SUMMARY
Mislocated enzymatic activity of DOT1L has been proposed as a driver of leukemogenesis in mixed lineage leukemia (MLL). The characterization of EPZ004777, a potent, selective inhibitor of DOT1L is reported. Treatment of MLL cells with the compound selectively inhibits H3K79 methylation and blocks expression of leukemogenic genes. Exposure of leukemic cells to EPZ004777 results in selective killing of those cells bearing the MLL gene translocation, with little effect on non-MLL-translocated cells. Finally, in vivo administration of EPZ004777 leads to extension of survival in a mouse MLL xenograft model. These results provide compelling support for DOT1L inhibition as a basis for targeted therapeutics against MLL.
INTRODUCTION
Mixed lineage leukemia (MLL) is a genetically distinct form of acute leukemia that constitutes over 70% of infant leukemias and approximately 10% of adult acute myeloid leukemias (AML) (Hess, 2004; Krivtsov and Armstrong, 2007). MLL represents a particularly aggressive form of leukemia. Patients with this disease generally have poor prognoses and often suffer from early relapse after treatment with current therapies. There is thus a great and present need for new treatment modalities for patients suffering with MLL.
A universal hallmark of MLL disease is a chromosomal translocation affecting the MLL gene on chromosome 11q23 (Hess, 2004; Krivtsov and Armstrong, 2007). Normally, the MLL gene encodes for a SET domain histone methyltransferase that catalyzes the methylation of lysine 4 of histone H3 (H3K4) at specific gene loci (Milne et al., 2002; Nakamura et al., 2002). Gene localization is conferred by specific interactions with recognition elements within MLL, external to the SET domain (Ayton et al., 2004; Slany et al., 1998; Zeleznik-Le et al., 1994). In the disease-linked translocations, the catalytic SET domain is lost and the remaining MLL protein is fused to a variety of partners, including members of the AF and ENL family of proteins such as AF4, AF9, AF10, and ENL (Hess, 2004; Krivtsov and Armstrong, 2007; Slany, 2009). These fusion partners are capable of interacting directly, or indirectly, with another histone methyltransferase, DOT1L (Bitoun et al., 2007; Mohan et al., 2010; Mueller et al., 2007, 2009; Okada et al., 2005; Park et al., 2010; Yokoyama et al., 2010; Zhang et al., 2006). As a result, translocation products retain gene-specific recognition elements within the remainder of the MLL protein, but also gain the ability to recruit DOT1L to these locations (Monroe et al., 2010; Mueller et al., 2007, 2009; Okada et al., 2005).
DOT1L catalyzes the methylation of H3K79, a chromatin modification associated with actively transcribed genes (Feng et al., 2002; Steger et al., 2008). The ectopic H3K79 methylation that results from MLL fusion protein recruitment of DOT1L leads to enhanced expression of leukemogenic genes, including HOXA9 and MEIS1 (Guenther et al., 2008; Krivtsov et al., 2008; Milne et al., 2005; Monroe et al., 2010; Mueller et al., 2009; Nguyen et al., 2011; Okada et al., 2005; Thiel et al., 2010). Hence, while DOT1L is not genetically altered in the disease per se, its mislocated enzymatic activity is a direct consequence of the chromosomal translocation affecting MLL patients. Studies in model systems have demonstrated that DOT1L is required for the transforming activity of MLL fusion proteins and DOT1L has therefore been proposed to be a catalytic driver of leukemogenesis in this disease (Chang et al., 2010; Jo et al., 2011; Krivtsov et al., 2008; Monroe et al., 2010; Mueller et al., 2007; Nguyen et al., 2011; Okada et al., 2005; Yokoyama et al., 2010).
As briefly summarized above, there is evidence to suggest that the enzymatic activity of DOT1L is critical to pathogenesis in MLL. Therefore, it has been proposed that inhibition of DOT1L may provide a pharmacologic basis for therapeutic intervention in this disease. However, in the absence of selective DOT1L inhibitors, it has not been possible to address this hypothesis directly. Toward this ultimate goal, we report here the development of a small-molecule inhibitor of DOT1L.
RESULTS
Identification and Biochemical Characterization of EPZ004777, a Potent and Selective Inhibitor of DOT1L
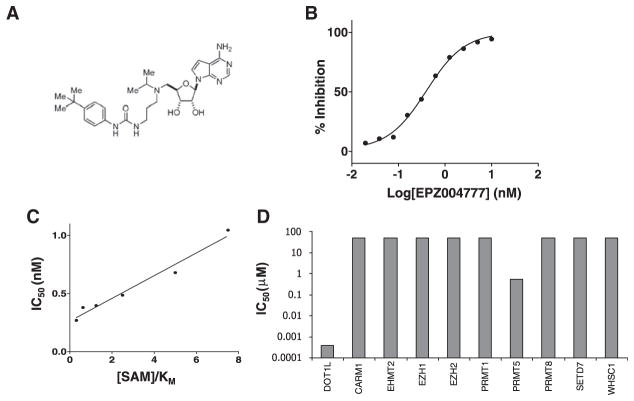
Based on the chemical structures of the S-adenosylmethionine (SAM) substrate and S-adenosylhomocysteine (SAH) product, the reaction mechanism of DOT1L catalysis and the published crystal structure of the DOT1L active site, medicinal chemistry design tenets were established to facilitate mechanism-guided inhibitor discovery; chemical analogs thus designed were synthesized and tested as inhibitors of DOT1L enzymatic activity. From these efforts, EPZ004777 (Figure 1A) was identified. This compound demonstrates potent, concentration-dependent inhibition of DOT1L enzyme activity with an IC50 of 400 ± 100 pM, as illustrated in Figure 1B. The chemical structure of this compound retains the nucleoside core of the SAM substrate, and SAH product. As such it was designed to bind to the enzyme within the SAM binding pocket. Steady-state kinetic analysis confirms that the compound binds to the enzyme competitively with SAM. For example, a distinguishing feature of competitive inhibition is a linear increase in the apparent IC50 of the compound as a function of substrate concentration; EPZ004777 displays this pattern when assayed as a function of SAM concentration relative to the KM of SAM (Figure 1C). SAM is a common methyl group donator that is used by all histone methyltransferases (HMTs). Despite the universality of SAM utilization by HMTs, EPZ004777 displays remarkable selectivity for inhibition of DOT1L over other HMTs, as summarized in Table 1 and Figure 1D. Thus, the compound displays a minimum selectivity of >1000-fold for DOT1L relative to all HMTs that have been tested.
Figure 1. Biochemical Characterization of EPZ004777 Inhibition of DOT1L.
(A) Chemical structure of EPZ004777.
(B) Inhibition of recombinant human DOT1L by EPZ004777. Each data point represents the mean of two replicates at each specified concentration of compound. The data are fit to a standard Langmuir isotherm equation for inhibition (Copeland, 2005).
(C) Plot of IC50 values of EPZ004777 as a function of SAM concentration relative to the Km of S-adenosylmethionine (SAM) ([SAM]/Km). These values display a linear relationship as expected for SAM-competitive inhibition with a Ki of 0.3 ± 0.03 nM (± error represents standard error of the mean [SEM]).
(D) Plot of IC50 values for EPZ004777 against a panel of human HMT enzymes.
Table 1.
Inhibition of Human Histone Methyltransferases by EPZ004777
| Enzyme | IC50 (nM)a | Fold Selectivityb |
|---|---|---|
| DOT1L | 0.4 ± 0.1 | 1.0 |
| CARM1 | >50,000 | >100,000 |
| EHMT2 | >50,000 | >100,000 |
| EZH1c | >50,000 | >100,000 |
| EZH2c | >50,000 | >100,000 |
| PRMT1 | >50,000 | >100,000 |
| PRMT5 | 521 ± 137 | 1280 |
| PRMT8 | >50,000 | >100,000 |
| SETD7 | >50,000 | >100,000 |
| WHSC1 | >50,000 | >100,000 |
IC50 is the concentration of compound resulting in 50% inhibition of enzyme activity. ± error represents the 95% confidence interval.
Fold selectivity is relative to DOT1L, and is calculated as the ratio of the IC50 for the enzyme under study over the IC50 for DOT1L.
Measured in the context of the PRC2 multiprotein complex.
