Iron is an essential chemical element for all forms of life. It serves as a cofactor for many proteins and enzymes involved in oxygen transport, energy metabolism and DNA synthesis[1, 2]. Mammalian cells need to maintain a sufficient amount of iron to support the synthesis of proteins that require iron as a co-factor. Iron is imported into the cells through the circulating iron transporter transferrin[3, 4]. Iron-loaded transferrin binds to cell surface transferrin receptor, resulting in the endocytosis of transferrin and delivery of the iron cargo into the cells[4]. Excess iron in the cells is stored in the cytosolic protein ferritin[4]. Iron regulatory proteins IRP1 and IRP2 maintain the homeostasis of iron through post-translational regulation of the expression of transferrin receptor and ferritin[2, 5]. In iron-replete cells IRP2 is ubiquitinated and degraded by the proteasome. The F-box and leucine-rich repeat containing protein FBXL5 serves as a cytosolic iron sensor that regulates the ubiquitination of IRP2 by the SKP1-CUL1-FBXL5 (SCF) E3 ubiquitin ligase complex[6, 7]. FBXL5 also plays critical roles in sensing oxygen in the cytosol[6, 7]. Knock out of FBXL5 in mice result in embryonic mortality due to excess accumulation of iron[8].
FBXL5 contains a hemerythrin (Hr) like domain at its N-terminus, an F-box-domain in the middle, and a leucine-rich repeat (LRR) domain at the C-terminus. The Hr like domain, which is related to a family of iron and oxygen binding proteins in invertebrates and bacteria[9, 10], is responsible for iron and oxygen sensing[6, 7]. The F-box domain interacts with SKP1, which serves as a bridge between FBXL5 and Cullin-1 in the SCF E3 complex[6, 7, 11, 12]. The LRR domain is proposed to associate with the substrate IRP2. Under iron deficient or hypoxic conditions FBXL5 is not stable, presumably due to the unfolding of the Hr like domain[6, 7]. Since FBXL5 Hr only shows limited sequence homology to invertebrate or bacterial Hr and two of the seven iron ligands are not conserved in the sequence of FBXL5, it is not clear how FBXL5 binds iron. Unlike invertebrate and bacterial Hr, which serve as oxygen binding molecules and have stable diiron centers[9, 13], it is not clear how FBXL5 binds iron reversibly and how it senses oxygen. To understand the structural basis of iron and oxygen sensing by FBXL5, we have conducted extensive biochemical and biophysical characterizations of the hemerythrin like domain of FBXL5 (referred to as FBXL5 Hr) and determined the crystal structures of the protein in oxidized and reduced forms.
We have expressed and purified the Hr domain of human FBXL5 (residues 1–161, Supplementary Methods). The protein is monomeric under reducing conditions, but tends dimerize under aerobic conditions, presumably through the formation of a disulfide bond between the free cysteine Cys159. The C159S mutant of the protein exists as a stable monomer. Elemental analysis by ICP-AES shows two iron atoms bind to each molecule of FBXL5 Hr. UV-visible spectroscopy of the protein under oxidized conditions shows FBXL5 Hr exhibits no absorption at 500 nm (Supplementary Figure 1), which is characteristic of the oxygen-bound form of hemerythrin-like proteins[9, 10, 14]. These data indicate that FBXL5 Hr does not bind oxygen directly. Electron paramagnetic resonance (EPR) spectra of fully reduced and oxidized FBXL5 Hr show no signal (Figure 1A), likely due to the antiferromagnetic coupling of the two Fe ions, indicating the two irons are ferrous under reduced conditions and ferric under oxidized conditions. In contrast, clear EPR signals (at g = 1.95, 1.82 and 1.77), which are characteristic of mixed-valence [FeII, FeIII] site, are observed from a partially reduced sample (Figure 1A). Comparison of 1H-15N HSQC NMR spectra of FBXL5 Hr purified under aerobic conditions and the reduced sample (Figure 1B) shows that the protein exists in a stable equilibrium of two conformations under aerobic conditions. Reduction of the protein with sodium dithionite (Na2S2O4) under anaerobic conditions results in a homogeneous sample corresponding to one of the two conformations (Figure 1B). Although FBXL5 Hr purified under aerobic conditions is stable, it is much less stable under reducing conditions. The reduction of FBXL5 Hr with Na2S2O4 under aerobic conditions results in the denaturation and aggregation of the protein (Figure 1C), likely due to the collapse of the Hr domain after the dissociation of the iron. The addition of EDTA under reducing conditions results in faster denaturation and aggregation of the protein (Figure 1C). In contrast, the protein is stable under anaerobic conditions even in presence of dithionite or EDTA (Figure 1C). These results indicate that the reduced FBXL5 Hr loses the iron readily under aerobic conditions, confirming previous observations that FBXL5 is sensitive to cellular iron and oxygen levels[6, 7].
