901–908. [DOI] [PubMed] [Google Scholar]
- Pollack A. L.; Runyan R. B.; Mostov K. E. (1998) Morphogenetic mechanisms of epithelial tubulogenesis: MDCK cell polarity is transiently rearranged without loss of cell–cell contact during scatter factor/hepatocyte growth factor-induced tubulogenesis. Dev. Biol. 204, 64–79. [DOI] [PubMed] [Google Scholar]
- Balkovetz D. F.; Gerrard E. R. Jr.; Li S.; Johnson D.; Lee J.; Tobias J. W.; Rogers K. K.; Snyder R. W.; Lipschutz J. H. (2004) Gene expression alterations during HGF-induced dedifferentiation of a renal tubular epithelial cell line (MDCK) using a novel canine DNA microarray. Am. J. Physiol. Renal Physiol. 286, F702–710. [DOI] [PubMed] [Google Scholar]
- Isalan M.; Santori M. I.; Gonzalez C.; Serrano L. (2005) Localized transfection on arrays of magnetic beads coated with PCR products. Nat. Methods 2, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinnin M.; Ihn H.; Mimura Y.; Asano Y.; Yamane K.; Tamaki K. (2005) Matrix metalloproteinase-1 up-regulation by hepatocyte growth factor in human dermal fibroblasts via ERK signaling pathway involves Ets1 and Fli1. Nucleic Acids Res. 33, 3540–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Zhao X.; Fang Y.; Jiang X.; Duong T.; Fan C.; Huang C. C.; Kain S. R. (1998) Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 273, 34970–34975. [DOI] [PubMed] [Google Scholar]
- Hellman N. E.; Spector J.; Robinson J.; Zuo X.; Saunier S.; Antignac C.; Tobias J. W.; Lipschutz J. H. (2008) Matrix metalloproteinase 13 (MMP13) and tissue inhibitor of matrix metalloproteinase 1 (TIMP1), regulated by the MAPK pathway, are both necessary for Madin–Darby canine kidney tubulogenesis. J. Biol. Chem. 283, 4272–4282. [DOI] [PubMed] [Google Scholar]
- Eden E.; Geva-Zatorsky N.; Issaeva I.; Cohen A.; Dekel E.; Danon T.; Cohen L.; Mayo A.; Alon U. (2011) Proteome half-life dynamics in living human cells. Science 331, 764–768. [DOI] [PubMed] [Google Scholar]
- Belle A.; Tanay A.; Bitincka L.; Shamir R.; O’Shea E. K. (2006) Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. U.S.A. 103, 13004–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan S.; Shen C. J.; Desai R. A.; Sniadecki N. J.; Nelson C. M.; Chen C. S. (2010) Decoupling diffusional from dimensional control of signaling in 3D culture reveals a role for myosin in tubulogenesis. J. Cell Sci. 123, 2877–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isalan M. (2009) Gene networks and liar paradoxes. Bioessays 31, 1110–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAteer J. A.; E. A.; Vance E. E.; Gardner K. D. Jr. (1986) MDCK cysts: An in vitro model of epithelial cyst formation and growth. J Tiss Cult Meth 10, 245–248. [Google Scholar]
- Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.