EPZ004777 Selectively Inhibits Cellular H3K79 Methylation
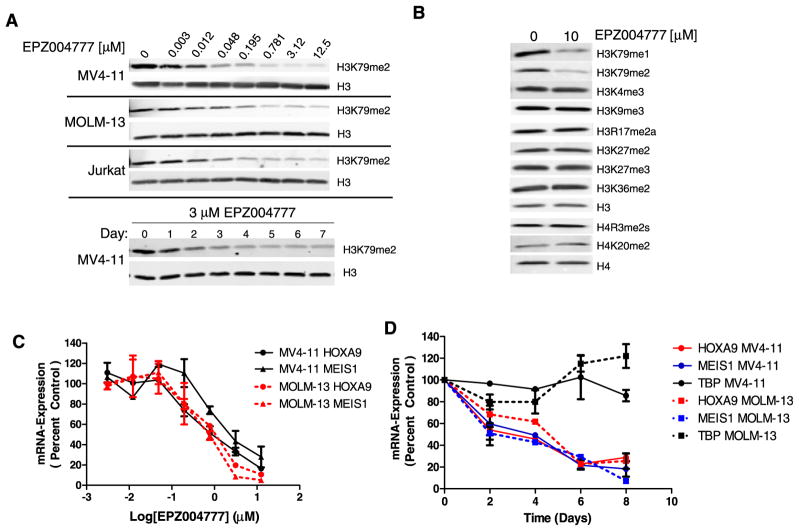
Having established that EPZ004777 is a potent and highly selective DOT1L inhibitor in biochemical assays, we next tested the ability of EPZ004777 to inhibit DOT1L in cells by immunoblot analysis of extracted histones using an antibody specific for dimethylated H3K79 (H3K79me2). Treatment of human cell lines derived from MLL-rearranged acute myeloid leukemia (AML) (MOLM-13, MLL-AF9), MLL-rearranged biphenotypic leukemia (MV4-11, MLL-AF4), or non-MLL-rearranged T cell acute leukemia (Jurkat) with EPZ004777 led to a concentration-dependent reduction in global H3K79me2 levels (Figure 2A, upper panel). To understand the kinetics of EPZ004777-mediated cellular H3K79me2 depletion, we performed a time course analysis in MV4-11 cells incubated with 3 μM EPZ004777, a concentration sufficient for maximal cellular DOT1L inhibition (Figure 2A, lower panel). A modest reduction in H3K79me2 levels was apparent within 1 day of treatment, but full depletion took 4–5 days. There is no known histone demethylase enzyme specific for H3K79, so the decline in methylation at this residue following DOT1L inhibition is presumably due to incorporation into chromatin of unmethylated H3 through histone turnover and replacement.
Figure 2. EPZ004777 Inhibits Cellular H3K79 Methylation and MLL Fusion Target Gene Expression.
(A) Upper panel: immunoblot analysis of H3K79me2 levels in MV4-11 (MLL-AF4), MOLM-13 (MLL-AF9), and Jurkat human leukemia cell lines following 4 days of treatment with EPZ004777 at indicated concentrations. Lower panel: time course of depletion of cellular H3K79me2 levels in MV4-11 cells treated with 3 μM EPZ004777.
(B) Immunoblot analysis of histones extracted from MV4-11 cells treated with 10 μM EPZ004777 with a panel of methyl-lysine and methyl-arginine residue specific antibodies.
(C) Quantitative real-time PCR analysis of HOXA9 and MEIS1 mRNA levels in MV4-11 and MOLM-13 cells following 6 day incubation with EPZ004777. Relative mRNA expression levels are plotted as a percentage of those in vehicle-treated control cells.
(D) Time course of HOXA9, MEIS1 and TBP mRNA expression in MV4-11 and MOLM-13 cells over 8 days of incubation with 3 μM EPZ004777 as measured by quantitative real-time PCR. Relative mRNA expression levels are plotted as a percentage of those at day 0. Error bars: standard deviation (SD).
We saw a persistent residual western blot signal with the anti-H3K79me2 antibody, even at the highest EPZ004777 concentrations, and with extended compound incubation periods (Figures 2A and 2B; data not shown). One possibility is that the residual signal is background caused by a low level of cross-reactivity of the antibody with other methylated residues on histone H3.
Histone H3K79me1 levels were also evaluated and found to be reduced by EPZ004777 treatment (Figure 2B, top panel). We were unable to evaluate H3K79me3 levels because commercially available anti-H3K79me3 antibodies demonstrated strong cross-reactivity with unrelated methyl marks (data not shown).
To assess the specificity of EPZ004777 inhibitory activity in cells, we immunoblotted histones extracted from EPZ004777-treated MV4-11 cells with a panel of methyl-lysine and methyl-arginine residue specific antibodies. As shown in Figure 2B, the only methyl marks affected by EPZ004777 treatment were H3K79me1 and H3K79me2. Importantly, we saw no changes in H4R3me2, a residue targeted by PRMT5, the only HMT, other than DOT1L, for which EPZ004777 gave a biochemical IC50 of less than 50 μM. Furthermore, EPZ004777 treatment did not change the methylation state of residues targeted by PRDM and SMYD family members (H3K9 and H3K4, respectively). This is significant because we were unable to assess EPZ004777’s ability to inhibit these HMTs directly due a lack of robust biochemical assays. These data are consistent with EPZ004777 being a highly specific DOT1L inhibitor in a cellular context.
EPZ004777 Blocks MLL Fusion Target Gene Expression
We next tested whether EPZ004777 was able to inhibit expression of key MLL fusion target genes. HOXA9 and MEIS1 overexpression is a hallmark of MLL-rearranged leukemias (Armstrong et al., 2002; Ferrando et al., 2003; Ross et al., 2003, 2004; Rozovskaia et al., 2001; Yeoh et al., 2002). Furthermore, both genes are bound by MLL fusion proteins, hypermethylated at H3K79 and downregulated by DOT1L depletion in MLL fusion-expressing cells, including the MV4-11 cell line (Chang et al., 2010; Guenther et al., 2008; Jo et al., 2011; Krivtsov et al., 2008; Lin et al., 2010; Milne et al., 2005; Monroe et al., 2010; Mueller et al., 2009; Nguyen et al., 2011; Okada et al., 2005; Thiel et al., 2010; Yokoyama et al., 2010). We used quantitative real-time PCR to examine the effect of EPZ004777 on HOXA9 and MEIS1 transcript levels in MOLM-13 and MV4-11 cells (expression of these transcripts in non-MLL-rearranged Jurkat cells was below the limit of real-time PCR quantitation). Treatment with EPZ004777 led to a concentration-dependent decrease of both transcripts in each cell line with IC50s of approximately 700 nM (Figure 2C). We evaluated the kinetics of this decrease by measuring HOXA9 and MEIS1 mRNA levels over time in cells treated with 3 μM EPZ004777 (Figure 2D). Levels of both transcripts were significantly decreased within 48 hr of compound addition, and were maximally reduced after 6–8 days of EPZ004777 treatment (fitting of these data yielded estimated half-lives of 2.3 and 3.3 days for HOXA9 and MEIS1 inhibition, respectively). This decrease was not due to a general inhibitory effect on gene expression since transcript levels of the housekeeping gene TBP were unaffected (Figure 2D).
