Abstract
Strain typing using variable-number tandem repeats of mycobacterial interspersed repetitive units (MIRU-VNTR) is a powerful tool for studying the epidemiology and genetic relationships of Mycobacterium tuberculosis isolates. For this study, isolates from 291 patients in Singapore were genotyped by this method. One hundred sixty-six distinct MIRU-VNTR patterns were detected. One hundred sixty-two strains were grouped into 1 of 35 different MIRU-VNTR clusters and 131 isolates were unique. In this sample collection, 9 of the 12 MIRU-VNTR loci were moderately or highly discriminative according to their allelic diversities. The Hunter-Gaston discriminatory index was 0.975, indicating the high power of discrimination of MIRU-VNTR typing. By direct comparisons with previously typed MIRU-VNTR patterns and by genetic relationship analyses, we could identify and clearly define four epidemic groups of M. tuberculosis in our sample, corresponding to the W/Beijing, East-Africa-Indian, Haarlem, and Delhi genotype families. Furthermore, MIRU-VNTR typing was able to clearly distinguish ancestral and modern M. tuberculosis strains as defined by TbD1 genomic deletion analysis. These results indicate that MIRU-VNTR typing can be a useful first-line tool for studying the genetic diversity of M. tuberculosis isolates in a large urban setting such as Singapore.
Tuberculosis remains a major infectious disease and causes high morbidity and mortality worldwide. Mycobacterium tuberculosis genotyping is a useful tool for the epidemiological surveillance and control of disease transmission. Although a large number of DNA-fingerprinting methods for typing M. tuberculosis isolates have been developed (15), only IS6110 restriction fragment length polymorphism (RFLP) typing (5) and spacer oligonucleotide typing (spoligotyping) (14) have gained wide acceptance.
IS6110 typing is the most commonly used strain-typing method for M. tuberculosis. This method is based on variations among IS6110 insertion elements between strains. IS6110 typing has been shown to be reproducible and highly discriminatory (15). However, there are several drawbacks of IS6110 typing. Firstly, it has poor discriminatory power for M. tuberculosis strains with low copy numbers of IS6110 (7, 15, 17). Secondly, it requires relatively large quantities of purified DNA. Thirdly, it is time-consuming to perform and analyze. Finally, although a standardized methodology has been recommended (29), it is difficult to compare IS6110 RFLP results between laboratories. This hinders global studies of M. tuberculosis epidemiology and transmission. Spoligotyping is a PCR-based method which detects the presence or absence of 43 spacers in the direct repeat locus of the M. tuberculosis genome. This has advantages over IS6110 typing in that it is faster and easier to perform, it is reproducible, and it requires only small quantities of DNA. However, the major drawback of spoligotyping is that it has a lower discriminatory power than IS6110 typing. In particular, this method is not informative in areas where the W/Beijing genotype family is prevalent, as most strains in this family share the same typical spoligotypes (3, 31). Therefore, this method is of limited use as a first-line typing method (7, 15).
A technique for strain typing of M. tuberculosis based on variable-number tandem repeats (VNTRs) of mycobacterial interspersed repetitive units (MIRU) has recently been introduced (18, 25). This method makes use of the length variation at 12 independent minisatellite-like loci scattered throughout the M. tuberculosis genome. A number of studies have proven that MIRU-VNTR typing is a reliable and reproducible typing method that enables a level of discrimination between strains that is comparable to that of IS6110 typing (7, 18, 24, 26). The ease with which this PCR-based method can be performed, its adaptability to high-throughput automation (26), and the digital format of MIRU-VNTR patterns make this method suitable for the global study of the molecular epidemiology of M. tuberculosis. MIRU-VNTR typing also has a high discriminatory power for M. tuberculosis strains with a low copy number of IS6110 or that are devoid of IS6110 (7, 17, 24, 26). Moreover, preliminary studies indicated that this method could also efficiently discriminate W/Beijing strains sharing the same spoligotypes (26; also unpublished data). Therefore, MIRU-VNTR typing may be particularly useful in geographical regions such as East and Southeast Asia where these strains are prevalent (1, 6, 8, 19-22, 28, 31, 32).
An analysis of different groups of epidemiologically linked M. tuberculosis isolates has indicated that the stability of the MIRU-VNTR loci is adequate for the tracking of outbreak episodes, identification of laboratory cross-contamination, and discrimination between relapses and reinfections (12, 18, 23). One comparison study has suggested that MIRU-VNTR typing may be more suitable for defining epidemiologically related isolates than are IS6110 typing and spoligotyping (16).
Singapore is a Southeast Asian country with a heterogeneous population in which the majority is of Chinese origin and a substantial minority is of Indian origin. In order to evaluate the usefulness of MIRU-VNTR typing in Singapore, we genotyped M. tuberculosis isolates from 291 patients with tuberculosis in Singapore. Our results clearly demonstrate that MIRU-VNTR typing is a suitable approach for studies of the genetic diversity of M. tuberculosis in this region of the world.
MATERIALS AND METHODS
Clinical isolates and DNA samples.
M. tuberculosis clinical isolates were collected from the Central Tuberculosis Laboratory in Singapore between August 1994 and June 1995. The incidences of tuberculosis in 1994 and 1995 were, respectively, 49 and 52 per 100,000 people for a population of approximately 3,000,000 people (Communicable Disease Surveillance Report 1997, the Department of Clinical Epidemiology, Tan Tock Seng Hospital). M. tuberculosis DNAs were extracted from cultures according to the protocol of van Soolingen et al. (30). Three hundred three M. tuberculosis DNA samples from 291 patients were typed by investigators who did not know the clinical origins of the samples. Mycobacterium bovis BCG Pasteur and M. tuberculosis H37Rv were used as controls. To test the reproducibility of MIRU-VNTR typing, we blindly typed 24 isolates in two independent experiments.
MIRU-VNTR PCR.
