Summary
While many studies have explored the growth of Pseudomonas aeruginosa, comparatively few have focused on its survival. Previously, we reported that endogenous phenazines support the anaerobic survival of P. aeruginosa, yet the physiological mechanism underpinning survival was unknown. Here, we demonstrate that phenazine redox cycling enables P. aeruginosa to oxidize glucose and pyruvate into acetate, which promotes survival by coupling acetate and ATP synthesis through the activity of acetate kinase. By measuring intracellular NAD(H) and ATP concentrations, we show that survival is correlated with ATP synthesis, which is tightly coupled to redox homeostasis during pyruvate fermentation but not during arginine fermentation. We also show that ATP hydrolysis is required to generate a proton-motive force using the ATP synthase complex during fermentation. Together, our results suggest that phenazines enable maintenance of the proton-motive force by promoting redox homeostasis and ATP synthesis. This work demonstrates the more general principle that extracellular redox-active molecules, such as phenazines, can broaden the metabolic versatility of microorganisms by facilitating energy generation.
Keywords: Pseudomonas aeruginosa, survival, phenazines, anaerobic metabolism, pyruvate fermentation
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that forms both acute and chronic infections. It is well known to form biofilms in diverse environments, and microbial stasis within biofilms is thought to complicate the treatment of many persistent infections. For example, cells at the base of P. aeruginosa biofilms enter a dormant state with decreased levels of transcription and translation, and they undergo physiological adaptations to hypoxic conditions (Alvarez-Ortega and Harwood, 2007; Williamson et al., 2012). These cells are substantially more resistant to antibiotics than actively growing cells (Williamson et al., 2012). Moreover, nutrient deprivation triggers active responses for P. aeruginosa that increase antibiotic resistance in growth-arrested cells (Nguyen et al., 2011). Even under active growth, many bacterial species, including P. aeruginosa, maintain a small population of phenotypically resistant cells called persisters (Lewis, 2010). The persister state is thought to be controlled by stochastic changes in cell physiology and metabolism (Keren et al., 2004; Lewis, 2010; Allison et al., 2011). Together, these subpopulations of resistant non-replicating cells can serve as reservoirs for recurrent infections (Lewis, 2010). Our incomplete understanding of the metabolic changes underpinning microbial quiescence thus limits our ability to treat chronic infections.
In addition to forming biofilms and persister cells during infections, pseudomonads synthesize a class of redox-active pigments collectively termed phenazines (Price-Whelan et al., 2006; Mavrodi et al., 2006). Phenazines are important virulence factors (Lau et al., 2005) that serve as antibiotics towards microbial competitors (Baron and Rowe, 1981) and damage mammalian cells (Britigan et al., 1992). Phenazines can benefit P. aeruginosa by serving as signaling molecules (Dietrich et al., 2006), regulating persister cell formation (Möker et al., 2010), influencing colony morphology (Dietrich et al., 2008; Dietrich et al., 2013), and promoting iron acquisition and biofilm development (Wang et al., 2011).
We previously reported that micromolar concentrations of phenazines can support anaerobic survival by transferring electrons to an extracellular oxidant (Wang et al., 2010). Although the physiological basis for this survival mechanism was unknown, phenazine redox cycling was correlated with a more oxidized intracellular environment for planktonic cultures (Price-Whelan et al., 2007; Sullivan et al., 2011) and colony biofilms (Dietrich et al., 2013), suggesting that phenazines alter metabolism. P. aeruginosa is thought to form biofilms within the lungs of cystic fibrosis patients (Singh et al., 2000; Worlitzsch et al., 2002), and in many patients, the lung sputum contains phenazines at concentrations that can support anaerobic survival (Wilson et al., 1988; Hunter et al., 2012; Wang et al., 2010). Phenazines are positively correlated with the progression of P. aeruginosa lung infections (Hunter et al., 2012), and so they may directly alter the physiology of P. aeruginosa infections in situ.
Here, we sought to elucidate the physiological basis for phenazine-promoted anaerobic survival in P. aeruginosa. We discovered that phenazines facilitate the conversion of glucose and pyruvate into acetate. Our data support a model where phenazines dissipate excess reducing equivalents, allowing for the coupling of acetate and ATP synthesis through the activity of acetate kinase. ATP is required for survival, in part to maintain the proton-motive force through the ATP synthase complex. Our results highlight the interplay between redox homeostasis and ATP synthesis in the context of long-term survival, and illuminate the important role extracellular redox active metabolites can play in facilitating core metabolism.
Results
Phenazine redox cycling requires the activity of acetate kinase to promote survival
Phenazine redox cycling occurs when a phenazine molecule undergoes alternating reduction and oxidation reactions, resulting in the transfer of electrons from the reductant (e.g. NAD(P)H) to the oxidant (e.g. oxygen). Phenazines can cross cell membranes, enabling the transfer of reducing equivalents from inside the cell to the extracellular environment. We have previously shown that this transfer of reducing equivalents can promote anaerobic survival (Wang et al., 2010). We found that a carbon source, specifically glucose, is required for anaerobic survival in the presence of oxidized phenazines (Wang et al., 2010). To understand how phenazines promote anaerobic survival, we began by exploring the role of glucose.
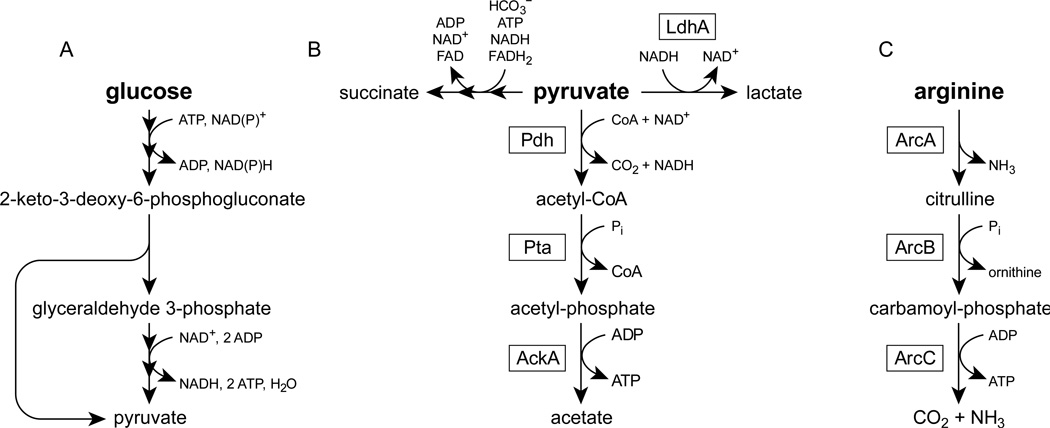
To metabolize glucose, P. aeruginosa uses the Entner-Doudoroff pathway to convert one glucose molecule into two pyruvate molecules (Figure 1a). This process is coupled to the synthesis of one net ATP molecule and two excess reducing equivalents (NAD(P)H) (Figure 1a). Pyruvate can be further converted into acetate to produce additional ATP and reducing equivalents (Figure 1b). To sustain anaerobic glycolysis, P. aeruginosa must regenerate the oxidants (i.e. NAD+ and NADP+) that were consumed during pyruvate and acetate synthesis. We therefore hypothesized that phenazines, by dissipating excess reducing equivalents, might promote the oxidation of glucose into acetate during anaerobic glycolysis. In this model, acetate synthesis is coupled to ATP synthesis through the activity of acetate kinase (Figure 1b), and it is the ATP synthesis that we expected to promote survival.