EPZ004777 Selectively Inhibits Proliferation of MLL-Rearranged Cells
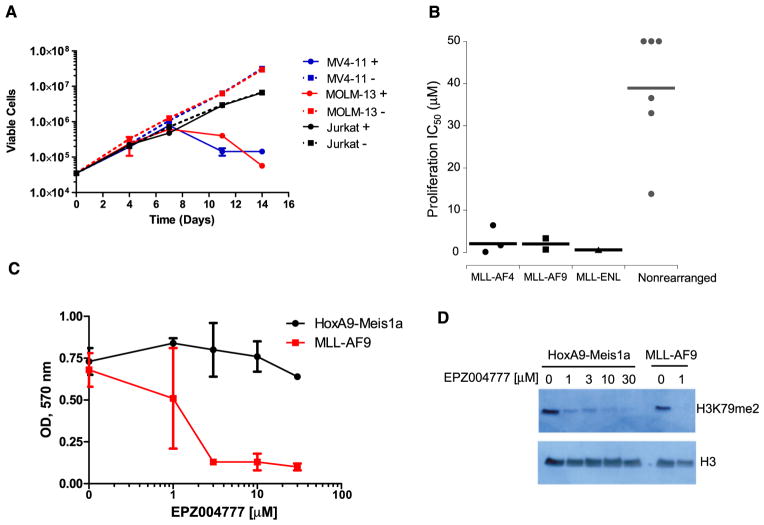
Having established that EPZ004777 can inhibit H3K79 methylation and block MLL fusion target gene expression, we investigated whether this translated into antiproliferative activity in MLL-rearranged leukemic cells. We performed proliferation assays over several days with MV4-11 and MOLM-13 cells in the presence or absence of 3 μM EPZ004777. Jurkat cells were included as a non-MLL-rearranged cell line control. As shown in Figure 3A, the effect of extended EPZ004777 treatment was remarkably specific for the MLL-rearranged cell lines. The number of viable MV4-11 and MOLM-13 cells was dramatically reduced by EPZ004777, whereas the growth of Jurkat cells was unaffected. A small population of MV4-11 cells remained viable in the presence of EPZ004777, but their number remained constant when growth curves were tracked over longer periods indicating that they had ceased to divide (see Figure S1 available online). The lack of effect on Jurkat cells was not due to differences in the ability of EPZ004777 to inhibit DOT1L in these cells as measured by immunoblot for cellular H3K79me2 levels (Figure 2A, upper panel). This analysis also revealed a significant delay before the antiproliferative effects of EPZ004777 became apparent; both MLL-rearranged cell lines continued to proliferate at a normal rate for several days after exposure to the inhibitor. This may reflect the time required to reverse fully the aberrant expression of MLL fusion target genes following DOT1L inhibition, a process that presumably involves depletion of methylated H3K79, followed by decreased mRNA expression and reduced levels of gene products critical for leukemogenic growth. To expand our analysis of the differential sensitivity of MLL-rearranged cell lines to EPZ004777, we determined IC50 values for inhibition of proliferation in a panel of six MLL-rearranged and six nonrearranged human leukemia cell lines. The MLL-rearranged panel (Table 2) included human cell lines derived from ALL, AML and biphenotypic leukemias harboring MLL-AF4, MLL-AF9, or MLL-ENL fusions. As shown in Figure 3B, and Table 2, IC50 values for MLL-rearranged cell lines were in the nanomolar to low micromolar range, whereas IC50s for non-MLL-rearranged cell lines were always above 10 μM or undetermined due to lack of inhibition at the highest concentration tested (reported as IC50 > 50 μM in Table 2 and represented as 50 μM for illustrative purposes in Figure 3B). Concentrations above 50 μM were not tested because they are above the solubility limit for EPZ004777, and because efficient loss of H3K79me2 was achieved at concentrations below 1 μM (Figure 2A). We next determined whether these results would extend to primary murine hematopoietic progenitors transformed by retroviral expression of an MLL-AF9 fusion protein. As shown in Figure 3C, the proliferation of MLL-AF9-transformed cells was strongly inhibited by EPZ004777 at concentrations of 3 μM or greater. In contrast, the survival of hematopoietic progenitors transformed by coexpression of HoxA9 and Meis1a was unaffected by EPZ004777 at concentrations of up to 30 μM. Immunoblot analysis of histones using an anti-H3K79me2 antibody confirmed that EPZ004777 inhibited H3K79 methylation in both transformed cell populations (Figure 3D).
Figure 3. EPZ004777 Selectively Inhibits Proliferation of MLL-Rearranged Cell Lines and MLL-AF9-Transformed Murine Hematopoietic Cells.
(A) Growth of MV4-11 (MLL-AF4), MOLM-13 (MLL-AF9), and Jurkat (non-MLL-rearranged) cells during several days incubation with 3 μM EPZ004777. Viable cells were counted every 3 to 4 days in the presence of EPZ004777 (+) or DMSO vehicle control (−) and results plotted on a logarithmic scale. See also Figure S1. Error bars represent standard deviation.
(B) Effect of EPZ004777 on the proliferation of leukemia cell lines bearing MLL-AF4, MLL-AF9, and MLL-ENL fusions, or cell lines lacking an MLL rearrangement. Cell lines were maintained in the presence of increasing concentrations of EPZ004777 up to 50 μM. Viable cells counts were used to derive IC50 values after 14 days of treatment for all cell lines apart from THP-1 cells, which were treated for 18 days. Fifty percent inhibition of growth was not achieved in HL60, Jurkat and U937 cells even at the highest EPZ004777 concentration and so IC50 values are given as >50 μM.
(C) Effect of EPZ004777 on the proliferation of primary murine hematopoietic progenitors transformed by retroviral expression of MLL-AF9, or coexpression of HoxA9 and Meis1a. Retrovirally transformed cells were maintained in the presence of increasing concentrations of EPZ004777 up to 30 μM for 10 days, then plated in 96-well plates for MTT assay at day 12. Following the MTT assay, plate ODs were read at 570 nM and plotted on the y axis. Results from two independent transductions are shown for each retroviral construct. Error bars represent standard deviation.
(D) Inhibition of cellular H3K79me2 levels in MLL-AF9 or Hoxa9-Meis1a transformed hematopoietic progenitors following 10 days of treatment with the indicated concentrations of EPZ004777 as measured by immunoblot analysis of extracted histones with an anti-H3K79me2 antibody.
Table 2.
Concentration-Dependent Inhibition of Cell Proliferation by EPZ004777 in Various Cell Types
| MLL Gene Fusion | Cell Type | IC50 (μM)a | 95% Confidence Interval for IC50 (μM) |
|---|---|---|---|
| MLL-AF4 | RS4;11 | 6.47 | 4.22–9.94 |
| MLL-AF4 | SEM | 1.72 | 1.21–2.46 |
| MLL-AF4 | MV4-11 | 0.17 | 0.15–.19 |
| MLL-AF9 | THP-1 | 3.36 | 2.51–3.80 |
| MLL-AF9 | MOLM-13 | 0.72 | 0.65–0.80 |
| MLL-ENL | KOPN-8 | 0.62 | 0.49–0.79 |
| Nonrearranged | REH | 13.90 | 8.25–23.46 |
| Nonrearranged | Kasumi-1 | 32.99 | 30.03–36.24 |
| Nonrearranged | 697 | 36.57 | 34.77–38.46 |
| Nonrearranged | HL-60 | >50 | NA |
| Nonrearranged | Jurkat | >50 | NA |
| Nonrearranged | U937 | >50 | NA |
IC50 is the concentration of compound that results in a half-maximal degree of inhibition.
These results demonstrate that DOT1L methyltransferase activity is required for proliferation of MLL-rearranged cells and MLL fusion-mediated transformation, but is not essential for proliferation and viability of non-MLL-rearranged cells in culture.
EPZ004777 Causes Differentiation and Apoptosis in MLL-Rearranged Cells
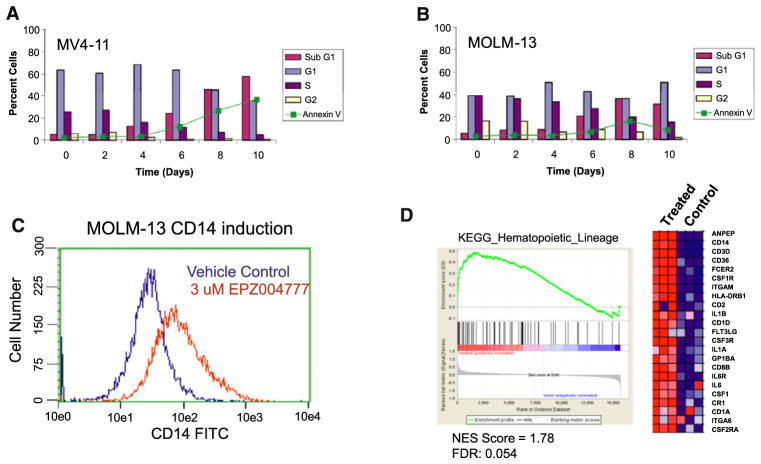
To explore the mechanism of cell killing in more detail, we determined effects of EPZ004777 on the cell cycle and apoptosis in MV4-11 and MOLM-13 cells by flow cytometry for DNA content and Annexin V staining. The results are represented graphically in Figures 4A and 4B. In MV4-11 cells, a modest increase in G0/G1 phase, and a decrease in S-phase cells were apparent after 4 days of incubation with 3 μM EPZ004777. This was followed by an increase in sub-G1 and Annexin-positive cells over the next 6 days, consistent with apoptotic cell death (Figure 4A). Compound treatment also led to caspase activation (Figure S2) providing further evidence for an apoptotic component to EPZ004777-mediated cell killing. Similar results were obtained in MOLM-13 cells, although the percentage of Annexin-positive cells was significantly lower (Figure 4B). We next analyzed whether EPZ004777 induced differentiation prior to cell death. MOLM-13 cells were treated with 3 μM EPZ004777 and monitored for cell surface expression of the myeloid differentiation marker CD14 by flow cytometry. As shown in Figure 4C, expression of CD14 was induced following 12 days of EPZ004777 treatment. Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) of genes upregulated by EPZ004777 treatment of MOLM-13 cells (see below) also demonstrated significant enrichment for hematopoietic cell lineage markers, including CD14, (normalized enrichment score [NES] = 1.78, false discovery rate [FDR] = 0.054) (Figure 4D). This provides further evidence that small-molecule inhibition of DOT1L promotes some degree of differentiation prior to cell killing.