We used fluorescently labeled PCR primers flanking each polymorphic MIRU-VNTR locus that were described by Supply et al. (26), except that the MIRU 4 reverse primer was changed to 5′-GCG CAG CAG AAA CGC CAG C-3′ (17). Each MIRU-VNTR locus was individually amplified in a 50-μl reaction volume in a 96-well PCR plate. The reaction mixture contained a 0.4 μM concentration of each primer, a 0.2 mM concentration (each) of dATP, dCTP, dGTP, and dTTP, 1.5 mM MgCl2 , 1× Q solution, 1× PCR buffer, 1 U of HotStartTaq DNA polymerase (Qiagen, Hilden, Germany), and approximately 5 to 50 ng of DNA. Thermocycling parameters were as described by Supply et al. (26). PCR products were diluted and pooled together according to the sets described by Supply et al. (26). However, those samples that were not properly analyzed in the pooled sets were then diluted for individual capillary electrophoresis.
MIRU-VNTR analysis.
One microliter of diluted PCR products was mixed with 10 μl of deionized formamide and 0.5 μl of a Genescan ROX-2500 DNA size standard. Denatured samples were subjected to electrophoresis through a 36-cm-long POP-4 polymer capillary array with an ABI PRISM 3100-Avant genetic analyzer. The key parameters for capillary electrophoresis were as follows: run temperature, 60°C; prerun voltage, 15 kV; prerun time, 180 s; sample injection voltage, 1 kV; sample injection time, 15 s; run voltage, 13 kV; and run time, 2,600 s. The PCR products were sized by GeneScan and Genotyper software (Applied Biosystems) using the custom macro program of Supply et al. (26). The tables used for MIRU-VNTR allele scoring are available at http://www.ibl.fr/mirus/mirus.html.
Phylogenetic analysis.
PAUP (phylogenetic analysis using parsimony and other methods) software, beta version 4.0 (Sinauer Associates, Inc., Sunderland, Mass.), was used to perform phylogenetic analysis by the unweighted pair group method using arithmetic averages (UPGMA) algorithm and by treating alleles as categorical characters. TreeView (available at http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) was used to print out the dendrogram generated by PAUP analysis.
The MIRU-VNTR allelic diversity (h) at each of the 12 loci was calculated by the equation h = 1 − Σxi2, where xi is the frequency of the ith allele at the locus (11).
The Hunter-Gaston discriminatory index (HGDI) described by Hunter and Gaston (13) was used as a numerical index for MIRU-VNTR discriminatory power. HGDI was calculated by using the following formula:
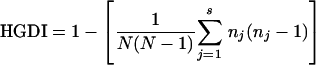 |
(1) |
where N is the total number of strains in the typing scheme, s is the total number of different MIRU-VNTR patterns, and nj is the number of strains belonging to the jth pattern.
TbD1 PCR.
The PCR primer sequences flanking the M. tuberculosis specific deletion 1 (TbD1) region have been described by Brosch et al. (4). PCRs were performed in 50- μl reaction volumes in 96-well PCR plates. The reaction mixture contained the same PCR reagents as did the MIRU-VNTR PCR, except for the primers, and the reagents were used at the same concentrations as in the MIRU-VNTR PCR, except that 2.5 U of HotStartTaq DNA polymerase was used. The thermocycling conditions were as follows: after an initial denaturation at 95°C for 3 min, a touch-down program was used, including 10 cycles of denaturation at 94°C for 15 s, annealing at 59°C for 15 s with a 1°C decrease per cycle (to 49°C), and extension at 72°C for 4 min, followed by 30 cycles of 94°C for 15 s, 49°C for 15 s, and 72°C for 4 min. The reaction was terminated at 72°C for 10 min. PCR products were analyzed in 1% agarose gels. The amplicons were 2,638- and 485-bp long for the M. tuberculosis strains in which the TbD1 region was present and for the strains in which the TbD1 region was absent, respectively.
RESULTS
Genetic diversity of M. tuberculosis in Singapore.
Using MIRU-VNTR typing, we analyzed 303 M. tuberculosis isolates from 291 patients. Twelve patients provided two samples. For each of the 12 pairs of duplicate samples, the IS6110 RFLP patterns were identical, and 10 pairs of duplicate samples also had the same MIRU-VNTR patterns (data not shown). For the remaining two pairs, distinct MIRU-VNTR patterns differing by one or two loci (pattern numbers 36, 37, 78, and 83 in Table 1) were obtained. This was confirmed by a blind repeat experiment. Patterns 36 and 37 corresponded to IS6110 low-copy-number fingerprints, while patterns 78 and 83 corresponded to IS6110 high-copy-number fingerprints (data not shown). Among several explanations, these results might have been due to laboratory cross-contamination or to reinfection by a closely related strain in the two patients. Therefore, we considered that a total of 293 isolates were typed; 166 distinct MIRU-VNTR patterns, including 35 cluster patterns and 131 unique patterns, were identified (Table 1); 162 isolates fell into one of the 35 clusters and each of the cluster patterns was shared by 2 to 39 isolates. The discriminatory power of MIRU-VNTR typing was high (HGDI = 0.975) for this M. tuberculosis sample collection.
TABLE 1.