Figure 1.
Metabolic pathways in P. aeruginosa. Multiple arrowheads indicate that several reaction steps have been condensed for clarity. Boxes indicate the enzyme that catalyzes each reaction. (A) Glucose oxidation via the Entner-Doudoroff pathway. For clarity, the pathway is abbreviated to the key intermediates of glycolysis. (B) Pyruvate fermentation (Eschbach et al., 2004). LdhA, lactate dehydrogenase; Pdh, pyruvate dehydrogenase; Pta, phosphate acetyltransferase; AckA, acetate kinase. (C) The arginine deiminase pathway (Vander Wauven et al., 1984). ArcA, arginine deiminase; ArcB, ornithine carbamoyltransferase; ArcC, carbamate kinase.
To address this hypothesis, we tested mutants with markerless deletions of pyruvate metabolism genes in a phenazine redox cycling survival assay. In this experiment, early stationary phase cells were resuspended in anoxic MOPS-buffered minimal medium containing glucose as the sole carbon and energy source. To control the concentration of phenazines, we used strains with deletions in both phenazine biosynthetic operons (ΔphzA1-G1 ΔphzA2-G2, abbreviated Δphz1/2) (Dietrich et al., 2006) and added phenazine-1-carboxylic acid (PCA) back to the culture at a physiologically relevant concentration (75 µM) (Hunter et al., 2012). We chose PCA because it is the biosynthetic precursor of all other phenazines in P. aeruginosa and other pseudomonads (Mavrodi et al., 2001; Mavrodi et al., 2013), and also because it is synthesized whether oxygen is present or not. While oxygen and ferric iron are likely oxidants of phenazines in infection environments (Wang and Newman, 2008), we instead supplied an electrode poised at a potential sufficient to oxidize phenazines in our culture vessels (E0 = +207 mV).
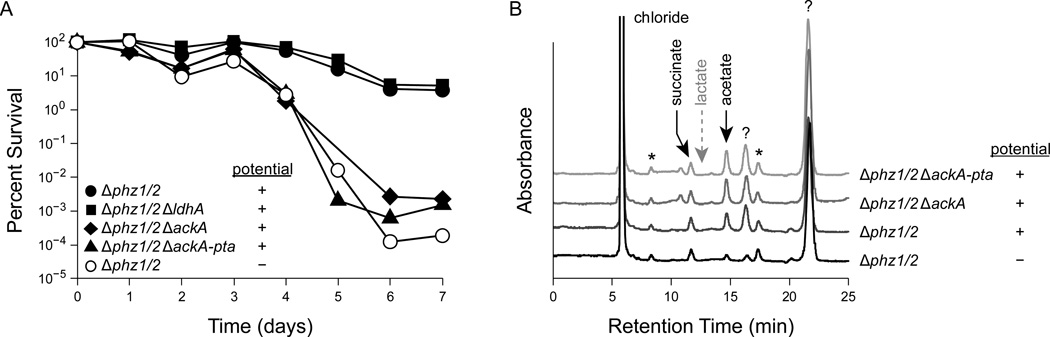
Consistent with the fact that P. aeruginosa is not known to ferment glucose, cultures declined to about 0.0001% viability after seven days when provided with glucose alone (Figure 2a). As previously reported (Wang et al., 2010), the addition of phenazines and an extracellular oxidizing potential improved anaerobic survival to 10% after seven days (Figure 2a). A strain lacking lactate dehydrogenase, ΔldhA, was not impaired in survival relative to the Δphz1/2 parent strain (Figure 2a). However, strains lacking acetate kinase that are unable to generate ATP by converting pyruvate into acetate, ΔackA and ΔackA-pta, failed to survive relative to the parent strain, with only about 0.001% of cells remaining viable after seven days (Figure 2a).
Figure 2.
Phenazine redox cycling in P. aeruginosa PA14 ∆phz1/2. Cells were grown aerobically in LB to an OD500 of 2.8 and then pelleted, washed, and resuspended in anoxic MOPS-buffered minimal medium with 20 mM glucose. Where noted in the ‘potential’ column of the legend, cultures also included 75 µM PCA and an electrode poised at a potential to oxidize any reduced PCA. Results are typical of at least three independent experiments. (A) Survival of strains with deletions of pyruvate metabolism genes. 100% survival represents approximately 7 × 108 CFU/mL. (B) HPLC traces from culture supernatants after three days of phenazine cycling as in panel A. Peaks were identified by comparing retention times to known standards. The dashed gray arrow indicates the retention time of lactate, which was not observed in this experiment. Question marks (?) indicate unidentified products. Asterisks (*) indicate trace contaminants that were present from the start of the experiment.
We detected current through the extracellular electrode only when phenazines were present (Supplemental Figure 1), consistent with the model that phenazines mediate electron transfer from inside the cells to the electrode. After three days of survival, before we observed a loss in viability (Figure 2a), the Δphz1/2 strain transferred a total charge of 11 C to the electrode. Using the relationship Q = 2F(acv), where Q is the total charge transferred (11 C), F is Faraday’s constant (96,485 C mole−1), c is the concentration of PCA (7.5 × 10−5 M), v is the reaction volume (0.1 L), and a is the average number of redox cycles, we calculated an average of 8 redox cycles per molecule of PCA, a value similar to previous measurements (Wang et al., 2010).
To test if acetate is produced during phenazine redox cycling, we used high performance liquid chromatography (HPLC) to directly measure the secreted metabolic products in our experiment. We detected the production of acetate, succinate, and two additional unknown products whose identification is a work in progress. Consistent with our hypothesis, cultures with oxidized phenazines produced more acetate than did cultures without phenazines (Figure 2b). After three days, in duplicate measurement, the Δphz1/2 strain produced 350 µM (5.9 × 10−8 nanomoles CFU−1) and 330 µM (5.0 × 10−8 nanomoles CFU−1) of acetate with phenazine redox cycling but only 78 µM (1.3 × 10−8 nanomoles CFU−1) and 130 µM (2.1 × 10−8 nanomoles CFU−1) of acetate without phenazine redox cycling. Succinate and one unknown compound were produced in approximately equal amounts irrespective of phenazines (Figure 2a), with the Δphz1/2 strain producing 2.4 × 10−8 nanomoles CFU−1 of succinate after three days with oxidized phenazines. The other unknown product was produced in greater amounts in cultures with oxidized phenazines than in cultures without phenazines (Figure 2a).