Figure 1.
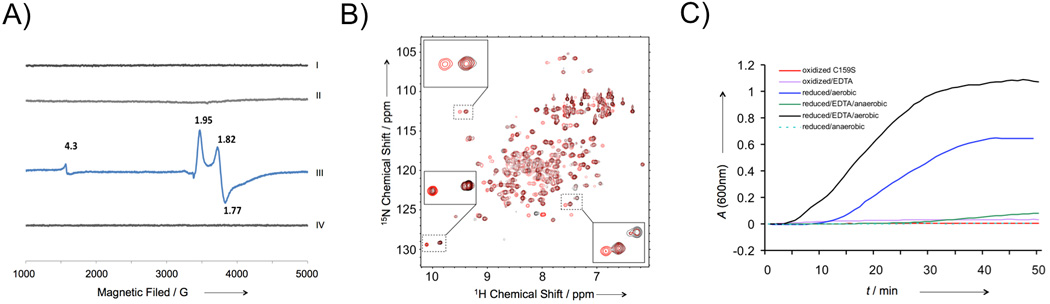
Characterization of the FBXL5 hemerythrin-like domain. A) EPR spectra of oxidized (I), aerobically purified (II), partially reduced (III), and fully reduced (IV) FBXL5 Hr. B) 1H-15N HSQC NMR spectra of FBXL5 Hr purified under aerobic conditions (red) and the reduced protein under anaerobic conditions (black). The insets show the cross-peaks that correspond to two different conformations. C) Stability of FBXL5 Hr under different conditions. The denaturation and aggregation of FBXL5 Hr was monitored spectrophotometrically at 600 nm. The concentration of the samples used is 40 µM.
To understand the structural basis of iron sensing by FBXL5, we have determined the 1.60 Å resolution structure of the oxidized protein using Se-Met derivative protein (Supplementary Table 1). The crystallographic asymmetric unit contains two FBXL5 Hr molecules, forming a head to tail dimer related by noncrystallographic two-fold symmetry. Anomalous difference maps calculated from native data collected using home source and Se-Met derivative data collected at Fe peak wavelength showed four high peaks that correspond to the four iron atoms bound to the two protein molecules (Supplementary Figure 2). As predicted by previous proteomic analysis[6, 7], FBXL5 Hr folds into a long up-and-down helical bundle structure similar to the structure of hemerythrin and related proteins (Figure 2A). Unlike hemerythrin, however, the N-terminus of helix α3 contains a flexible loop (loop 23, residues 74 to 82) that is poorly defined in the electron density map except for residue His80, which coordinates with Fe(1). The diiron center is located near one end of the helical bundle (Figure 2A). The overall structure of the diiron center of FBXL5 Hr is similar to the diiron center of deoxy hemerythrin (Supplementary Figure 3)[9, 15]. Four histidine and three glutamate residues coordinate with the two Fe ions (Figure 2B). His15 and His57 from helices α1 and α2 coordinate with Fe (2). His80 in the flexible loop23, His126 from helix α4, and Glu58 from helix α2 coordinate with Fe(1). The two iron atoms are bridged bidentately by the carboxylate of two glutamate residues Glu61 and Glu130. In addition, a μ-hydroxo group bridges the two Fe atoms. The average Fe-μ-O bond length is 2.2 Å (Figure 3). The average Fe-μ-O-Fe bound angle