MIRU-VNTR fingerprinting results for 293 M. tuberculosis isolates
| Pattern no. | MIRU-VNTR pattern at locus
|
No. of isolates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 4a | 10 | 16 | 20 | 23 | 24 | 26 | 27 | 31 | 39 | 40 | ||
| 1 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 1 | 1 | 3 | 4 | 1 |
| 2 | 2 | 7 | 4 | 3 | 2 | 6 | 2 | 2 | 2 | 1 | 4 | 4 | 1 |
| 3 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 2 | 3 | 4 | 1 |
| 4 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 4 | 3 | 4 | 2 |
| 5 | 2 | 5 | 4 | 3 | 2 | 7 | 2 | 2 | 3 | 4 | 3 | 4 | 1 |
| 6 | 2 | 7 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 4 | 3 | 4 | 1 |
| 7 | 2 | 3 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 4 | 3 | 4 | 1 |
| 8 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 4 | 4 | 4 | 1 |
| 9 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 5 | 4 | 4 | 1 |
| 10 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 5 | 1 | 4 | 1 |
| 11 | 2 | 5 | 4 | 1 | 2 | 6 | 2 | 2 | 3 | 5 | 3 | 4 | 1 |
| 12 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 2 | 3 | 2 | 1 |
| 13 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 4 | 3 | 2 | 10 |
| 14 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 4 | 3 | 1 | 2 |
| 15 | 2 | 4 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 4 | 3 | 1 | 1 |
| 16 | 2 | 5′ | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 4 | 2 | 2 | 1 |
| 17 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 4 | 2 | 2 | 3 |
| 18 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 1 | 4 | 2 | 2 | 1 |
| 19 | 2 | 5 | 4 | 1 | 2 | 6 | 2 | 2 | 3 | 4 | 2 | 2 | 1 |
| 20 | 2 | 5 | 4 | 2 | 2 | 6 | 2 | 2 | 3 | 4 | 2 | 5 | 1 |
| 21 | 2 | 5 | 4 | 3 | 2 | 2 | 2 | 2 | 3 | 4 | 1 | 2 | 1 |
| 22 | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 4 | 1 | 2 | 1 |
| 23 | 2 | 5 | 4 | 3 | 2 | 1 | 2 | 2 | 3 | 4 | 1 | 4 | 1 |
| 24 | 2 | 0 | 4 | 3 | 2 | 6 | 2 | 2 | 2 | 4 | 3 | 2 | 1 |
| 25 | 2 | 0 | 4 | 3 | 1 | 6 | 2 | 2 | 3 | 4 | 3 | 2 | 1 |
| 26 | 2 | 0 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 4 | 3 | 2 | 1 |
| 27 | 2 | 0 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 5 | 3 | 2 | 1 |
| 28 | 2 | 4 | 4 | 3 | 2 | 6 | 2 | 2 | 1 | 5 | 3 | 2 | 1 |
| 29 | 2 | 6 | 4 | 3 | 2 | 6 | 2 | 2 | 4 | 6 | 3 | 5 | 1 |
| 30 | 2 | 7 | 4 | 2 | 2 | 4 | 2 | 2 | 3 | 4 | 3 | 4 | 1 |
| 31 | 2 | 2 | 4 | 2 | 2 | 6 | 2 | 2 | 3 | 4 | 3 | 2 | 1 |
| 32 | 2 | 5 | 4 | 3 | 2 | 4 | 2 | 7 | 3 | 4 | 3 | 2 | 1 |
| 33 | 2 | 5 | 5 | 3 | 2 | 6 | 2 | 5 | 3 | 4 | 3 | 2 | 1 |
| 34 | 2 | 2 | 4 | 3 | 2 | 4 | 2 | 2 | 3 | 4 | 2 | 2 | 1 |
| 35 | 2 | 5 | 4 | 3 | 2 | 4 | 2 | 2 | 3 | 4 | 2 | 2 | 1 |
| 36 | 2 | 5 | 4 | 3 | 2 | 4 | 2 | 2 | 3 | 5 | 2 | 2 | 1 |
| 37 | 2 | 0 | 4 | 3 | 2 | 4 | 2 | 2 | 3 | 5 | 2 | 2 | 1 |
| 38 | 2 | 5 | 4 | 3 | 1 | 12 | 2 | 2 | 3 | 4 | 2 | 3 | 1 |
| 39 | 2 | 5 | 4 | 3 | 2 | 4 | 2 | 2 | 3 | 5 | 2 | 3 | 1 |
| 40 | 2 | 5 | 4 | 3 | 2 | 10 | 2 | 2 | 3 | 3 | 3 | 3 | 1 |
| 41 | 2 | 5 | 4 | 3 | 2 | 3 | 2 | 2 | 3 | 5 | 3 | 3 | 1 |
| 42 | 2 | 1′ | 4 | 3 | 2 | 4 | 2 | 2 | 3 | 5 | 3 | 3 | 1 |
| 43 | 2 | 0 | 4 | 3 | 2 | 1 | 2 | 2 | 3 | 5 | 1 | 3 | 1 |
| 44 | 2 | 5 | 3 | 3 | 2 | 4 | 2 | 2 | 3 | 6 | 3 | 3 | 1 |
| 45 | 2 | 5 | 4 | 2 | 2 | 5 | 3 | 2 | 3 | 2 | 3 | 3 | 1 |
| 46 | 2 | 5 | 4 | 2 | 2 | 5 | 2 | 2 | 3 | 5 | 3 | 3 | 1 |
| 47 | 3 | 0 | 4 | 2 | 2 | 5 | 2 | 2 | 3 | 4 | 3 | 3 | 1 |
| 48 | 3 | 2 | 4 | 2 | 2 | 5 | 2 | 2 | 3 | 5 | 3 | 3 | 1 |
| 49 | 2 | 0 | 4 | 2 | 1 | 5 | 2 | 2 | 3 | 5 | 3 | 2 | 1 |
| 50 | 2 | 6 | 4 | 2 | 2 | 5 | 2 | 2 | 3 | 5 | 2 | 3 | 2 |
| 51 | 2 | 5 | 6 | 3 | 2 | 6 | 2 | 7 | 3 | 5 | 1 | 3 | 1 |
| 52 | 2 | 6 | 5 | 1 | 2 | 5 | 1 | 2 | 3 | 5 | 3 | 3 | 1 |
| 53 | 2 | 6 | 4 | 1 | 2 | 5 | 1 | 2 | 3 | 5 | 3 | 3 | 1 |
| 54 | 2 | 2 | 4 | 1 | 2 | 5 | 1 | 2 | 3 | 5 | 3 | 3 | 1 |
| 55 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 2 | 3 | 5 | 3 | 3 | 2 |
| 56 | 2 | 0 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 3 | 1 |
| 57 | 2 | 0 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 2 | 3 | 1 |
| 58 | 2 | 0 | 3 | 3 | 2 | 5 | 1 | 10 | 3 | 5 | 3 | 3 | 1 |