We expected that acetate would be synthesized by acetate kinase (Figure 1b), which couples acetate synthesis to ATP synthesis. To our surprise, the ΔackA and ΔackA-pta mutants produced similar amounts of acetate as the parent strain during phenazine redox cycling with glucose (Figure 2b), suggesting that acetate kinase is not the only active acetate-producing enzyme in our experiment. The P. aeruginosa PA14 genome contains the gene poxB, which codes for a putative pyruvate oxidase that converts pyruvate to acetate and CO2 without directly generating ATP or NADH (Chang and Cronan, 1983). Using quantitative reverse-transcription PCR, we detected poxB transcripts during exponential growth, early stationary phase, and after resuspension in anaerobic MOPS-buffered minimal medium with glucose and 75 µM oxidized PCA. We also measured an increase in the relative abundance of pyruvate oxidase transcripts to control gene transcripts (pyruvate dehydrogenase and the housekeeping genes clpX and recA) after resuspension in minimal medium when compared to exponential growth (Supplemental Figure 2). Pyruvate oxidase, encoded by poxB, may therefore account for the observed acetate synthesis in the ΔackA and ΔackA-pta mutants during phenazine redox cycling. Pyruvate oxidase may also account for some acetate synthesis in the WT strain.
Together, the survival defect of the ΔackA and ΔackA-pta mutants (Figure 2a) demonstrates that the activity of acetate kinase is essential for long-term survival in our phenazine redox cycling experiment. As the ΔackA and ΔackA-pta mutants still produce acetate (Figure 2b), it is unlikely that acetate itself is sufficient for survival. Instead, our data are consistent with a model where ATP synthesis by acetate kinase is required for anaerobic survival.
Phenazine redox cycling promotes ATP synthesis during pyruvate fermentation
In keeping with the fact that the Δphz1/2 ΔldhA mutant survived as well as the parent strain Δphz1/2 (Figure 2a), we did not observe lactate synthesis during phenazine redox cycling with glucose (Figure 2b). This suggests that lactate dehydrogenase is not used to maintain redox homeostasis during anaerobic glycolysis. In contrast, lactate dehydrogenase is required for survival during pyruvate fermentation (Eschbach et al., 2004). During pyruvate fermentation, P. aeruginosa converts pyruvate into succinate, lactate, and acetate (Figure 1b). This process supports anaerobic survival but not growth (Eschbach et al., 2004) and uses acetate kinase to synthesize ATP (Figure 1b). If phenazine redox cycling is a general mechanism for maintaining redox homeostasis, we predicted that phenazine redox cycling should support redox homeostasis and ATP synthesis during pyruvate fermentation.
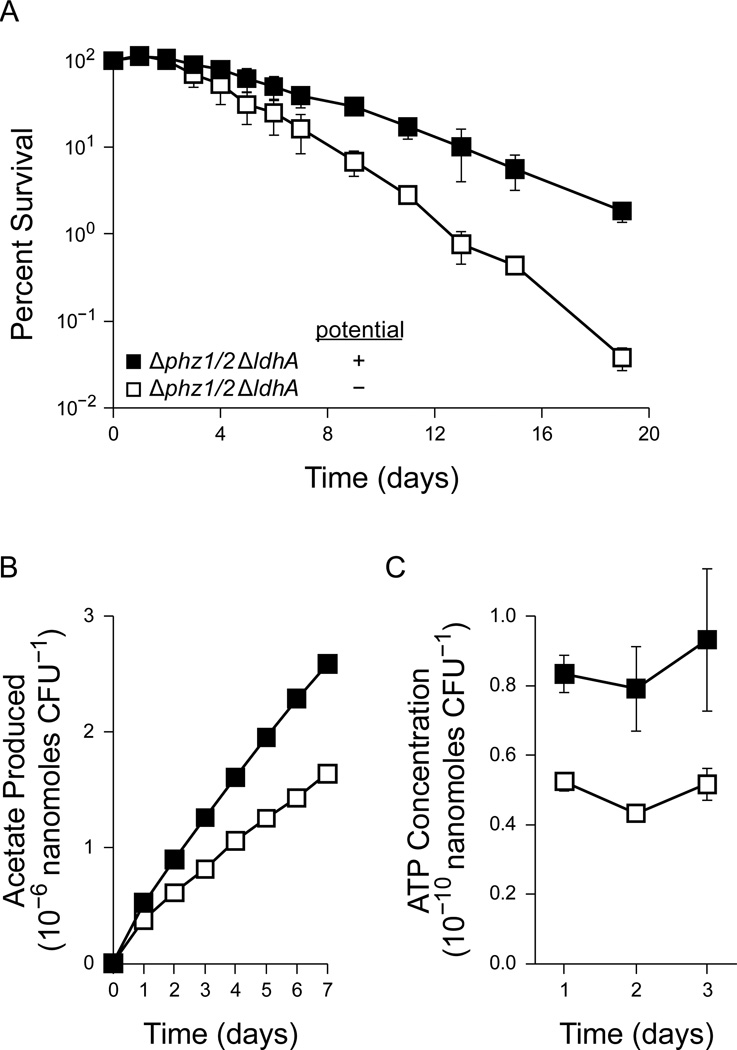
To test this prediction, we measured the anaerobic survival of the Δphz1/2 ΔldhA strain using pyruvate as the sole carbon and energy source. As expected, this strain was unable to survive by fermenting pyruvate (Figure 3a). Consistent with our prediction, however, the addition of PCA and an extracellular oxidizing potential improved the survival of the Δphz1/2 ΔldhA strain during pyruvate fermentation (Figure 3a).
Figure 3.
Pyruvate fermentation in the PA14 ∆phz1/2 ∆ldhA strain with 40 mM pyruvate. Cultures were incubated without phenazines or an oxidizing potential (open squares) or with 75 µM PCA and an oxidizing potential (closed squares). Error bars indicate the standard error of three independent experiments. (A) Survival of the ∆phz1/2 ∆ldhA strain during pyruvate fermentation. 100% survival represents approximately 1.4 × 109 CFU/mL. (B) Acetate produced during pyruvate fermentation in the ∆phz1/2 ∆ldhA strain. The amount of acetate produced in each experiment was normalized to the CFU measurement at day 0. Error bars are present but smaller than the data points. (C) ATP concentrations during pyruvate fermentation in the ∆phz1/2 ∆ldhA strain.
In our model of survival, redox homeostasis allows for additional acetate and ATP synthesis through the oxidative branch of the pyruvate fermentation pathway (Figure 1b). To further confirm this model, we used HPLC to measure acetate synthesis during pyruvate fermentation. The Δphz1/2 ΔldhA strain produced more acetate when provided with phenazines and an extracellular oxidizing potential (Figure 3b). We observed a difference in acetate production before the Δphz1/2 ΔldhA cultures began to lose viability (on day 3), suggesting that the difference in acetate synthesis was not simply due to a difference in survival.