Figure 4. EPZ004777 Causes Apoptosis and Differentiation in MLL-Rearranged Cell Lines.
(A and B) MV4-11 (A) and MOLM-13 (B) cells were treated with 3 μM EPZ004777 for up to 10 days and analyzed by flow cytometry every 2 days for DNA content, and Annexin V staining. See also Figure S2.
(C) MOLM-13 cells were incubated in the presence of 3 μM EPZ004777 for 12 days and analyzed by flow cytometry for cell surface expression of CD14. Similar results were obtained in two independent experiments.
(D) Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) of genes upregulated by EPZ004777 treatment of MOLM-13 cells as compared with members of the KEGG hematopoietic cell lineage gene set. The heat map shows genes comprising the leading edge of the GSEA plot. Red indicates high expression; blue indicates low expression.
EPZ004777 Reverses the MLL-Rearranged Gene Signature
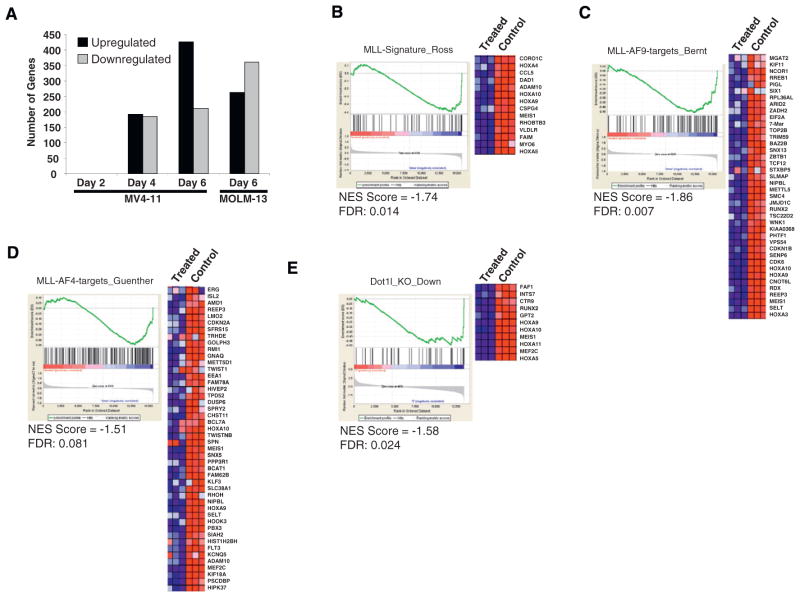
To determine effects of EPZ004777 treatment on gene expression in MLL-rearranged leukemia cell lines, RNA was isolated from MV4-11 cells and MOLM-13 cells treated with 3 μM EPZ004777 for up to 6 days, amplified, and hybridized to Affymetrix microarrays. Statistically significant changes in gene expression (probes with statistically significant changes (q < 0.15) and up or downregulated at least 2-fold; see Experimental Procedures) were not observed until 4 days after inhibitor treatment, consistent with the relatively delayed effects of EPZ004777 on H3K79 methylation and proliferation (Figure 5A). Among the genes downregulated in MV4-11 and MOLM-13 cells following 6 days of EPZ004777 treatment are several that have been previously implicated in MLL fusion-mediated leukemogenesis including multiple HOXA genes, MEIS1 and MEF2C (Figures 5B–5E). GSEA of genes downregulated following 6 day EPZ004777 treatment of MOLM-13 cells demonstrated strong enrichment (NES = −1.74, FDR = 0.014) for genes overexpressed in MLL-rearranged human acute leukemias as compared with MLL-germline acute leukemias (Ross et al., 2004) (Figure 5B). This indicates that small-molecule inhibition of DOT1L is able to reverse the MLL-rearranged gene expression signature in MLL-rearranged cell lines. We next used GSEA to compare genes downregulated following 6 day EPZ004777 treatment of MOLM-13 (MLL-AF9) or MV4-11 (MLL-AF4) cells with genes identified as direct targets of MLL-AF9 (see Experimental Procedures and paper by Bernt et al., 2011 [this issue of Cancer Cell]), or MLL-AF4 (Guenther et al., 2008) through genome-wide chromatin immunoprecipitation coupled with large scale sequencing (ChIP-seq). Genes downregulated by EPZ004777 in MOLM-13 cells were significantly enriched for direct MLL-AF9 targets (NES = −1.86, FDR: 0.007, Figure 5C), whereas genes downregulated by EPZ004777 in MV4-11 cells were enriched for direct MLL-AF4 targets (NES = −1.51, FDR: 0.081, Figure 5D). Both results indicate that small-molecule inhibition of DOT1L decreases the expression of direct MLL fusion targets. Finally, we compared gene expression changes caused by EPZ004777 treatment of MOLM-13 (MLL-AF9) cells with those caused by genetic knockout of Dot1l in a mouse model of MLL-AF9 leukemia (see Experimental Procedures and paper by Bernt et al., 2011 [this issue of Cancer Cell]). We found significant overlap between these gene expression changes (NES = −1.58, FDR: 0.024, Figure 5E) indicating that EPZ004777 treatment and genetic ablation of Dot1l cause cell killing of MLL-rearranged cells through similar pathways.
Figure 5. Effect of EPZ004777 Treatment on MLL-Rearranged Gene Expression.
(A) Gene expression changes following treatment of MV4-11 cells with EPZ004777 for 2, 4, or 6 days and MOLM-13 cells for 6 days. Two-group comparison expression data were collapsed to unique gene symbols. Changes in gene expression were filtered by log2 change > 1 or < −1 and by false discovery rate q values < 0.15.
(B) GSEA of genes downregulated by EPZ004777 treatment of MOLM-13 cells as compared with genes overexpressed in MLL-rearranged human acute leukemias (Ross et al., 2004). The heat map shows genes comprising the leading edge of the GSEA plot. Red indicates high expression; blue indicates low expression.
(C) GSEA of genes downregulated by EPZ004777 treatment of MOLM-13 cells as compared with genes bound by MLL-AF9 in MLL-AF9-transformed murine hematopoietic progenitors (see Experimental Procedures and Bernt et al., 2011 [this issue of Cancer Cell]). The heat map shows genes comprising the leading edge of the GSEA plot.
(D) GSEA of genes downregulated by EPZ004777 treatment of MV4-11 cells as compared with genes bound by MLL-AF4 in human SEM cells (Guenther et al., 2008). The heat map shows genes comprising the leading edge of the GSEA plot.
(E) GSEA of genes downregulated by EPZ004777 treatment of MOLM-13 cells as compared with genes downregulated following genetic knockout of Dot1l in a murine MLL-AF9 leukemia model (see Experimental Procedures and Bernt et al., 2011 [this issue of Cancer Cell]). The heat map shows genes comprising the leading edge of the GSEA plot.