| 59 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 6 | 3 | 5 | 3 | 3 | 11 |
| 60 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 6 | 2 | 5 | 3 | 3 | 1 |
| 61 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 6 | 3 | 5 | 3 | 4 | 1 |
| 62 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 5 | 3 | 5 | 3 | 3 | 2 |
| 63 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 8 | 3 | 5 | 3 | 3 | 4 |
| 64 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 1 | 3 | 5 | 3 | 3 | 1 |
| 65 | 2 | 2 | 3 | 3 | 1 | 5 | 1 | 7 | 3 | 3 | 3 | 3 | 1 |
| 66 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 6 | 3 | 3 | 2 |
| 67 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 4 | 3 | 3 | 5 |
| 68 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 4 | 3 | 4 | 3 | 3 | 2 |
| 69 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 5 | 3 | 4 | 3 | 3 | 1 |
| 70 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 5 | 3 | 4 | 3 | 4 | 1 |
| 71 | 2 | 2 | 7 | 4 | 2 | 5 | 1 | 1 | 3 | 4 | 3 | 4 | 1 |
| 72 | 2 | 2 | 2 | 4 | 2 | 5 | 1 | 1 | 3 | 4 | 3 | 4 | 1 |
| 73 | 2 | 2 | 3 | 3 | 2 | 9 | 1 | 1 | 3 | 4 | 3 | 3 | 1 |
| 74 | 2 | 2 | 7 | 2 | 2 | 5 | 1 | 7 | 3 | 4 | 3 | 3 | 3 |
| 75 | 2 | 2 | 7 | 2 | 2 | 5 | 1 | 7 | 3 | 4 | 3 | 4 | 1 |
| 76 | 2 | 2 | 6 | 2 | 2 | 5 | 1 | 7 | 3 | 4 | 3 | 3 | 1 |
| 77 | 2 | 2 | 5 | 2 | 2 | 5 | 1 | 7 | 3 | 4 | 3 | 3 | 2 |
| 78 | 2 | 2 | 5 | 2 | 2 | 5 | 1 | 7 | 3 | 4 | 2 | 3 | 1 |
| 79 | 2 | 2 | 8 | 2 | 2 | 5 | 1 | 7 | 3 | 4 | 4 | 3 | 1 |
| 80 | 2 | 3′ | 8 | 2 | 2 | 1 | 1 | 7 | 3 | 4 | 4 | 3 | 1 |
| 81 | 2 | 2 | 8 | 2 | 2 | 5 | 1 | 7 | 3 | 4 | 2 | 2 | 1 |
| 82 | 2 | 2 | 3 | 2 | 2 | 5 | 1 | 7 | 3 | 4 | 3 | 3 | 1 |
| 83 | 2 | 2 | 5 | 2 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 3 | 1 |
| 84 | 2 | 2 | 3 | 2 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 3 | 2 |
| 85 | 2 | 2 | 3 | 2 | 2 | 5 | 1 | 5 | 1 | 5 | 3 | 3 | 2 |
| 86 | 2 | 2 | 3 | 4 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 3 | 1 |
| 87 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 3 | 39 |
| 88 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 3 | 6 |
| 89 | 2 | 2 | 1 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 3 | 1 |
| 90 | 2 | 2 | 1 | 3 | 2 | 7 | 1 | 7 | 3 | 5 | 3 | 4 | 1 |
| 91 | 2 | 2 | 3 | 3 | 2 | 6 | 1 | 7 | 3 | 5 | 3 | 3 | 2 |
| 92 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 3 | 3 | 1 | 1 |
| 93 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 6 | 3 | 1 | 1 |
| 94 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 1 | 5 | 3 | 1 | 3 |
| 95 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 7 | 1 | 5 | 4 | 1 | 1 |
| 96 | 2 | 3 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 2 | 1 |
| 97 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 2 | 5 |
| 98 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 2 | 3 | 5 | 2 | 2 | 1 |
| 99 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 2 | 2 | 1 |
| 100 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 6 | 3 | 5 | 3 | 2 | 2 |
| 101 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 8 | 3 | 5 | 3 | 2 | 1 |
| 102 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 4 | 3 | 2 |
| 103 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 5 | 3 | 1 |
| 104 | 2 | 4 | 3 | 3 | 2 | 5 | 1 | 6 | 3 | 2 | 1 | 3 | 1 |
| 105 | 2 | 2 | 3 | 3 | 2 | 2 | 1 | 7 | 3 | 2 | 1 | 3 | 1 |
| 106 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 6 | 3 | 5 | 1 | 3 | 1 |
| 107 | 2 | 2 | 3 | 3 | 2 | 1 | 1 | 7 | 3 | 5 | 1 | 3 | 1 |
| 108 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 1 | 3 | 2 |
| 109 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 1 | 2 | 3 | 1 |
| 110 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 2 | 2 | 1 | 1 |
| 111 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 2 | 2 | 3 | 1 |
| 112 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 2 | 3 | 7 |
| 113 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 7 | 3 | 3 | 4 | 3 | 1 |
| 114 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 4 | 3 | 2 |
| 115 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 9 | 3 | 5 | 4 | 3 | 2 |