To confirm that phenazine redox cycling provides an energetic benefit to P. aeruginosa, we also measured the ATP concentrations of cultures with and without phenazine redox cycling. Consistent with a coupling between acetate and ATP synthesis, Δphz1/2 ΔldhA cultures with oxidized phenazines maintained higher ATP concentrations than those without phenazines (Figure 3c). This difference was again observed before the cultures began to lose viability, suggesting that the difference in ATP concentration was not simply a consequence of cell death.
ATP synthesis is limited by redox homeostasis during pyruvate fermentation
Our model suggests that, by helping to maintain redox homeostasis, phenazines promote the conversion of pyruvate to acetate through the activity of acetate kinase. This model predicts that redox homeostasis is the limiting factor for ATP synthesis during pyruvate fermentation. From the stoichiometry of pyruvate fermentation, acetate and lactate synthesis are coupled through the common cofactor NAD(H) (Figure 1b). Sustained pyruvate oxidation to acetate, and thus ATP synthesis, requires the regeneration of NADH to NAD+, which can be accomplished by the activity of lactate dehydrogenase. To determine whether this is indeed the case, we compared the concentration of relevant metabolites during pyruvate fermentation for wild type (WT) P. aeruginosa PA14 and different mutant strains.
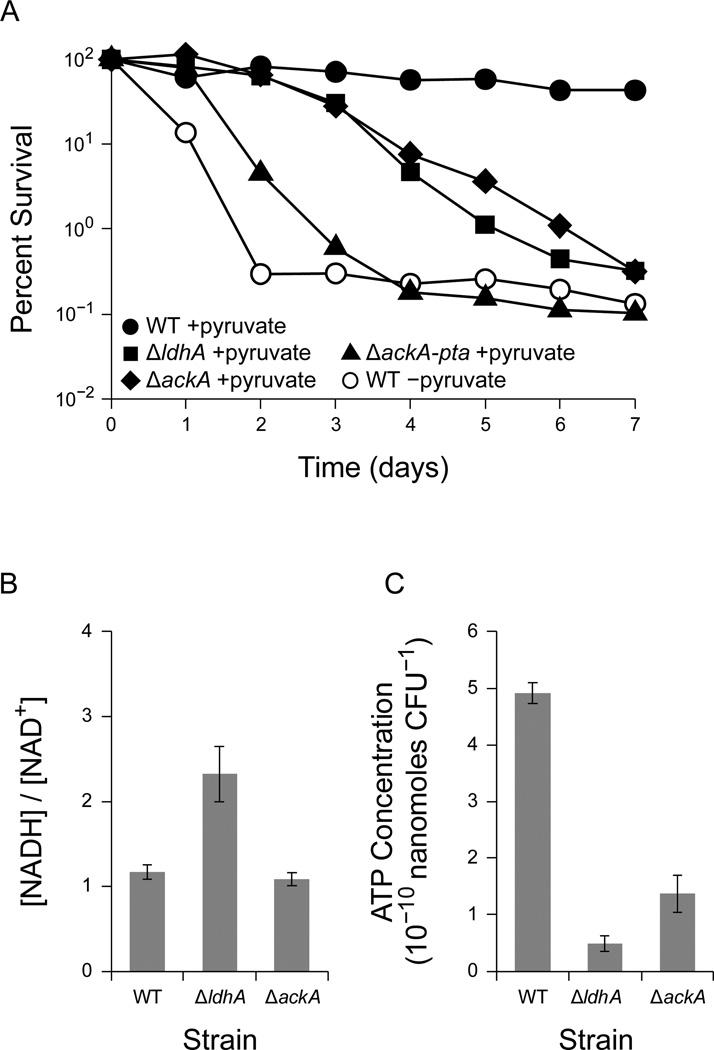
To initiate pyruvate fermentation, we cultured PA14 using pyruvate as the sole carbon source aerobically, shifted the cells to an anoxic environment, and washed and resuspended the cells in fresh anoxic medium. As expected, anaerobic WT cultures with pyruvate survived for at least seven days, while cultures without pyruvate quickly declined in viability (Figure 4a). In contrast to previous studies with PAO1 (Eschbach et al., 2004; Schreiber et al., 2006), which demonstrated nearly complete survival, in our study we observed only 50% survival after 7 days of anaerobic incubation with pyruvate.
Figure 4.
Survival with pyruvate fermentation in P. aeruginosa. Where shown, error bars indicate the standard error of three independent experiments. (A) Anaerobic survival for wild type PA14 (WT), PA14 ΔldhA (ΔldhA), PA14 ΔackA (ΔackA), and PA14 ΔackA-pta (ΔackA-pta) in the presence of 40 mM pyruvate (+pyruvate, closed points). WT PA14 without pyruvate is shown for comparison (−pyruvate, open points). 100% survival represents approximately 4 × 108 CFU/mL. Results are typical of least three independent experiments. (B) The [NADH]/[NAD+] ratio after 24 hours of pyruvate fermentation in PA14 WT, ∆ldhA, and ∆ackA. (C) ATP concentrations from the same experiments shown in panel B.
As expected, mutants deficient in the conversion of pyruvate to acetate, ΔackA and ΔackA-pta, displayed a survival defect during pyruvate fermentation (Figure 4a). A mutant deficient in the conversion of pyruvate to lactate, ΔldhA, had a survival defect similar to that of the ΔackA mutant (Figure 4a). To our surprise, a mutant unable to convert acetyl-CoA to acetyl-phosphate, ΔackA-pta, was more defective in survival than the ΔackA mutant (Figure 4a). The survival difference between the ΔackA and ΔackA-pta mutants might result from a difference in intracellular acetyl-phosphate concentrations, as acetyl-phosphate can serve as a phosphate donor in signaling pathways (Klein et al., 2007). Alternatively, the ΔackA-pta mutant might accumulate acetyl-CoA, starving other essential pathways that require coenzyme A. Similar results have been observed in E. coli, where the expression of a pathway to consume acetyl-CoA enhances the survival of a Δpta mutant in stationary phase (Chang et al., 1999).
We used ion chromatography to confirm the biosynthesis of acetate, lactate, and succinate from pyruvate as previously reported (Eschbach et al., 2004) (Supplemental Figure 3). As expected, both the ΔackA-pta and ΔldhA mutants produced significantly less acetate than the WT strain (Supplemental Figure 3), suggesting that the ΔldhA mutant is limited in its ability to generate acetate and ATP using acetate kinase.
To confirm that the ΔldhA strain is impaired in redox homeostasis and ATP synthesis, we measured the NAD+, NADH, and ATP concentrations in cultures fermenting pyruvate. After 24 hours of anaerobic incubation with pyruvate as the sole carbon and energy source, both the WT strain and ∆ackA mutant maintained an [NADH]/[NAD+] ratio of approximately 1 (Figure 4b). In contrast, the ∆ldhA mutant contained a more reduced intracellular environment with an [NADH]/[NAD+] ratio of approximately 2 (Figure 4b). The ΔldhA mutant also contained significantly less total NAD(H) than the WT strain and ΔackA mutant (Supplemental Figure 4). In support of our model, both the ∆ldhA and ∆ackA mutants contained significantly less ATP than the WT strain (Figure 4c).