is 94.0°. Compared to hemerythrin diiron centers[9], a conserved bridging aspartic residue (Asp106 in Hr) is replaced by Glu130, and a histidine ligand (His77 in Hr) to Fe(1) is replaced by Glu58 (Supplementary Figure 3). The sidechain carboxylate of Glu58 coordinates monovalently with Fe(1). It also forms a hydrogen bond with the bridging hydroxo group (Oε2 to μ-O distance 2.6 Å). Similar to the deoxy hemerythrin[9, 15], the five ligands of Fe(2) arrange as trigonal bipyramidal coordination geometry. The six ligands of Fe(1) form an octohederal coordination sphere (Figure 2B and 3). Unlike the structure of oxidized hemerythrin (Supplementary Figure 3), no oxygen ligand is observed near Fe(2) in the electron density map, providing structural evidence that FBXL5 does not bind oxygen. The distance between the two Fe atoms is 3.21±0.01Å in the refined structure (Figure 3B), in agreement with the distances between the iron peaks in the anomalous difference map (3.17 Å, Supplementary Figure 2). One of the major differences between the diiron center of hemerythrin and FBXL5 Hr is the replacement of Fe(1) ligand His77 by Glu58 (Supplementary Figure 3). In hemerythrin, two adjacent histidine residues in helix α3 (His73 and His77) make significant contributions to stabilizing the structure of helix α3 by coordinating with Fe(1) simultaneously[15, 16]. The replacement His77 in hemerythrin by Glu58 in helix α2 instead of helix α3 in FBXL5 Hr likely contributes to the flexibility of loop23 and may regulates the reversible binding of iron by the protein. Fe(1) of FBXL5 will be exposed to solvent when its ligand His80 is removed. Indeed, the replacement of His80 with alanine reduced the yield and stability of FBXL5 Hr. The H80A mutant of FBXL5 Hr aggregates faster than the wild type protein under reducing conditions in presence or in absence of EDTA (Supplementary Figure 4), indicating the Fe ions dissociate faster from the H80A mutant than the wild type protein under these conditions.
Figure 2.
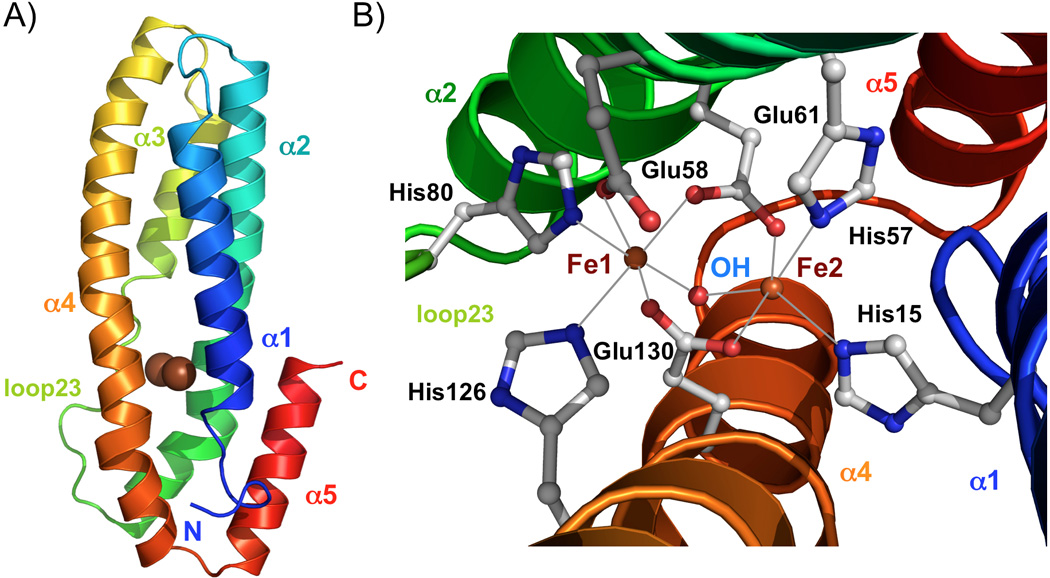
Structure of oxidized FBXL5 hemerythrin-like domain. A) Ribbon representation of the FBXL5 Hr structure. The two iron atoms are shown with the brown spheres. B) Close up of the diiron center. Iron ligands of the irons are shown in ball-and-stick representation. The red sphere respresents the μ-hydroxo bridge.