| 116 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 6 | 3 | 5 | 4 | 3 | 1 |
| 117 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 4 | 3 | 16 |
| 118 | 2 | 2 | 2 | 2 | 2 | 5 | 1 | 7 | 3 | 5 | 4 | 3 | 1 |
| 119 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 2 | 3 | 2 |
| 120 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 4 | 3 | 2 | 2 | 3 | 1 |
| 121 | 2 | 2 | 5 | 3 | 2 | 5 | 1 | 4 | 3 | 5 | 2 | 3 | 1 |
| 122 | 2 | 2 | 5 | 1 | 2 | 5 | 1 | 1 | 3 | 1 | 2 | 2 | 1 |
| 123 | 2 | 2 | 5 | 1 | 2 | 5 | 1 | 1 | 1 | 2 | 2 | 2 | 1 |
| 124 | 2 | 2 | 5 | 1 | 2 | 5 | 1 | 1 | 3 | 3 | 2 | 2 | 2 |
| 125 | 2 | 2 | 2 | 1 | 2 | 5 | 1 | 1 | 3 | 3 | 2 | 2 | 1 |
| 126 | 2 | 2 | 7 | 2 | 2 | 5 | 1 | 1 | 3 | 3 | 2 | 2 | 1 |
| 127 | 2 | 2 | 2 | 2 | 2 | 5 | 1 | 1 | 3 | 3 | 2 | 2 | 1 |
| 128 | 2 | 2 | 2 | 2 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 2 | 1 |
| 129 | 2 | 2 | 6 | 4 | 2 | 3 | 1 | 5 | 3 | 3 | 2 | 4 | 1 |
| 130 | 2 | 2 | 2 | 4 | 2 | 6 | 1 | 5 | 3 | 3 | 2 | 2 | 1 |
| 131 | 2 | 2 | 2 | 4 | 2 | 5 | 1 | 4 | 3 | 3 | 2 | 4 | 1 |
| 132 | 2 | 2 | 2 | 4 | 2 | 5 | 1 | 6 | 3 | 3 | 2 | 2 | 1 |
| 133 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 2 | 1 |
| 134 | 2 | 4 | 2 | 3 | 2 | 4 | 1 | 5 | 3 | 3 | 2 | 2 | 1 |
| 135 | 2 | 4 | 2 | 3 | 2 | 5 | 1 | 2 | 2 | 2 | 2 | 2 | 1 |
| 136 | 2 | 4 | 2 | 3 | 2 | 5 | 1 | 3 | 2 | 2 | 2 | 2 | 1 |
| 137 | 2 | 4 | 2 | 3 | 2 | 5 | 1 | 5 | 2 | 2 | 2 | 1 | 1 |
| 138 | 2 | 4 | 2 | 3 | 2 | 5 | 1 | 5 | 2 | 2 | 2 | 2 | 1 |
| 139 | 2 | 4 | 2 | 3 | 2 | 5 | 1 | 5 | 2 | 3 | 2 | 2 | 3 |
| 140 | 2 | 4 | 2 | 3 | 2 | 4 | 1 | 4 | 2 | 3 | 2 | 2 | 1 |
| 141 | 2 | 4 | 2 | 3 | 2 | 4 | 1 | 1 | 2 | 3 | 2 | 2 | 1 |
| 142 | 2 | 4 | 2 | 3 | 2 | 4 | 1 | 5 | 2 | 3 | 2 | 2 | 2 |
| 143 | 2 | 4 | 2 | 3 | 2 | 5 | 1 | 2 | 2 | 3 | 2 | 2 | 1 |
| 144 | 2 | 4 | 2 | 3 | 2 | 5 | 1 | 4 | 2 | 4 | 2 | 2 | 1 |
| 145 | 2 | 3 | 2 | 3 | 2 | 5 | 1 | 3 | 2 | 3 | 2 | 2 | 1 |
| 146 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 5 | 2 | 3 | 2 | 2 | 1 |
| 147 | 2 | 4 | 2 | 3 | 2 | 5 | 1 | 4 | 2 | 3 | 2 | 3 | 1 |
| 148 | 2 | 1 | 2 | 2 | 2 | 5 | 1 | 1 | 2 | 3 | 2 | 3 | 1 |
| 149 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | 5 |
| 150 | 2 | 3′ | 2 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | 2 |
| 151 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 5 | 1 | 3 | 2 | 3 | 1 |
| 152 | 2 | 2 | 6 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | 1 |
| 153 | 2 | 2 | 5 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | 2 |
| 154 | 2 | 3 | 4 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | 1 |
| 155 | 2 | 2 | 4 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | 1 |
| 156 | 2 | 2 | 5 | 3 | 2 | 5 | 1 | 4 | 3 | 3 | 2 | 3 | 1 |
| 157 | 2 | 2 | 4 | 3 | 2 | 5 | 1 | 4 | 3 | 3 | 2 | 3 | 1 |
| 158 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 6 | 3 | 3 | 2 | 3 | 1 |
| 159 | 2 | 2 | 4 | 3 | 2 | 5 | 1 | 6 | 3 | 3 | 2 | 4 | 1 |
| 160 | 2 | 2 | 4 | 4 | 2 | 5 | 1 | 2 | 3 | 3 | 2 | 3 | 1 |
| 161 | 1 | 2 | 4 | 4 | 2 | 5 | 1 | 8 | 3 | 5 | 2 | 4 | 1 |
| 162 | 2 | 2 | 2 | 2 | 2 | 4 | 1 | 5 | 3 | 2 | 2 | 3 | 1 |
| 163 | 2 | 2 | 3 | 3 | 1 | 6 | 1 | 5 | 3 | 2 | 2 | 6 | 1 |
| 164 | 2 | 3 | 2 | 3 | 2 | 1 | 1 | 6 | 3 | 3 | 2 | 1 | 1 |
| 165 | 2 | 2 | 3 | 2 | 2 | 5 | 1 | 5 | 2 | 5 | 2 | 2 | 1 |
| 166 | 2 | 2 | 7 | 3 | 2 | 6 | 1 | 1 | 2 | 4 | 2 | 2 | 1 |
The prime symbol for MIRU numbers in locus 4 designates that the invariable 3′-terminal 53-bp unit of locus 4 which is present in nearly all isolates is absent from that isolate(s).
By direct comparison, a number of the MIRU-VNTR patterns shown in Table 1, e.g., patterns 4, 5, 10, 13, 62, 63, 66, 76, 86, 87, 97, 102, 149, 152, 153, and 155, were identical to one of the previously typed MIRU-VNTR patterns of the representative East-Africa-Indian (EAI), W/Beijing, and Haarlem family strains (15, 24, 26). In addition, patterns 71 and 72 from Table 1 were identical to the novel “Delhi” M. tuberculosis strains identified in the Delhi region of India (2) that were recently typed by MIRU-VNTR typing (P. Supply et al., unpublished data).