In Vivo Efficacy of EPZ004777 in a Mouse Xenograft Model of MLL
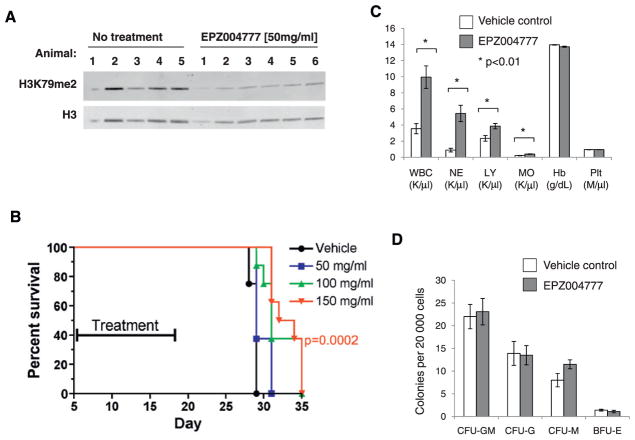
We next tested the efficacy of EPZ004777 in a mouse xenograft model of MLL. As a first step, we determined whether EPZ004777 was capable of inhibiting DOT1L activity in vivo. We chose to do this in a mouse MV4-11 subcutaneous xenograft model, since tumor tissue can be readily harvested and assessed for changes in H3K79 methylation levels. The poor pharmacokinetic properties of EPZ004777 precluded conventional dosing methods. Instead, compound was delivered via subcutaneously implanted mini-osmotic pumps capable of continuous infusion for a period of 7 days. Six female nude mice bearing MV4-11 xenograft tumors of sizes ranging from 300 to 400 mm3 were implanted with pumps loaded with a 50 mg/ml solution of EPZ004777. A control group of five tumor bearing mice did not receive pumps. After 6 days, all mice were sacrificed and tumors were harvested. Immunoblot analysis of histones extracted from MV4-11 tumors revealed that H3K79me2 levels were significantly decreased in tumors from mice treated with EPZ004777 relative to untreated controls (Figure 6A). Analysis of plasma samples taken during this experiment revealed that treated mice were exposed to average circulating levels of 0.55 ± 0.12 μM EPZ004777.
Figure 6. In Vivo Activity of EPZ004777.
(A) H3K79me2 levels were evaluated by immunoblot in subcutaneous MV4-11 tumors derived from untreated animals or animals implanted with pumps containing a 50 mg/ml solution of EPZ004777 for a period of 6 days.
(B) EPZ004777 extends survival of NSG mice after intravenous injection of MV4-11 cells. MV4-11 cells were injected into the tail vein of NSG mice, and animals were implanted with pumps containing vehicle or 50, 100, or 150 mg/ml EPZ004777. Pumps were exchanged once to give a total of 14 days of exposure as indicated. See also Figure S3A.
(C) Complete blood count analysis in C57BL/6 mice treated for 14 days with EPZ004777 or vehicle control. WBC, white blood cell count; NE, neutrophils; LY, lymphocytes; MO, monocytes; Hb, Hemoglobin; Plt, platelets; K/μl, thousands per microliter; M/μl, millions per microliter. N = 5 mice per group, *p < 0.01, all others not significant (p > 0.05).
(D) Effect of 14 days of EPZ004777 treatment on colony formation of hematopoietic progenitors. Five mice per group, all differences not significant (p > 0.05). Error bars represent standard error of the mean. See also Figures S3B–S3D.
Having demonstrated that EPZ004777 is able to inhibit DOT1L in vivo, we next tested the efficacy of EPZ004777 in a more therapeutically relevant model of MLL in which MV4-11 cells are injected into the tail vein of immunodeficient mice resulting in disseminated leukemic disease. Female NSG mice were injected via tail vein with 1 × 106 MV4-11 cells stably expressing the firefly luciferase gene. Leukemia engraftment was confirmed by bioluminescence imaging 5 days after inoculation, at which point animals were divided into treatment and control groups (n = 8 per group). Optimization of EPZ004777 formulation enabled us to increase solubility to 150 mg/ml. Mice were therefore implanted with pumps loaded with vehicle alone, or containing EPZ004777 at concentrations of 50, 100, or 150 mg/ml. Pilot experiments indicated that mini-pumps containing 100 and 150 mg/ml EPZ004777 solutions could maintain average steady-state plasma concentrations of 0.64 ± 0.48 and 0.84 ± 0.45 μM, respectively. Osmotic pumps were replaced once to achieve a total of 14 days of in vivo compound exposure (from day 5 to day 19 postinoculum, see Figure 6B). Local irritation at the pump implant site in the 100 and 150 mg/ml groups precluded a second pump replacement (see Experimental Procedures). Despite the limited 14 day duration of EPZ004777 exposure achieved, we observed a dose-dependent and statistically significant increase in median survival (p = 0.0002, 0.0007 and 0.0285 for the 150, 100, and 50 mg/ml dose groups, respectively, log-rank test, Figure 6B). Histopathological analysis of similarly xenografted mice confirmed that animals were dying of leukemic disease (data not shown). EPZ004777 administration was well tolerated, and no significant weight loss was observed (Figure S3). Together, these results demonstrate that EPZ004777 has both pharmacodynamic and antitumor efficacy in a mouse xenograft model of MLL leukemia.
Efficacious Doses of EPZ004777 Are Well Tolerated in Normal Mice
To further explore whether a therapeutic window for small-molecule DOT1L inhibition exists, we treated normal C57BL/6 mice with mini-pumps loaded with vehicle or 150 mg/ml EPZ004777 for 14 days and monitored for signs of toxicity. We focused on effects on the hematopoietic system, in particular, because DOT1L has been shown to play a role in normal hematopoietic development and homeostasis. As in the previous experiments, EPZ004777 was well tolerated and no overt toxicity was observed. Complete blood count analysis after 14 days of continuous exposure to EPZ004777 revealed a statistically significant increase in the total white blood cell count, which resulted from an increase in neutrophils, monocytes, and lymphocytes (Figure 6C). The etiology of this effect is not clear. We did not find any significant differences in the colony forming potential of bone marrow progenitors after 14 days of in vivo exposure (Figure 6D), and bone marrow cellularity was preserved (Figure S4A). Further analysis of more primitive hematopoietic compartments revealed a statistically significant decrease in committed progenitors (Lin−Sca−1−c-Kit+), that was most apparent in the common myeloid progenitors (CMP, Il-7R− Lin−Sca−1−c-Kit+CD34+FcγRII/IIIlo) and megakaryocyte/erythroid progenitors (MEP, Il-7R−Lin−Sca−1−c-Kit+CD34−FcγRII/III−). Granulocyte/monocyte progenitors (GMP, Il-7R−Lin−Sca−1−c-Kit+CD34+FcγRII/III+) were not significantly affected (Figures S4B and S4C). Similarly, flow cytometric analysis of populations that are highly enriched for hematopoietic stem cells (Lin−Sca−1+c-Kit+ = LSK; long-term repopulating cells defined as Lin−Sca-1+c−Kit+CD34−flk2−; and Lin−Sca-1+c-Kit+CD34−flk2−CD48−CD150+) did not reveal any difference between mice treated with EPZ004777 and control mice (Figures S4B and S4C). These results demonstrate that it is possible to achieve a therapeutically meaningful level of small-molecule DOT1L inhibition, without overt toxicity or severe hematopoietic side effects.
DISCUSSION
The small-molecule inhibitor EPZ004777 provides a powerful tool for understanding the biological role of DOT1L enzymatic activity. As we demonstrate here, this compound is an extremely potent and selective inhibitor of its target enzyme, thus providing sufficient pharmacologic specificity with which to probe the cellular consequences of DOT1L inhibition in MLL-transformed and nontransformed cells. We find that inhibition of DOT1L by EPZ004777 results in robust and selective ablation of cellular H3K79 methylation, which, in turn, leads to diminished transcription of key genes associated with leukemogenesis in MLL disease, such as HOXA9 and MEIS1. Strikingly, the inhibition of DOT1L-catalyzed H3K79 methylation is cytotoxic to leukemic cells bearing the MLL translocation, but has little effect on proliferation of nontranslocated cells. A particularly powerful demonstration of this selectivity is the finding that hematopoietic progenitors transformed by MLL-AF9 expression are sensitive to EPZ004777 treatment, whereas those transformed by HoxA9 and Meis1, a combination that presumably bypasses the requirement for DOT1L activity, are not.