The reduced acetate synthesis and depleted ATP concentration in the ∆ldhA mutant suggest that the ∆ldhA mutant is unable to synthesize ATP through acetate kinase. The elevated [NADH]/[NAD+] ratio suggests that acetate and ATP synthesis are limited by redox homeostasis, corroborating the fact that phenazine redox cycling improves survival in the ∆phz1/2 ∆ldhA mutant during pyruvate fermentation (Figure 3a).
Arginine supports anaerobic ATP synthesis and survival independently of redox state
The survival defects of the ΔackA and ∆ldhA mutants during pyruvate fermentation suggest that ATP synthesis is an essential component of survival in P. aeruginosa. To test this hypothesis using an independent anaerobic metabolism that produces ATP, we investigated survival using arginine as the sole carbon and energy source. Arginine is used as an ATP source during anaerobic growth through the arginine deiminase pathway (Vander Wauven et al., 1984) (Figure 1c). Previous work has shown that mutants in the arginine deiminase pathway have reduced anaerobic survival in undefined complex media (Schreiber et al., 2006), but to our knowledge there have been no reports of arginine alone serving as a sufficient energy source for survival.
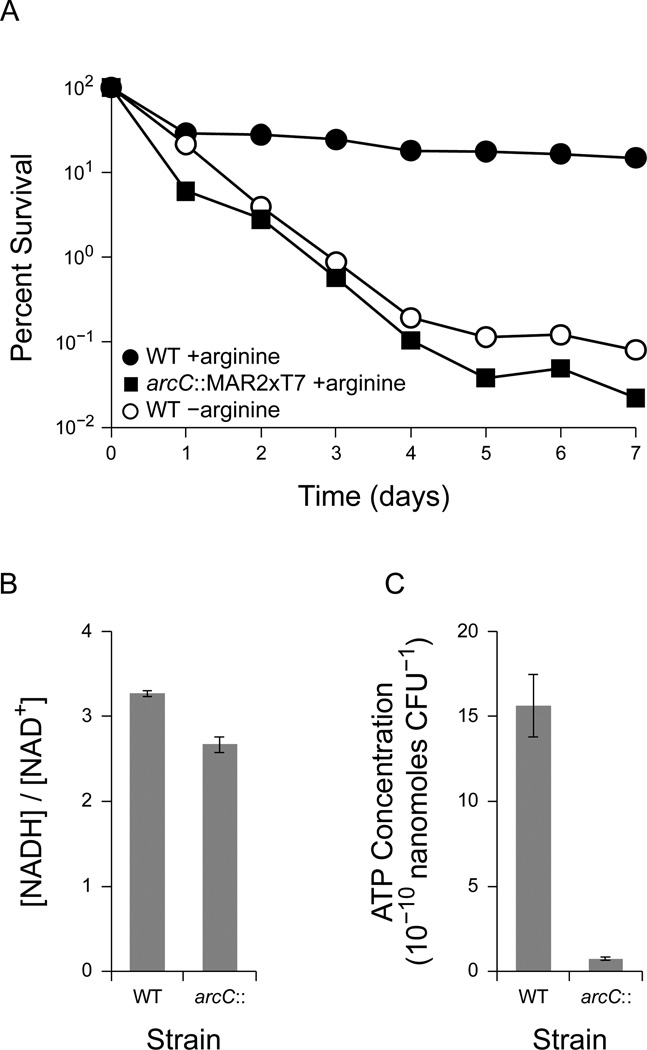
To determine whether arginine is sufficient for anaerobic survival, we cultured P. aeruginosa PA14 aerobically using arginine as the sole carbon source, shifted the cells to an anoxic environment, and washed and resuspended the cells in fresh anoxic medium with arginine. As with pyruvate, arginine alone was sufficient as an energy source to promote anaerobic survival (Figure 5a). WT PA14 dropped to 30% viability after one day of anaerobic incubation and slowly decreased in viability to 15% after seven days. In contrast, cells incubated without arginine declined to 0.1% viability after seven days (Figure 5a). A transposon insertion mutant in the arginine deiminase pathway, arcC::MAR2×T7 (Liberati et al., 2006), rapidly declined in viability to 0.02% after seven days despite the presence of arginine (Figure 5a). Similar results were obtained with a second independent transposon insertion mutant, arcA::MAR2×T7, disrupting another gene in the same pathway (Figure 1c) (data not shown). This demonstrates that arginine is sufficient to promote anaerobic survival through the arginine deiminase pathway.
Figure 5.
Survival with the arginine deiminase pathway in P. aeruginosa. Where shown, error bars indicate the standard error of three independent experiments (A) Anaerobic survival for wild type PA14 (WT) and PA14 arcC::MAR2×T7 (arc::) in the presence of 40 mM arginine. WT PA14 without arginine is shown for comparison (−arginine, open points). 100% survival represents approximately 4 × 108 CFU/mL. Results are typical of at least three independent experiments. (B) The [NADH]/[NAD+] ratio after 24 hours of anaerobic incubation with arginine in PA14 WT and arcC::MAR2×T7. (C) ATP concentrations from the same experiments shown in panel B.
To confirm that disrupting the arginine deiminase pathway also disrupted ATP synthesis, we measured NAD+, NADH, and ATP concentrations in cultures incubated with arginine. After 4 hours of anaerobic incubation with arginine as the sole carbon and energy source, both the WT strain and arcC::MAR2×T7 mutant maintained similarly high [NADH]/[NAD+] ratios of approximately 3 (Figure 5b). However, the arcC::MAR2×T7 mutant contained significantly less ATP than the WT strain (Figure 5c), consistent with the disruption of the ATP-generating step in the arginine deiminase pathway. The ability of the WT strain to anaerobically generate ATP from arginine, despite an elevated [NADH]/[NAD+] ratio, is consistent with the fact that the arginine deiminase pathway does not involve redox reactions (Figure 1c).
Together, these measurements are consistent with the hypothesis that ATP synthesis, rather than the [NADH]/[NAD+] ratio itself, is essential for long-term anaerobic survival in P. aeruginosa. Although redox homeostasis is essential when it is coupled to ATP synthesis, as in glycolysis and pyruvate fermentation, survival correlates most directly with ATP synthesis itself.
ATP synthesis is required to maintain the proton-motive force using the F1FO-ATPase complex during fermentation
The requirement for ATP suggests that P. aeruginosa consumes ATP during long-term survival. During fermentation in many bacteria, the F1FO-ATPase complex, also known as ATP synthase, hydrolyses ATP to extrude protons from the cytoplasm to the periplasm, thereby generating a proton-motive force (PMF) (Krulwich et al., 2011). Previous studies have highlighted numerous reasons why a PMF might be required for survival; for example, the PMF is required for the exchange of metabolites and other ions (Krulwich et al., 2011), the localization of cell division and cytoskeletal proteins (Strahl and Hamoen, 2010), protonation of the peptidoglycan layer (Calamita et al., 2001), and the regulation of autolysis (Jolliffe et al., 1981). We therefore hypothesized that P. aeruginosa requires ATP to maintain its PMF during fermentation.