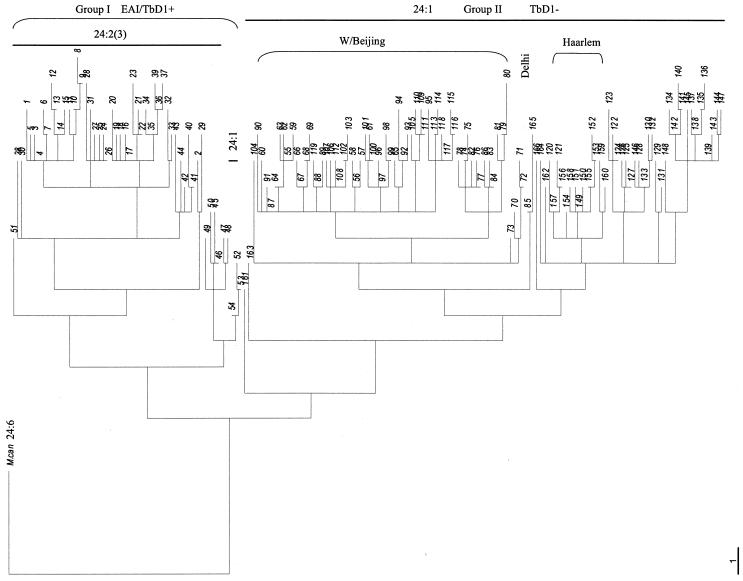
The dendrogram (Fig. 1) . generated by using the UPGMA algorithm based on the MIRU-VNTR patterns shown in Table 1 describes the genetic relationships of the 293 isolates. As shown in Fig. 1, all strains were divided into one of two main groups, groups I and II. Interestingly, group I and group II isolates could be distinguished by merely using the allelic status of locus 24. All of the isolates that contained at least two repeats went into group I, and all but three of the isolates that contained one repeat went into group II. The four representative MIRU-VNTR patterns of EAI strains (patterns 4, 5, 10, and 13) fell into group I. The representative MIRU-VNTR patterns of W/Beijing strains (patterns 62, 63, 66, 76, 86, 87, 97, and 102), Haarlem strains (patterns 149, 152, 153, and 155), and Delhi strains (patterns 71 and 72) fell into two subgroups of group II. This is consistent with previous studies that showed that M. tuberculosis EAI strains contain at least two alleles in locus 24, whereas the rest of the M. tuberculosis family strains, such as W/Beijing, Haarlem, Africa, Central Asia 1 (CAS1), X, and Latin American and Mediterranean strains, contain one allele in locus 24 (7, 24, 26).
FIG. 1.
Genetic relationships of the 293 M. tuberculosis isolates. This dendrogram was generated by using the UPGMA algorithm based on the 166 distinct MIRU-VNTR patterns (Table 1) which represented the 293 isolates. M. canettii (26) was used as an outgroup. The M. tuberculosis EAI, W/Beijing, Delhi, and Haarlem families are indicated. Group I includes only the ancestral EAI strains which were TbD1+, and group II includes all of the modern M. tuberculosis strains which were TbD1−. The allele number in locus 24 is indicated as 24:n. The linkage distance is indicated at the left bottom corner.
To confirm this grouping, we analyzed the TbD1 region by PCR. Brosch et al. (4) have reported that M. tuberculosis complex strains can be divided into ancestral or modern strains based on the presence or absence, respectively, of the TbD1 region. The TbD1 region is present in ancestral M. tuberculosis strains (such as EAI strains) but is absent from modern M. tuberculosis strains (such as W/Beijing, Haarlem, and Africa strains). Strikingly consistent with this, our TbD1 PCR results showed that all of the isolates in group I had the TbD1 region (TbD1+) and all of the isolates in group II did not have the TbD1 region. Taken together, these results indicate that all of the isolates in group I correspond to ancestral EAI strains and all of the isolates in group II correspond to modern M. tuberculosis strains.
The ancestral EAI isolates accounted for 68 of the 293 strains (23.2%) and included 54 MIRU-VNTR patterns. Therefore, each of the EAI MIRU-VNTR patterns was shared by a mean of 1.3 isolates (68/54), and the HGDI for these isolates was 0.978, suggesting a high discrimination of MIRU-VNTR typing for EAI strains. The EAI isolates also appeared highly heterogeneous by an analysis of MIRU-VNTR genetic distances (Fig. 1). Only 5 of the 12 loci (42%) had the same alleles between some isolates.
Among the modern M. tuberculosis isolates, W/Beijing and Haarlem isolates appeared well grouped by the MIRU-VNTR analysis (Fig. 1). The eight representative MIRU-VNTR patterns of W/Beijing strains were scattered into two subgroupings which represented 161 of the 293 isolates. The isolates within each of the two groupings had the same alleles in at least 7 of the 12 loci (58%), while at least 6 loci (50%) had the same MIRU-VNTR alleles between all isolates in the two groupings, except for isolate 80. The four representative MIRU-VNTR patterns of Haarlem strains were included in a grouping which contained 20 isolates. These isolates had the same MIRU-VNTR alleles in at least 8 of the 12 loci (67%).
Allelic diversities of the MIRU-VNTR loci.
Table 2 shows the allelic diversities of the 12 MIRU-VNTR loci based on this collection of samples. Loci 10, 26, 31, and 39 were highly discriminative (≥0.6), loci 4, 16, 23, 24, and 40 were moderately discriminative (≥0.3), and loci 2, 20, and 27 were poorly discriminative (<0.3) when ranked according to the method of Sola et al. (24). Overall, the allelic diversity of our sample was fairly high compared to two local collections of M. tuberculosis (7, 27), but it was lower than that of a global collection which included 104 M. tuberculosis isolates (24). This is consistent with our sample's origin from a heterogeneous population.
TABLE 2.