Killing of MLL-translocated cells by EPZ004777 involves an apoptotic component and is preceded by phenotypic and gene expression changes that suggest attempted hematopoietic differentiation. Gene expression analysis of inhibitor treated MLL-rearranged cells demonstrates a reversal of an MLL-rearranged gene signature, previously derived through transcriptomic analysis of primary human acute leukemia samples (Ross et al., 2004). This includes downregulation of genes known to be critical for MLL fusion-mediated leukemogenesis such as members of the HOXA gene cluster and MEIS1 (Ayton and Cleary, 2003; Faber et al., 2009; Kumar et al., 2009; Wong et al., 2007; Zeisig et al., 2004). These data are concordant with results seen for DOT1L message ablation using genetic knockdown methodologies, a manipulation that inhibits MLL fusion-driven tumor progression in vitro and in vivo as detailed in the paper by Bernt et al. (2011) (this issue of Cancer Cell). Indeed, our gene profiling results demonstrate overlapping transcriptomic impact of both methods of DOT1L inhibition.
In vivo administration of EPZ004777 results in an extension of survival in a mouse xenograft model of MLL. Although this extension in survival is significant, its duration is relatively modest. This may reflect the limited length of exposure achieved in this experiment, suboptimal circulating levels of EPZ004777, or a combination of both factors. For example, while the circulating levels of EPZ004777 are well above IC50 values for inhibition of proliferation of cultured MV4-11 cells, factors such as plasma protein binding may limit the effective free fraction of EPZ004777 available for DOT1L inhibition in tumor cells. In normal mice, therapeutically active doses of EPZ004777 do not give rise to overt toxicity. Careful hematologic analysis reveals a mild to moderate decrease in myeloid progenitor compartments. This did not translate into a significant decrease in peripheral blood counts over the 2 week dosing period, but raises the possibility that extended courses of treatment with a DOT1L inhibitor could affect normal hematopoiesis. Overall, however, the absence of severe hematopoietic side effects with EPZ004777 is encouraging.
The present paper and the paper by Bernt et al. (2011) (this issue of Cancer Cell) demonstrate clearly that ablation of DOT1L enzymatic activity, either by genetic knockdown of the entire protein or blockade of the catalytic active site, selectively kills cells bearing the MLL translocation in culture and has anti-tumor activity in in vivo models of MLL. The effect of DOT1L inhibition by EPZ004777 in cell culture and in a mouse xenograft model of disease provides strong support for the use of DOT1L selective inhibitors as a molecular basis for targeted therapeutics against MLL-rearranged leukemias. Furthermore, the selectivity for MLL-rearranged cells in culture, and our ability to demonstrate efficacy in a mouse model of MLL at doses that were well tolerated suggests that a therapeutic strategy, based on inhibition of DOT1L, may demonstrate an exploitable therapeutic index.
In summary, we have designed and characterized a potent, selective DOT1L inhibitor as a starting point toward the development of personalized medicines for the treatment of patients suffering from MLL. The poor pharmacokinetic properties of EPZ004777 preclude its clinical development; however, we are currently developing related inhibitors with improved potency and drug-like properties for this purpose. The data presented here demonstrate the utility of DOT1L inhibition as an approach to targeted therapy for this disease. In a broader context, the current results also provide validation for small-molecule inhibition of HMTs as a therapeutic modality in cancer and other diseases where genetic alterations to these epigenetic enzymes drive pathogenesis. Continued research into pharmacologically tractable mechanisms of HMT inhibition should yield novel medicines for patients suffering from such diseases.
EXPERIMENTAL PROCEDURES
EPZ004777
EPZ004777 was synthesized by Epizyme. Stock solutions (50 or 10 mM) were prepared in DMSO and stored at −20°C. Serial dilutions of stock solutions were carried out just prior to use in each experiment and final DMSO concentrations were kept at, or below 0.2%. For experiments involving RS4;11 and Kasumi-1 cells, stock solutions were prepared in water.
Determination of Inhibitor IC50 Values
Preparation of recombinant purified human DOT1L protein is described in Supplemental Experimental Procedures. Avian (chicken) erythrocyte oligonucleosomes were purified as previously described (Fang et al., 2004). EPZ004777 was serially diluted 3-fold in DMSO for a total of ten concentrations, beginning at 1 μM. A 1 μl aliquot of each inhibitor dilution was plated in a 384-well microtiter plate. The 100% inhibition control consisted of 2.5 μM final concentration of the product inhibitor S-adenosyl-L-homocysteine, (SAH). Compound was incubated for 30 min with 40 μl per well of 0.25 nM DOT1L(1-416) in assay buffer (20 mM TRIS [pH 8.0] 10 mM NaCl, 0.002% Tween 20, 0.005% Bovine Skin Gelatin, 100 mM KCl, and 0.5 mM DTT). 10 μl per well of substrate mix comprising assay buffer with 200 nM 3H-SAM (American Radiolabeled Chemicals: 80 Ci/mmol), 600 nM unlabeled SAM, and 20 nM nucleosomes were added to initiate the reaction (both substrates were present in the final reaction mixture at their respective KM values, an assay format referred to as “balanced conditions” (Copeland, 2003). Reactions were incubated for 120 min and quenched with 10 μl per well of 800 μM SAM. Incorporation of radioactivity into nucleosome substrate was measured in a flashplate similar to previously described studies (Sneeringer et al., 2010). IC50 values for enzymes in the histone methyltransferase panel were determined under similar balanced assay conditions with both SAM and protein/peptide substrate present at concentrations equal to their respective KM values. See Supplemental Experimental Procedures for more information.
Determination of SAM Competition
Experimental conditions were similar to IC50 experiment with the following exceptions. EPZ004777 was serially diluted 2-fold in DMSO for a total of ten concentrations, beginning at 10 nM. SAM was titrated over a range between 250 and 8000 nM. To monitor the reaction, a radioactive tracer 3H-SAM was used in each reaction, equivalent to 200 nM (which was accounted for in the total SAM concentration). Each SAM concentration had equivalent wells with no enzyme present in the reaction as negative controls. Reactions were incubated for 120 min and quenched with 10 μl per well of 800 μM SAM.
Cell Culture
Human leukemia cell lines MV4-11 (CRL-9591), THP-1 (TIB-202), RS4;11 (CRL-1873), Kasumi-1 (CRL-2724), HL-60 (CCL-240), REH (CRL-8286), and Jurkat (TIB-152) were obtained from the ATCC. SEM (ACC 546), KOPN-8 (ACC 552), 697 (ACC 42), and MOLM-13 (ACC 554) cells were obtained from DSMZ. All cell culture reagents were purchased from Invitrogen Life Technologies and cells were maintained in a humidified incubator set to 37°C, 5% CO2. RS4;11, Kasumi-1, MOLM-13, KOPN-8, REH, 697, and Jurkat cells were grown in RPMI with 10% FBS. THP-1 cells were grown in RPMI plus 10% FBS supplemented with 1× β-mercaptoethanol. MV4-11, SEM, and HL-60 cell lines were grown in IMDM with 10% FBS.
Generation of MLL-AF9- and HoxA9-Meis1a-Transformed Murine Hematopoietic Progenitors
Mouse fetal liver cells were transduced with MSCV-based MLL-AF9-IRES-GFP or Hoxa9-IRES-GFP and Meis1a-Puromycin retroviral vectors and propagated as described in Supplemental Experimental Procedures.
Analysis of Cell Proliferation and Viability
For assessment of cell proliferation and viability in human cell lines, exponentially growing cells were plated, in triplicate, in 96-well plates at a density of 3 × 104 cells/well in a final volume of 150 μl. Cells were incubated in the presence of 3 μM (proliferation curve), or increasing concentrations (IC50 determination) of EPZ004777 up to 50 μM. Viable cell number was determined every 3–4 days for up to 18 days using the Guava Viacount assay (Millipore # 4000-0040) and analyzed on a Guava EasyCyte Plus instrument (Millipore) according to the manufacturer’s protocol. On days of cell counts, growth media and EPZ004777 were replaced and cells split back to a density of 5 × 104 cells/well. Total cell number is expressed as split-adjusted viable cells per well. For each cell line, IC50 values were determined from concentration-dependence curves at each time point using Graphpad Prism software. Experiments to determine IC50 values continued until IC50 values stabilized (day 18 for THP-1 cells, day 14 for all other cell lines).
For assessment of the effect of EPZ004777 treatment on transformed murine hematopoietic progenitors, cells from two independent transductions for each virus were plated in 24-well plates at a density of 0.5–1 × 105 cell/well in 1 ml media in 24-well plates and exposed to increasing concentrations of EPZ004777 up to 30 μM. Cells were counted and replated at equal cell numbers in fresh media with fresh compound every 3–4 days. For MTT assays, cells from serial replatings were harvested on day 10 and plated, in triplicate at 2 × 104 cells/well in 100 μl media with the appropriate concentration of EPZ004777. Cells were incubated for 2.5 days, the exposed to 10 μl MTT-reagent for 3 hr, and lysed over night in 100 μl MTT- solubilization buffer (both from Cell Proliferation Kit I [MTT], Roche Diagnostics, Indianapolis, IN).