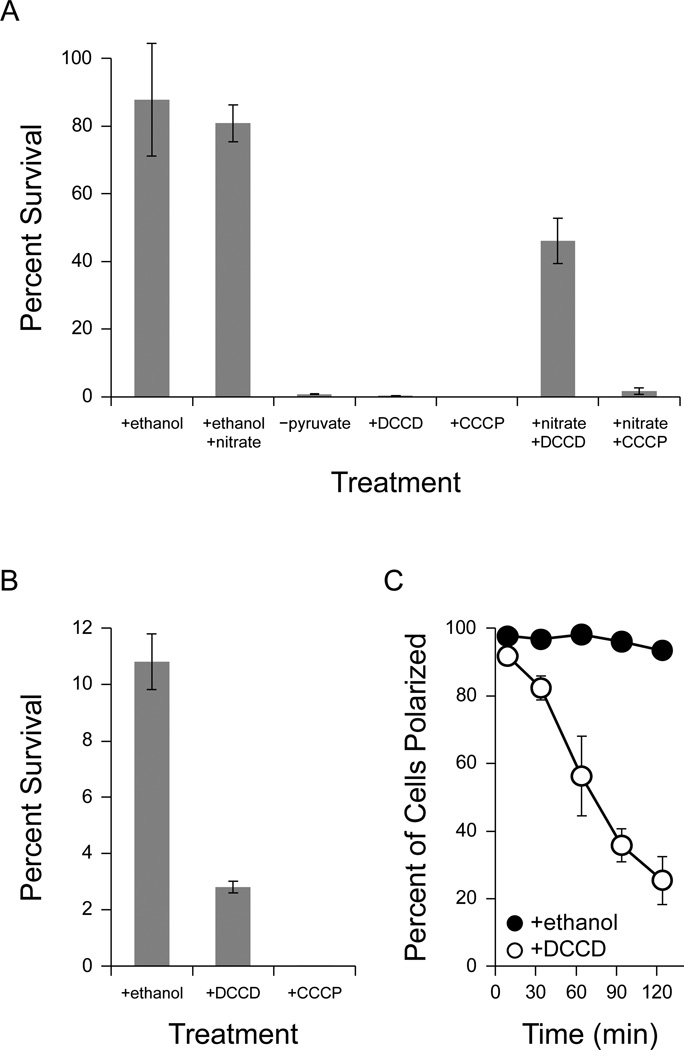
To test this hypothesis, we incubated pyruvate-fermenting cells anaerobically with N,N′-dicyclohexylcarbodiimide (DCCD), an inhibitor of the proton-translocation channel in the FO subunit of ATP synthase (Sebald et al., 1980). DCCD (dissolved in ethanol) was efficient at killing anaerobic P. aeruginosa, reducing viability to 0.16% ± 0.07% after two days (Figure 6a). Ethanol from the addition of DCCD did not significantly affect survival, as a control culture with 0.2% ethanol maintained 88 ± 17% viability (Figure 6a). This suggests that the activity of ATP synthase is required for survival during pyruvate fermentation in P. aeruginosa.
Figure 6.
Response of P. aeruginosa PA14 to drugs that disrupt the proton-motive force. Error bars represent the standard error of at least three independent experiments. 100% survival represents approximately 4 × 108 CFU/mL. (A) Anaerobic survival of P. aeruginosa PA14 after two days of anaerobic incubation with 40 mM pyruvate. At the start of anaerobiosis, potassium nitrate (50 mM), DCCD (2 mM), and/or CCCP (100 µM) were added as indicated. DCCD and CCCP were dissolved in ethanol, and so as a control ethanol was added to a final concentration of 0.2% where indicated. A culture incubated without pyruvate is shown for comparison (−pyruvate). (B) Anaerobic survival after two days of anaerobic incubation with 40 mM arginine. At the start of anaerobiosis, ethanol (0.2%), DCCD (2 mM), or CCCP (100 µM) were added as indicated. (C) Effect of DCCD on membrane polarization. After 24 hours of anaerobiosis, 2 mM DCCD was added to cultures surviving anaerobically on 40 mM pyruvate (+DCCD, open circles). Since DCCD was dissolved in ethanol, an equivalent concentration of ethanol (0.2%) was added to control cultures (+ethanol, closed circles). Membrane polarization was monitored over time as described in Methods (for an example analysis, see Supplemental Figure 5). Time 0 indicates the addition of DCCD and ethanol to the cultures.
To confirm that the PMF is required for survival, we incubated cells with carbonyl cyanide 3-chlorophenylhydrazone (CCCP), an ionophore that equilibrates the proton concentration across cell membranes and dissipates the PMF (Hopfer et al., 1968). CCCP (dissolved in ethanol) reduced the viability of anaerobic cultures to below 0.01% after two days (Figure 6a), confirming that the PMF is required for survival.
If the PMF is required for survival, then restoring the PMF should recover survival even in the presence of DCCD. Although DCCD will inhibit oxidative phosphorylation, the cells can still synthesize ATP through pyruvate fermentation. To recover the PMF after DCCD treatment, we incubated cells anaerobically with nitrate, which serves as an alternate terminal electron acceptor for the electron transport chain in P. aeruginosa (Carlson and Ingraham, 1983; Van Alst et al., 2009). Cultures incubated with pyruvate, DCCD, and nitrate sustained 46% ± 7% viability after two days (Figure 6a), demonstrating that respiration mitigates the killing effects of DCCD. Nitrate also improved the survival of cultures that were depolarized with CCCP to 2% ± 1% (Figure 6a). We did not observe significant growth after two days in cultures incubated with pyruvate, 0.2% ethanol, and nitrate, although we did observe growth after three days (data not shown), suggesting a long lag phase for growth by anaerobic nitrate respiration.
Because both pyruvate fermentation and the arginine deiminase pathway support anaerobic ATP synthesis, we also tested the effects of DCCD and CCCP on cultures surviving anaerobically with arginine. Cultures treated with 0.2% ethanol maintained 11% ± 1% viability after two days (Figure 6b), consistent with the incomplete long-term survival we observed with arginine (Figure 5a). As with survival on pyruvate, DCCD inhibited the survival of cultures incubated with arginine, reducing viability to 2.8% ± 0.2% (Figure 6b), and 100 µM CCCP reduced viability to below 0.001% (Figure 6b). This suggests that maintenance of the PMF by the F1FO-ATPase complex is a general survival strategy used by P. aeruginosa during fermentation.