Allelic diversity of each MIRU-VNTR locus
| No. of alleles | No. of isolates with MIRU-VNTR locus
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 4 | 10 | 16 | 20 | 23 | 24 | 26 | 27 | 31 | 39 | 40 | |
| 0 | 11 | |||||||||||
| 1 | 1 | 1 | 2 | 10 | 5 | 5 | 228 | 14 | 11 | 4 | 12 | 12 |
| 2 | 290 | 202 | 70 | 34 | 288 | 2 | 64 | 71 | 22 | 15 | 89 | 72 |
| 3 | 2 | 5 | 123 | 240 | 2 | 1 | 2 | 259 | 49 | 160 | 182 | |
| 4 | 18 | 70 | 9 | 15 | 10 | 1 | 66 | 31 | 24 | |||
| 5 | 43 | 14 | 217 | 38 | 154 | 1 | 2 | |||||
| 6 | 5 | 4 | 47 | 22 | 5 | 1 | ||||||
| 7 | 3 | 7 | 2 | 127 | ||||||||
| 8 | 3 | 6 | ||||||||||
| 9 | 1 | 2 | ||||||||||
| 10 | 1 | 1 | ||||||||||
| 12 | 1 | |||||||||||
| 1a | 1 | |||||||||||
| 3a | 3 | |||||||||||
| 5a | 1 | |||||||||||
| Allelic diversityb | 0.02 | 0.50 | 0.71 | 0.31 | 0.03 | 0.42 | 0.35 | 0.73 | 0.21 | 0.64 | 0.60 | 0.54 |
The 53-bp unit at the 3′ terminus of locus 4 was absent.
Allelic diversity (h) was defined as h = 1 − Σ xi, where xi is the frequency of the ith allele at the locus (11).
Reproducibility of MIRU-VNTR typing.
In order to check the reproducibility of MIRU-VNTR typing in our setting, we performed two independent blind typing tests with 24 isolates. The results were identical between the two tests, indicating that MIRU-VNTR typing had 100% reproducibility under our conditions.
DISCUSSION
Using the novel MIRU-VNTR typing method, we studied the genetic diversity of M. tuberculosis strains in Singapore. Four families of M. tuberculosis strains were identified, although the lineages of quite a number of strains remain to be recognized. Three of the four family strains are major worldwide epidemic strains, including W/Beijing, EAI, and Haarlem strains. W/Beijing strains appear to account for the largest number of strains in Singapore, which is not surprising because the majority of the Singaporean population is of Chinese descent and because tuberculosis affects the elderly, many of whom are first- or second-generation immigrants from mainland China. W/Beijing strains are also dominant in other countries of East and Southeast Asia, in addition to China, such as Mongolia, Vietnam, South Korea, and Thailand (1, 6, 20-22, 31). In Malaysia and Indonesia, Singapore's closest geographical neighbors, the W/Beijing genotype represents 24 and 34% of the strains, respectively (8, 28).
Another highly prevalent M. tuberculosis genotype family in Singapore is the EAI family, which is known to contain a high frequency of IS6110 low-copy-number (or zero-copy-number) strains (4, 15). IS6110 low-copy-number strains, though not all belonging to the EAI family, are also prevalent in South India (9) and in other countries of Southeast Asia (8, 19, 32). The small number of IS6110 copies renders IS6110 typing inefficient for discriminating between strains of this family (15). Moreover, EAI strains have highly homogeneous spoligotypes. In a spoligotype database, EAI strains have been subdivided into only five subfamilies by five spoligotypes, each of which is shared by a mean of 129 isolates (10). This is in striking contrast with our MIRU-VNTR typing results, which showed that each of the EAI MIRU-VNTR patterns was shared only by a mean of less than two isolates. Consistently, spoligotyping analysis of EAI isolates from Singapore and India showed that conserved spoligotypes in this family are often split by MIRU-VNTR types (unpublished data). These results indicate that MIRU-VNTR types are better typing markers for evaluating the diversity of M. tuberculosis EAI strains than are IS6110 types or spoligotypes, although different populations should be compared with caution.
EAI strains are also called ancestral TbD1+ strains because of the presence of a specific region, called the TbD1 region, in their genomes. This region is also found in other M. tuberculosis complex species, such as M. canettii, M. bovis, M. africanum, and M. microti, but has been lost in other modern M. tuberculosis strains, which have been shown to include members of the W/Beijing, Haarlem, and Africa families (4). Fully consistent with this, we found that all of the isolates assigned to the EAI family by MIRU-VNTR analysis (group I) were positive for the TbD1 region and that all of the remaining isolates, including those assigned to known modern M. tuberculosis genotype families (group II), were negative for the TbD1 region. This systematic association supports an evolutionary scenario in which the TbD1 deletion occurred as a single event in a common progenitor rather than as events occurring independently or occasionally in individual strains (4).
Additionally, this perfect consistency between MIRU-VNTR typing and TbD1 typing for grouping ancestral and modern M. tuberculosis strains supports the claim that MIRU-VNTR types are useful genetic markers for phylogenetic analysis. Interestingly, all isolates that contained at least two alleles in locus 24 were included in the ancestral group, and all but three isolates that contained one allele in locus 24 were included in the modern group. Therefore, our results indicate that the use of locus 24 alone has an almost equal performance to the use of the TbD1 region as a marker for identifying ancestral and modern M. tuberculosis strains. This supports our previous contention that the selective use of some less-variable loci, such as this one, may be especially informative for evolutionary studies (26).
In conclusion, MIRU-VNTR typing based on a capillary electrophoresis system offers convenience, automation, a short turnaround time, high throughput, and perfect reproducibility. The digital format of the MIRU-VNTR pattern allowed us to readily compare our results with previous results (24, 26), illustrating the ease of interlaboratory comparisons by this method. Since there is a high proportion of EAI and W/Beijing strains in Singapore for which IS6110 RFLP and/or spoligotyping is poorly informative, MIRU-VNTR typing may be the method of first choice for M. tuberculosis strain typing in this country.