Western Blot Analysis
For western blot analysis in human cell lines, exponentially growing cells were plated in 12-well plates at 2 ×105 cells/well in a final volume of 2 ml. Cells were incubated in the presence of 3 μM (time course analysis), or increasing concentrations (IC50 determination) of EPZ004777 up to 12.5 μM. Cells (1–2 ×106) were harvested at the appropriate time point and histones were extracted as detailed in Supplemental Experimental Procedures. Histones (400 ng) were separated on 4%–20% Tris-Glycine gels (Invitrogen), transferred to 0.2 μM nitrocellulose membranes and probed with the appropriate primary antibodies (see Supplemental Experimental Procedures for a list of primary antibodies used) in Li-COR Odyssey blocking buffer (Li-Cor 927-40000) diluted 1:3 with water. Following primary antibody incubation, membranes were probed with IRDye 800CW Donkey-anti-mouse IgG (Li-COR 926-32212) or Alexa Fluor 680 goat anti-rabbit IgG (Invitrogen A-21076) secondary antibodies and signal was detected using the Li-COR Odyssey Infrared system.
For western blot analysis of subcutaneous MV4-11 xenograft tumors, tumors were harvested and snap-frozen in liquid nitrogen. Frozen tumor samples were cut into a 20 mg pieces and placed in 500 μL of ice cold lysis buffer (see Supplemental Experimental Procedures). A single 5 mm stainless steel bead (QIAGEN 69989) was added to each tube and samples homogenized using the TissueLyser II (QIAGEN 85210). Samples were homogenized by high speed shaking for 60 s at a frequency of 30 times/second. Histones were extracted and analyzed by immunoblotting as described above.
For western blot analysis in transformed murine hematopoietic progenitors, cells were incubated in the presence of increasing concentrations of EPZ004777 up to 30 μM for 10 days. Cells were counted and replated at equal cell numbers in fresh media with fresh compound every 3–4 days. Histones were extracted and analyzed by immunoblotting as detailed in Supplemental Experimental Procedures.
Quantitative Real-Time PCR
Exponentially growing MV4-11 and MOLM-13 cells were plated, in triplicate, in 96-well plates at 3 × 104 cells/well in a final volume of 150 μl. Cells were incubated in the presence of 3 μM (time course analysis), or increasing concentrations (IC50 determination) of EPZ004777 up to 12.5 μM. Compound and media were refreshed every 3–4 days during both the time course and concentration response experiments. RNA from cells was isolated using the 96-well Promega RNA isolation kit (Z3505) according to the manufacturer’s protocol. Approximately 30% of the total RNA isolated was reverse transcribed using a high capacity cDNA reverse transcription kit (Applied Biosystems 4368813) according to the manufacturer’s protocol. Predesigned labeled primer and probe sets for HOXA9 (Hs00365956), MEIS1 (Hs00180020), and TBP (4333769F) were purchased from Applied Biosystems. qPCR reactions contained 50 ng cDNA, 1× labeled primer and probe set, and 1× Taqman universal PCR master mix (Applied Biosystems 4304437). Samples were run on a 7900 HT Fast Real Time PCR machine (Applied Biosystems) with cycling conditions of 2 min at 50°C, 10 min at 95°C, 40 cycles at 15 sec at 95°C and 1 min at 60°C. Target gene cycle numbers were normalized to the house keeping gene β2-microglobulin to get a ΔCT value (Applied Biosystems 4333766). Percentage of vehicle-treated control was calculated with the Equation (2^−ΔΔCT)*100 where the ΔΔCT is the difference between normalized target gene and vehicle-treated control (ΔCT sample − ΔCT control = ΔΔCT).
Flow Cytometric Analysis of Cell Cycle and Annexin V
Exponentially growing MV4-11 and MOLM-13 cells were plated in 12-well plates at a density of 2 × 105 cells/ml. Cells were incubated with 3 μM EPZ004777 in a final volume of 2 mls for up to 10 days. For incubations longer than 4 days, media and compound were replaced on days 4 and 7 and cells were split back to a density of 2 × 105 cells/ml. Cells were harvested on days 0, 2, 4, 6, 8, and 10 and split to allow simultaneous analysis of cell cycle and Annexin V staining. Apoptosis was determined using the Guava Nexin Assay (Millipore 4500-0450) and cells prepared according to manufacturer’s recommendations. Cells treated for cell cycle analysis were pelleted by centrifugation at 200 × g for 5 min at 4°C. Pellets were washed twice with PBS then fixed with 70% ethanol on ice for 2 hr. Following fixation cells were washed once more with PBS and stained with the Guava cell cycle reagent (Millipore 4500-0220) for 20 min. Samples were analyzed using the Guava EasyCyte Plus System (Millipore).
Analysis of CD14 expression
Exponentially growing MOLM-13 cells were seeded in a 12-well plate at a density of 2 × 105 cells/ml in the presence of 3 μM EPZ004777 for 12 days. Media and compound were refreshed on days 4 and 8 and cells were split back to 2 × 105 cells/ml. Cells (3 × 105) were pelleted by centrifugation at 200 cells g and washed twice with PBS. Pellets were resuspended in 200 μl of 4% formaldehyde in PBS and fixed for 10 min at 37°C. Fixed cells were washed twice with blocking buffer (0.5% BSA, in PBS). After the last wash, cells were blocked with 1 ml of blocking buffer for 10 min at room temperature. Following the blocking step cells were incubated in blocking buffer containing either a mouse anti-CD14 labeled with FITC (1:50 dilution Chemicon CBL453F) or mouse IgG isotype control FITC-labeled (1:50 Millipore AP124F) antibody for 1 hr at room temperature. Cells were pelleted and resuspended in 200 μl of PBS and 10,000 events were analyzed using the Express Pro software on the Guava EasyCyte Plus System (Millipore).
RNA Amplification and Microarray Data Analysis
Exponentially growing MV4-11 and MOLM-13 cells were plated, in triplicate, in 6-well plates at a density of 4 × 105 cells/ml in 3 ml media in the presence or absence of 3 μM EPZ004777 for 2, 4, or 6 days. Cells were split back to 2 ×105 cells/ml and medium and compound were refreshed on day 4. Cells were pelleted by centrifugation at 200 × g, washed twice in PBS, snap-frozen on liquid nitrogen, and shipped to Expression Analysis, Inc. (Durham, NC) for RNA preparation and transcriptional profiling (see Supplemental Experimental Procedures). To determine whether genes targeted by MLL fusion proteins, or overexpressed in MLL-rearranged leukemias were suppressed by EPZ004777 treatment, Gene Set Enrichment Analysis (http://www.broadinstitute.org/gsea/index.jsp) was performed on control versus EPZ004777-treated expression files for each time point and cell line tested. The transcript lists used in this analysis were as follows. (1) MLL-AF9 targets –a list of 139 mouse genes determined to be direct targets of the MLL-AF9 fusion protein by ChiP-seq in MLL-AF9-transformed hematopoietic progenitors (see Bernt et al., 2011 [this issue of Cancer Cell]). The mouse gene list was mapped to 106 human orthologs using the NCBI Homologene database. (2) MLL-AF4 targets a list of human genes determined to be direct targets of the MLL-AF4 fusion protein by ChIP-seq in SEM cells. Genes were generated from Table S2 (Guenther et al., 2008) The “SEM fusion” was selected as positive (= 1). A total of 238 fusion targets were collapsed to 168 unique genes. (3) A common expression signature for MLL-rearranged acute leukemias, irrespective of lineage, (Ross et al., 2004) was generated from Table S13. The top 100 probes were used, excluding seven genes whose expression in leukemia with MLL fusion was below those in leukemia without the fusion. Ninety-three probe sets were collapsed into 73 unique genes. (4) Genes downregulated in in vivo established murine MLL-AF9 leukemia cells 5 days after deletion of Dot1l. Probes were selected by p value < 0.01 and fold change < −2 (see Bernt et al., 2011 [this issue of Cancer Cell]). A total of 70 mouse transcripts were mapped to 50 human orthologs.