Figure 3.
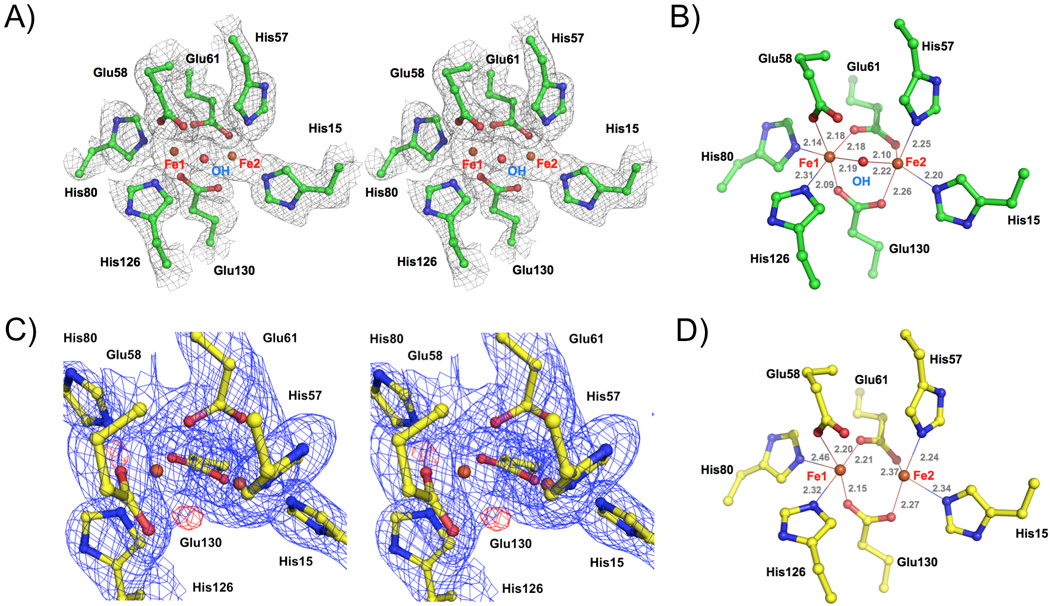
Structure of the diiron center of FBXL5 Hr. A) Stereo view of the experimental (SAD) electron density map of oxidized FBXL5 Hr diiron center. B) Geometry of the oxidized FBXL5 Hr diiron center showing the distances (in Å) between the iron atoms and their ligands. C) Stereo view of the electron density maps of the reduced FBXL5 Hr diiron center. The blue mesh shows the 2Fo − Fc map contoured at 1.1 σ and the red mesh shows the Fo − Fc map contoured at 3.5 σ. D) Geometry of the reduced FBXL5 Hr dirron center.