To validate that DCCD caused death by preventing the generation of a PMF, rather than by some non-specific mechanism, we used flow cytometry with the dye DiOC2(3) to assess the membrane potential of DCCD-treated cultures. DiOC2(3) is a green-excitable dye that accumulates intracellularly in polarized cells, where it undergoes a red fluorescence shift that is indicative of a membrane potential (Novo et al., 1999). Since this assay is not quantitative in Gram-negative bacteria (Shapiro and Nebe-von-Caron, 2004), we assessed the membrane potential qualitatively by comparing DCCD-treated samples to controls depolarized with CCCP (for an example analysis, see Supplemental Figure 5). As expected, pyruvate-fermenting cells maintained a detectable membrane potential. We observed a time-dependent depolarization of the cells after treatment with DCCD (Figure 6c), while an ethanol-treated control remained almost fully polarized (Figure 6c), demonstrating that DCCD prevents maintenance of the PMF and induces depolarization of the cell membrane.
These results indicate that P. aeruginosa uses the ATP generated during fermentation to maintain a PMF using the F1FO-ATPase complex, and that the PMF is required for long-term anaerobic survival. To integrate our findings, we measured the membrane potential of cells surviving anaerobically on glucose in our phenazine redox cycling experiment. After three days of survival, 38% ± 6% of cells maintained a detectable membrane polarization in the presence of oxidized phenazines, while less than 1% of cells maintained a detectable membrane potential in the absence of phenazines. Together, our results suggest that the ATP generated during phenazine redox cycling is used to maintain the PMF.
Discussion
In this study, we sought an explanation for how phenazines promote anaerobic survival in P. aeruginosa be it in planktonic (Wang et al., 2010) or biofilm (Dietrich et al., 2013) cultures. We found that phenazines help P. aeruginosa convert glucose and pyruvate into acetate (Figure 2b, 3b). This was an interesting result because P. aeruginosa is not known to ferment glucose or other sugars (Barnishan and Ayers, 1979; Eschbach et al., 2004). Previous studies have shown that small molecule electron carriers can couple fermentation to extracellular oxidants, leading to the synthesis of more oxidized fermentation products (Emde et al., 1989; Emde and Schink, 1990; Benz et al., 1998; Beck and Schink, 1995). Our results suggest that phenazines perform a similar role, enabling P. aeruginosa to generate ATP by oxidizing glucose into acetate. This anaerobic glycolysis promotes survival but not growth (Figure 2a), underscoring the importance of studying survival phenotypes independently from growth phenotypes.
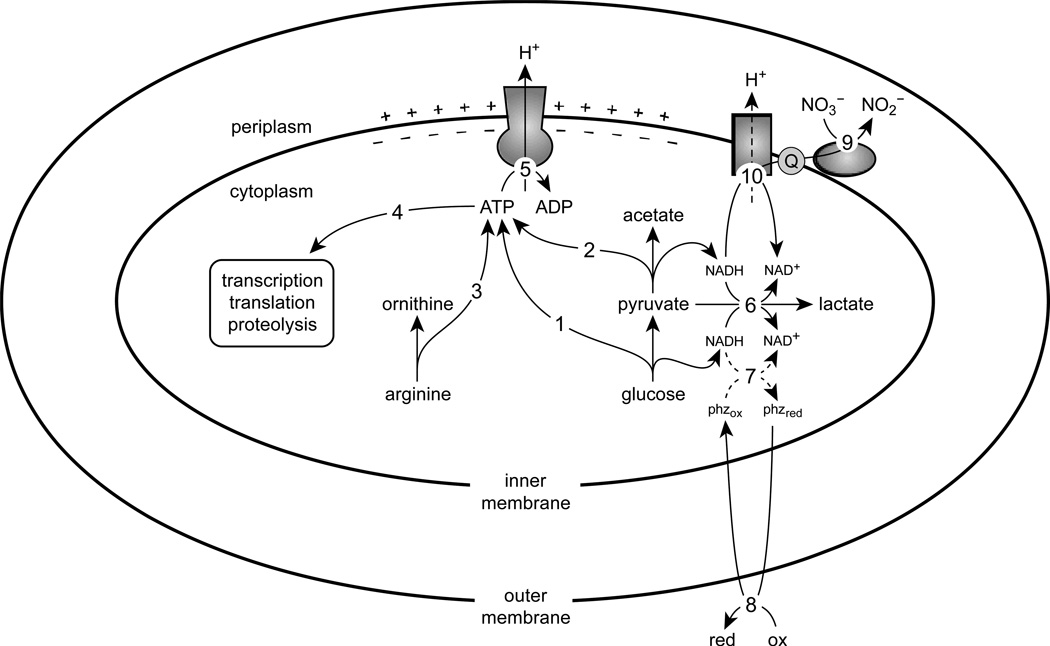
In Figure 7, we integrate the known survival pathways and processes in P. aeruginosa with a focus on redox homeostasis and ATP synthesis. By comparing phenazine redox cycling, pyruvate fermentation, and the arginine deiminase pathway, our combined results suggest that ATP synthesis is a key feature of long-term survival. The ATP synthesized during fermentation and phenazine redox cycling is required, in part, to maintain the PMF using the ATP synthase complex.
Figure 7.
A metabolic model of survival in P. aeruginosa. The ATP synthesized from glycolysis (1), pyruvate fermentation (2), and the arginine deiminase pathway (3) is used for essential processes including transcription, translation, and proteolysis (4). The F1FO-ATPase complex hydrolyses ATP to translocate protons across the inner membrane (5), thereby generating a proton-motive force. Excess reducing equivalents from pyruvate oxidation can be dispensed by converting pyruvate to lactate (6). Phenazines can also regenerate oxidants for metabolism (7); although NAD(P)H is a possible electron donor for phenazines in vivo (Price-Whelan et al., 2007), the dashed lines indicate that the nature of this interaction has not been elucidated. Phenazines are then oxidized extracellularly (8) and can be reused. Nitrate reductase can reduce nitrate to nitrite using electrons from the quinone pool (9), permitting oxidation of NADH via NADH dehydrogenase (10), which can result in proton translocation depending on which dehydrogenase complex is used.
During pyruvate fermentation, pyruvate dehydrogenase converts NAD+ to NADH in a pathway that allows acetate kinase to synthesize ATP (Figure 1b). NAD+ is regenerated by reducing pyruvate to lactate or succinate (Figure 1b). P. aeruginosa produces significantly more lactate than succinate during pyruvate fermentation (Supplemental Figure 3) (Eschbach et al., 2004), and so lactate synthesis by lactate dehydrogenase appears to be the primary pathway for NAD+ regeneration. This effectively couples the activities of acetate kinase and lactate dehydrogenase through the NAD(H) pool. As a result, the ΔldhA mutant has an elevated [NADH]/[NAD+] ratio during pyruvate fermentation (Figure 4b) and is also unable to maintain ATP levels as high as the WT strain (Figure 4c). Phenazine redox cycling can serve as an alternate pathway for redox homeostasis in the ΔldhA mutant and restore ATP synthesis (Figure 3c) and survival (Figure 3a).