The target lists were manually combined with MSigDB version 3, C2 KEGG gene set (http://www.broadinstitute.org/gsea/msigdb/index.jsp) as background. C2 KEGG was also used for assessment of enrichment of KEGG hematopoietic lineage genes. Due to small sample sizes, permutations by gene sets rather than by phenotypes were conducted to assess statistical significance of the gene set enrichment.
For determination of statistically significant changes in gene expression among each two-group comparison (EPZ004777 versus control), probe sets were collapsed to unique genes (by maximum probe signal). False discovery rate or q values were calculated based on t test p values, using qvalue package from R/Bioconductor (http://www.bioconductor.org/). Changes in gene expression were filtered for log2 difference > 1 or < −1 and q values < 0.15.
Pharmacokinetics
Blood samples were collected and EPZ004777 concentrations in plasma determined by an internal standard liquid chromatography tandem mass spectrometry method using protein precipitation and calibration standards prepared in blank mouse plasma. Reported concentrations are averaged from six mice in the 50 mg/ml mini-osmotic pump s.c. MV4-11 xenograft experiment, and five mice each in 100 and 150 mg/ml mini-pump pilot PK experiments.
MV4-11 Tumor Models
Animal experiments were performed at the Animal Research Facilities at Children’s Hospital Boston, the Dana-Farber Cancer Institute Boston, or the Piedmont Research Center North Carolina. Animal studies were approved by the Children’s Hospital, Piedmont or Dana-Farber Cancer Institute Animal Care and Use Committees. Experiments involving the subcutaneous MV4-11 xenograft model were performed at Piedmont Research Center (NC). Nine-week-old female nude mice (nu/nu, Harlan Laboratories, US) were injected subcutaneously with MV4-11 cells in the right flank (200 μl of a 5 × 107 cells/ml suspension in a 1:1 mixture of PBS and Matrigel (BD Biosciences)). Mice were randomized to treatment groups when tumor sizes reached 300–400 mm3. Six mice received subcutaneous implant of osmotic pumps (Alzet Model 2001, Durect Corp., Cupertino, CA), containing 50 mg/ml EPZ004777 in 10% ethanol, 90% water, and five control mice received no pump implant. Six days after pump implant, animals were sacrificed and tumor samples from treated and control animals were collected for immunoblot analysis. For the disseminated leukemia model, MV4-11 cells were transduced with the pMMP-LucNeo retrovirus as previously described (Armstrong et al., 2003). Eight-week-old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from Jackson Laboratories (Bar Harbor, ME). A total of 1 × 107 MV4-11-LucNeo cells were injected intravenously via the lateral tail vein. Engraftment of disseminate leukemia was determined by bioluminescence imaging (IVIS Spectrum, Caliper Life Sciences, Hopkinton, MA) after injection of 75 mg/kg of D-luciferin (Promega, Madison, WI), as previously described (Armstrong et al., 2003). Animals with documented leukemia were divided into treatment groups consisting of vehicle (15% ethanol, 50% PEG300, 35% water) loaded osmotic pumps, or EPZ004777 at 50, 100, or 150 mg/ml. Osmotic pumps were replaced after one week. Irritation caused by compound precipitation was observed in the 100 and 150 mg/ml dose groups, precluding additional pump replacements. Animals were monitored daily for clinical symptoms, and were euthanized when they displayed signs of distress consistent with terminal leukemic disease. Log-rank analysis was used to determine statistical significance of the survival curves (Prism, GraphPad Software, La Jolla, CA).
Hematologic Analysis of EPZ004777-Treated Mice
Eight-week-old C57BL/6 mice (Charles River Labs, Wilmington, MA) were implanted with osmotic pumps loaded with vehicle or EPZ004777 at 150 mg/ml, as described above. Osmotic pumps were replaced on day 7 to give 14 days of continuous EPZ004777 or vehicle exposure after which animals were euthanized and hematologic organs were collected. Blood for complete blood count analysis was obtained via cardiac puncture (terminal bleed). Bone marrow cell suspensions were prepared by crushing bones in a mortar after removal of muscle and connective tissues. Red blood cells were lysed on ice using red blood cells lysis buffer Puregene RBC Lysis Solution (Gentra Systems). Bone marrow lineage staining was performed as described in Supplemental Experimental Procedures. For colony forming assays, 20,000 nucleated bone marrow cells/plate were plated in duplicate in M3434 (Stem Cell Technologies, Vancouver, BC, Canada) methylcellulose with 50 U/ml Penicillin/Streptomycin (GIBCO, Invitrogen, Carlsbad, CA). Colonies were scored 8–9 days after plating.
Supplementary Material
Significance.
In mixed lineage leukemia (MLL), chromosomal translocations of the MLL gene result in recruitment of DOT1L to aberrant gene locations. This causes ectopic histone H3, lysine 79 (H3K79) methylation, and increased expression of genes involved in leukemogenesis. DOT1L has thus been proposed as a potential therapeutic target in MLL. Here, we report the development of EPZ004777, a potent, selective inhibitor of DOT1L. The compound blocks cellular H3K79 methylation, inhibits leukemogenic gene expression, and selectively kills cultured cells bearing MLL translocations. In addition, we show that EPZ004777 has antitumor activity in a mouse MLL xenograft model. This work represents a significant advance toward the goal of targeted therapeutics for MLL patients.
Acknowledgments
S.R.D., E.J.O., C.A.T., C.R.M., C.J.S., J.S., L.D.J., M.P.S., J.J.S., L.J., K.W.K., R.C., M.P.M., R.A.C., V.M.R., and R.M.P. are employees of Epizyme Inc. Y.X. and S.A.A. serve as consultants for Epizyme, Inc.
Footnotes
ACCESSION NUMBERS
The microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE29828 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29828).
Supplemental Information includes three figures and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.ccr.2011.06.009.
References
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, Pieters R, Kersey JH, Sallan SE, Fletcher JA, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(this issue):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Chang M-J, Wu H, Achille NJ, Reisenauer MR, Chou C-W, Zeleznik-Le NJ, Hemenway CS, Zhang W. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res. 2010;70:10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland RA. Mechanistic considerations in high-throughput screening. Anal Biochem. 2003;320:1–12. doi: 10.1016/s0003-2697(03)00346-4. [DOI] [PubMed] [Google Scholar]
- Copeland RA. A guide for medicinal chemists and pharmacologists. Hoboken, NJ: Wiley; 2005. Evaluation of enzyme inhibitors in drug discovery. [PubMed] [Google Scholar]
- Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, Zwaan CM, Kung AL, Armstrong SA. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Wang H, Zhang Y. Purification of histone methyltransferases from HeLa cells. Methods Enzymol. 2004;377:213–226. doi: 10.1016/S0076-6879(03)77012-8. [DOI] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, Look AT. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102:262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, Croce CM, Nakamura T, Canaani E, Young RA. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22:3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117:4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AR, Li Q, Hudson WA, Chen W, Sam T, Yao Q, Lund EA, Wu B, Kowal BJ, Kersey JH. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113:1756–1758. doi: 10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe SC, Jo SY, Sanders DS, Basrur V, Elenitoba-Johnson KS, Slany RK, Hess JL. MLL-AF9 and MLL-ENL alter the dynamic association of transcriptional regulators with genes critical for leukemia. Exp Hematol. 2010:18. doi: 10.1016/j.exphem.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, Slany RK. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, García-Cuéllar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011 doi: 10.1182/blood-2011-02-334359. Published online April 28 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Park G, Gong Z, Chen J, Kim JE. Characterization of the DOT1L network: implications of diverse roles for DOT1L. Protein J. 2010;29:213–223. doi: 10.1007/s10930-010-9242-8. [DOI] [PubMed] [Google Scholar]
- Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, Liu HC, Mahfouz R, Raimondi SC, Lenny N, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, Shurtleff SA, Pounds S, Cheng C, Ma J, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- Rozovskaia T, Feinstein E, Mor O, Foa R, Blechman J, Nakamura T, Croce CM, Cimino G, Canaani E. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4 : 11) abnormality. Oncogene. 2001;20:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94:984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slany RK, Lavau C, Cleary ML. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci USA. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA, Hua X. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig BB, Milne T, García-Cuéllar MP, Schreiner S, Martin ME, Fuchs U, Borkhardt A, Chanda SK, Walker J, Soden R, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeleznik-Le NJ, Harden AM, Rowley JD. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.