To understand why the FBXL5 becomes less stable and releases the iron readily under reducing conditions, we determined the 1.95 Å resolution structure of FBXL5 Hr using a crystal soaked in cryobuffer containing 2 mM Na2S2O4 (Supplementary Table 1). The structure revealed that one molecule of FBXL5 Hr in the asymmetric unit is reduced and another FBXL5 Hr molecule is partially reduced. Although the overall structures of the diiron center of oxidized and reduced FBXL5 Hr are similar (Supplementary Figure 3), reduction of FBXL5 Hr results in the removal of the μ-hydroxo bridge between the two iron atoms (Figure 3C and D). Both the 2Fo − Fc and Fo − Fc electron density maps (Figure 3C) clearly show that the hydroxo bridge is not present in the reduced form. Consistent with this observation, no hydroxo bridge is observed in another structure of reduced FBXL5 Hr refined at 2.45 Å resolution (Supplementary Table 1). The distance between the iron atoms is reduced from 3.21±0.01 Å in the oxidized form to 3.01±0.07 Å (the average of six sites in two structures) in reduced form (Figure 3D). His80, the ligand for Fe(1) in loop23, becomes poorly defined in the reduced form, indicating this ligand is not fully ordered. These structures provide insights into how FBXL5 binds iron reversibly under reducing conditions. The hydroxo bridge plays a critical role in regulating the iron binding. The removal of the hydroxo bridge seems require oxygen since the reduced protein is stable under anaerobic conditions (Figure 1C). Consistent with these observations, hemerythrin can be reconstituted in vitro from denatured apohemerythrin under reduced conditions with ferrous iron[17]. It is likely that FBXL5 exists in equilibrium between the oxidized and reduced forms in the cells. The oxygen concentration in the cytosol can influence the equilibrium between the two forms, allowing the cell to sense both iron and oxygen. The mechanism of how oxygen regulates iron binding by FBXL5 is not clear.
The overall length of the FBXL5 Hr helical bundle is about 54 Å, which is about 20 Å longer than the hemerythrin helical bundle (~ 35 Å). This large structural difference is due to the two major insertions between helices α1 and α2 (13 residues), and helices α3 and α4 (21 residues). Despite very low sequence homology (only 12% identity), the structure of the FBXL5 helical bundle is still similar to the hemerythrin helical bundle (rmsd of 1.65 Å for 47 structurally conserved Cα atoms, Supplementary Figure 5A)[15, 16]. The diiron centers are located at similar positions when the two structures are superimposed. The displacements between the two diiron centers in the two proteins is only about 1.2 Å. The overall structure of FBXL5 Hr is also similar to the bacterial hemerythrin like protein DcrH (rmsd of 1.96 Å of 61 structurally conserved Cα atoms, Supplementary Figure 5)[13]. Superposition of the structures FBXL5 Hr and oxy-hemerythrin indicates the oxygen binding pocket of FBXL5 Hr is restructured and is smaller than that of hemerythrin, explaining why FBXL5 does not bind oxygen. In addition, both hemerythrin and DcrH have an N-terminal extension that packs against helices α1 and α3 through hydrophobic interactions (Supplementary Figure 5). It was suggested that this extension might play a role in the oxygen sensing by DcrH[13]. In contrast, FBXL5 Hr does not have this N-terminal extension but has an extra helix at the C-terminus that interacts with helices α2 and α3 through hydrophobic interactions. Secondary structure prediction indicates residues 164 to 205 after this short helix form nearly continuous helical structures, so it is likely this helix will be extended in the full-length protein and form five-helical bundle in FBXL5. This helix may play a role in coupling the conformational change of FBXL5 induced by iron binding to its interactions with SKP1 or IRP2.
Based on results from our biochemical and structural studies of the FBXL5 Hr, we propose a mechanism of iron sensing by FBXL5. Depending on cytosolic iron and oxygen concentrations, FBXL5 exists in an equilibrium of the oxidized and reduced forms. Under high iron and high oxygen conditions, FBXL5 forms a stable complex with iron and exists mainly in the oxidized form that can associate with SKP1-CUL1 E3 ligase and mediates the ubiquitination of IRP2. In contrast, under low iron and low oxygen conditions, FBXL5 exists mainly in the reduce form and partially unfolded due to the desociation of the irons. The unfolded FBXL5 is ubiquitinated and degraded by the proteasome. Cytosolic oxygen plays a critical role in regulating the equilibrium between the oxidized and reduce states of FBXL5 and hence influcences the ability of FBXL5 to bind iron and SKP1-CUL1 E3 ligase.