Acknowledgments
We are grateful to Lynn L. H. Tang and Irene H. K. Lim for excellent technical assistance; Pablo Bifani for helpful discussions; and John T. Belisle, Colorado State University, NIH, NIAID contract N01 AI-75320 (TB Research Materials and Vaccine Testing contract), and Colorado State University for providing H37Rv genomic DNA. We also gratefully acknowledge the excellent administrative support by Bernard Peperstraete and the staff of the Infectious Diseases Research Centre.
Y. J. Sun is a research scientist supported by the National Medical Research Council, Singapore, Republic of Singapore. P. Supply is a research scientist of the Centre National de la Recherche Scientifique, France. This work was supported by a grant from the National Medical Research Council, Singapore, Republic of Singapore (NMRC/0557/2001).
REFERENCES
- 1.Anh, D. D., M. W. Borgdorff, L. N. Van, N. T. Lan, T. van Gorkom, K. Kremer, and D. van Soolingen. 2000. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6:302-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhanu, N. V., D. van Soolingen, J. D. van Embden, L. Dar, R. M. Pandey, and P. Seth. 2002. Predominance of a novel Mycobacterium tuberculosis genotype in the Delhi region of India. Tuberculosis (Edinburgh) 82:105-112. [DOI] [PubMed] [Google Scholar]
- 3.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 4.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cave, M. D., K. D. Eisenach, P. F. McDermott, J. H. Bates, and J. T. Crawford. 1991. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol. Cell. Probes 5:73-80. [DOI] [PubMed] [Google Scholar]
- 6.Chan, M. Y., M. Borgdorff, C. W. Yip, P. E. de Haas, W. S. Wong, K. M. Kam, and D. van Soolingen. 2001. Seventy percent of the Mycobacterium tuberculosis isolates in Hong Kong represent the Beijing genotype. Epidemiol. Infect. 127:169-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowan, L. S., L. Mosher, L. Diem, J. P. Massey, and J. T. Crawford. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dale, J. W., R. M. Nor, S. Ramayah, T. H. Tang, and Z. F. Zainuddin. 1999. Molecular epidemiology of tuberculosis in Malaysia. J. Clin. Microbiol. 37:1265-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, S., C. N. Paramasivan, D. B. Lowrie, R. Prabhakar, and P. R. Narayanan. 1995. IS6110 restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, south India. Tuber. Lung. Dis. 76:550-554. [DOI] [PubMed] [Google Scholar]
- 10.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. D. Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, G. Kallenius, E. Kassa-Kelembho, T. Koivula, H. M. Ly, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. de Waard, C. Sola, and N. Rastogi. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg. Infect. Dis. 8:1347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graur, D., and W.-H. Li. 2000. Dynamics of genes in populations, p. 58. In D. Graur and W.-H. Li (ed.), Fundamentals of molecular evolution. Sinauer Associates, Sunderland, Mass.
- 12.Hawkey, P. M., E. G. Smith, J. T. Evans, P. Monk, G. Bryan, H. H. Mohamed, M. Bardhan, and R. N. Pugh. 2003. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS6110 -based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J. Clin. Microbiol. 41:3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwara, A., R. Schiro, L. S. Cowan, N. E. Hyslop, M. F. Wiser, S. Roahen Harrison, P. Kissinger, L. Diem, and J. T. Crawford. 2003. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 41:2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, A. S., L. L. Tang, I. H. Lim, R. Bellamy, and S. Y. Wong. 2002. Discrimination of single-copy IS6110 DNA fingerprints of Mycobacterium tuberculosis isolates by high-resolution minisatellite-based typing. J. Clin. Microbiol. 40:657-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palittapongarnpim, P., P. Luangsook, S. Tansuphaswadikul, C. Chuchottaworn, R. Prachaktam, and B. Sathapatayavongs. 1997. Restriction fragment length polymorphism study of Mycobacterium tuberculosis in Thailand using IS6110 as probe. Int. J. Tuberc. Lung Dis. 1:370-376. [PubMed] [Google Scholar]
- 20.Park, Y. K., G. H. Bai, and S. J. Kim. 2000. Restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolated from countries in the Western Pacific region. J. Clin. Microbiol. 38:191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prodinger, W. M., P. Bunyaratvej, R. Prachaktam, and M. Pavlic. 2001. Mycobacterium tuberculosis isolates of Beijing genotype in Thailand. Emerg. Infect. Dis. 7:483-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian, L., J. D. van Embden, A. G. van der Zanden, E. F. Weltevreden, H. Duanmu, and J. T. Douglas. 1999. Retrospective analysis of the Beijing family of Mycobacterium tuberculosis in preserved lung tissues. J. Clin. Microbiol. 37:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savine, E., R. M. Warren, G. D. van der Spuy, N. Beyers, P. D. van Helden, C. Locht, and P. Supply. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4561-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sola, C., I. Filliol, E. Legrand, S. Lesjean, C. Locht, P. Supply, and N. Rastogi. 2003. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect. Genet. Evol. 3:125-133. [DOI] [PubMed] [Google Scholar]
- 25.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 26.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Supply, P., R. M. Warren, A. L. Banuls, S. Lesjean, G. D. van der Spuy, L. A. Lewis, M. Tibayrenc, P. D. van Helden, and C. Locht. 2003. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol. Microbiol. 47:529-538. [DOI] [PubMed] [Google Scholar]
- 28.van Crevel, R., R. H. Nelwan, W. de Lenne, Y. Veeraragu, A. G. van der Zanden, Z. Amin, J. W. van der Meer, and D. van Soolingen. 2001. Mycobacterium tuberculosis Beijing genotype strains associated with febrile response to treatment. Emerg. Infect. Dis. 7:880-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Soolingen, D., P. W. Hermans, P. E. de Haas, D. R. Soll, and J. D. van Embden. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuen, L. K., B. C. Ross, K. M. Jackson, and B. Dwyer. 1993. Characterization of Mycobacterium tuberculosis strains from Vietnamese patients by Southern blot hybridization. J. Clin. Microbiol. 31:1615-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]