Experimental Section
Experimental details of protein expression, purification, biochemical and biophysical characterization, and structural analysis are shown in supplementary materials online.
Supplementary Material
Acknowledgements
The diffraction data of the Se-Met crystal were collected at beamline 9.2 of the Stanford Synchrotron Radiation Lightsource (SSRL). The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health. This research is supported in part by grants from the National Institute of Health (Grant AI 087741) to P. L., and National Natural Science Foundation of China (No. 93013001) to X. T.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org.
Notes added in proof: While this manuscript was under review, the structure of FBXL5 hemerythrin like domain was published by the Bruick lab[18]. The structure is very similar to the oxidized FBXL5 Hr described in this work.
Accession codes: The atomic coordinates and structure factors of the oxidized and reduced FBXL5 Hr have been deposited with PDB under the accession codes 3U9J and 3U9M.
References
- 1.Muckenthaler MU, Galy B, Hentze MW. Annu Rev Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 2.Wallander ML, Leibold EA, Eisenstein RS. Biochim Biophys Acta. 2006;1763:668–689. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hentze MW, Muckenthaler MU, Andrews NC. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Pantopoulos K. Biochem J. 2011;434:365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rouault TA. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 6.Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, Spruck C, Leibold EA, Wohlschlegel JA. Science. 2009;326:718–721. doi: 10.1126/science.1176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, Bruick RK. Science. 2009;326:722–726. doi: 10.1126/science.1176326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moroishi T, Nishiyama M, Takeda Y, Iwai K, Nakayama KI. Cell Metab. 2011;14:339–351. doi: 10.1016/j.cmet.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Stenkamp RE. Chemical Reviews. 1994;94:715–726. [Google Scholar]
- 10.Xiong J, Kurtz DM, Jr, Ai J, Sanders-Loehr J. Biochemistry. 2000;39:5117–5125. doi: 10.1021/bi992796o. [DOI] [PubMed] [Google Scholar]
- 11.Duda DM, Scott DC, Calabrese MF, Zimmerman ES, Zheng N, Schulman BA. Curr Opin Struct Biol. 2011;21:257–264. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshaies RJ, Joazeiro CA. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 13.Isaza CE, Silaghi-Dumitrescu R, Iyer RB, Kurtz DM, Jr, Chan MK. Biochemistry. 2006;45:9023–9031. doi: 10.1021/bi0607812. [DOI] [PubMed] [Google Scholar]
- 14.Kurtz DM. Essays in Biochemistry, Vol 34, 1999. 1999;34:85–100. doi: 10.1042/bse0340085. [DOI] [PubMed] [Google Scholar]
- 15.Holmes MA, Le Trong I, Turley S, Sieker LC, Stenkamp RE. J Mol Biol. 1991;218:583–593. doi: 10.1016/0022-2836(91)90703-9. [DOI] [PubMed] [Google Scholar]
- 16.Holmes MA, Stenkamp RE. J Mol Biol. 1991;220:723–737. doi: 10.1016/0022-2836(91)90113-k. [DOI] [PubMed] [Google Scholar]
- 17.Zhang JH, Kurtz DM, Jr, Xia YM, Debrunner PG. Biochemistry. 1991;30:583–589. doi: 10.1021/bi00216a037. [DOI] [PubMed] [Google Scholar]
- 18.Thompson JW, Salahudeen AA, Chollangi S, Ruiz JC, Brautigam CA, Makris TM, Lipscomb JD, Tomchick DR, Bruick RK. J Biol Chem. 2012 doi: 10.1074/jbc.M111.308684. DOI:0.1074/jbc.M111.308684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