In contrast to pyruvate fermentation, ATP synthesis by the arginine deiminase pathway does not involve redox reactions (Figure 1c). During anaerobic incubation with arginine, both the WT strain and the arcC::MAR2×T7 mutant developed an [NADH]/[NAD+] ratio even higher than the ΔldhA mutant did during pyruvate fermentation (Figure 5b). Nonetheless, the WT strain maintained ATP levels considerably higher than the arcC::MAR2×T7 mutant (Figure 5c), demonstrating that the arginine deiminase pathway can proceed even with large intracellular redox imbalances. Despite maintaining a high [NADH]/[NAD+] ratio, cultures incubated with arginine were able to survive through the arginine deiminase pathway (Figure 5a). Pyruvate and arginine may therefore offer alternative sources of ATP during long-term anaerobic survival.
The requirement for ATP suggests that P. aeruginosa is not dormant during long-term survival. The ATP synthase inhibitor DCCD killed anaerobic PA14 cultures that were fermenting pyruvate or arginine (Figure 6a,b) and also depolarized cultures that were fermenting pyruvate (Figure 6c), demonstrating that P. aeruginosa actively maintains its PMF using the ATP synthase complex during fermentation. Previous studies have shown that survival also requires transcription (Hu and Coates, 1999), translation (Reeve et al., 1984), and proteolysis (Weichart et al., 2003), all of which consume ATP. By promoting ATP synthesis, phenazine redox cycling can therefore facilitate a variety of cellular processes that are required for long-term survival.
We do not yet know how native phenazines such as PCA are reduced in vivo. Phenazines oxidize NAD(P)H in vitro (Cox, 1986), and so PCA might directly oxidize NAD(P)H within the cell. Redox cycling compounds such as phenazines are oxidants of flavins and iron-sulfur clusters (Gu and Imlay, 2011), and so metabolic enzymes such as NADH dehydrogenase, pyruvate dehydrogenase, or pyruvate oxidase might also reduce phenazines. Synthetic electron shuttles such as anthraquinone-2,6-disulfonate and paraquat do not support anaerobic survival by redox cycling because they are reduced slowly by P. aeruginosa (Wang et al., 2010), suggesting that cells specifically reduce native phenazines. Our preliminary studies indicate PCA reduction is enzymatically catalyzed, and we are attempting to identify the proteins responsible for this activity.
Consistent with our finding that phenazines can alter metabolism, WT P. aeruginosa cultures grown aerobically with glucose secrete pyruvate in the oxygen-limited late stationary phase of growth, while phenazine-null mutant cultures do not (Price-Whelan et al., 2007). It was suggested that superoxide or pyocyanin radicals may inhibit pyruvate dehydrogenase, leading to pyruvate accumulation in the culture supernatant (Price-Whelan et al., 2007). Our results suggest an additional interpretation where glycolysis is limited by the availability of intracellular NAD+. By promoting redox homeostasis, phenazines might accelerate the conversion of glucose to pyruvate faster than pyruvate can be consumed. After the consumption of glucose, the secreted pyruvate can be fermented or oxidized in the tricarboxylic acid cycle, depending on the metabolic needs of the cell.
Together, this work underscores the importance of environmental metabolites that are not always accounted for in metabolic pathways. Extracellular electron shuttles, such as phenazines, may permit cells to utilize unexpected energy sources by alleviating redox constraints on a pathway. An understanding of environmental parameters that interface with metabolism is therefore essential to assess the true metabolic potential of some organisms. Furthermore, a distinction must be drawn between growth and survival when considering the environment. Numerous studies have demonstrated the metabolic versatility of P. aeruginosa (for example, (Behrends et al., 2009; Stover et al., 2000)), and here we have shown how several anaerobic pathways might enable survival under clinically relevant conditions. Characterizations of cystic fibrosis sputum indicate the presence of up to 100 µM phenazines (Hunter et al., 2012) and significant quantities of nitrate, arginine, and glucose (Palmer et al., 2007). Nitrate concentrations may be as high as 350 µM (Palmer et al., 2005) and oxygen levels vary widely (Worlitzsch et al., 2002; Kolpen et al., 2014), potentially resulting in extended periods of slow growth or quiescence. Given the metabolic heterogeneity of biofilms (Williamson et al., 2012) and the likely heterogeneity of infections, P. aeruginosa cells throughout infection environments might exhibit a range of growth rates, including survival without replication. As a result, understanding the multitude of survival mechanisms used by P. aeruginosa may be key to identifying new drug targets.
Materials and Methods
Bacterial strains and growth conditions
The strains and plasmids used in this study are shown in Supplemental Table 1. For routine growth, P. aeruginosa and E. coli were cultured at 37 °C in lysogeny broth (LB) containing 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, and optionally 15 g/L agar for solid medium. S. cerevisiae was cultured at 30 °C in YPD medium containing 10 g/L yeast extract, 20 g/L peptone, 20 g/L d-glucose, and optionally 20 g/L agar for solid medium. Specific experimental growth conditions are described where appropriate. For aerobic growth, liquid cultures were shaken at 250 rpm.
Construction and verification of P. aeruginosa mutants
Unmarked deletions in P. aeruginosa PA14 were constructed using a modification of previously described methods (Shanks et al., 2006). Briefly, PCR was used to amplify approximately 1-kilobase fragments flanking the target gene. Homologous recombination in yeast was used to assemble these fragments into the sacB-counterselectable suicide shuttle vector pMQ30. The knockout vector was transformed into E. coli DH5α, and triparental mating with DH5α and E. coli HB101/pRK2013 was used to conjugate the constructed knockout vector into PA14; merodiploids containing the chromosomally integrated vector were selected on cetrimide agar (HiMedia) containing 100 µg mL−1 gentamicin sulfate. Merodiploids were cultured overnight in nonselective LB, and resolved merodiploids were then selected on LB agar plates with 10% sucrose. Potential deletion mutants were screened using colony PCR with primers flanking the target gene, and clean deletions were confirmed by DNA sequencing of the PCR product (Retrogen). Transposon insertions were verified using colony PCR with primers flanking the annotated transposon position. Primer sequences for constructing and confirming the genetic mutants used in this study are listed in Supplemental Table 2.
Phenazine redox cycling survival
The assay for anaerobic survival enabled by phenazine redox cycling was performed as previously described (Wang et al., 2010). Briefly, strains of PA14 deleted in the core phenazine biosynthesis operons (∆phz1/2) (Dietrich et al., 2006) were grown overnight in LB. The overnight cultures were used to inoculate 250 mL of LB in 1-L flasks to an OD500 of 0.05. At early stationary phase (OD500 of 2.8) the cells were pelleted, washed twice aerobically in MOPS-buffered medium (100 mM 3-(N-morpholino)propanesulfonic acid (MOPS), 3.5 µM FeSO4, 43 mM NaCl, 3.7 mM KH2PO4, 9.3 mM NH4Cl at pH 7.2), and resuspended to a concentrated OD500 of 70 before being transferred to a nitrogen-only atmosphere (MBraun Unilab glove box). There, 1 mL of concentrated cells was added to sealed glass vessels containing 100 mL of anoxic MOPS-buffered medium amended with 75 µM phenazine-1-carboxylic acid and 20 mM d-glucose or 40 mM sodium pyruvate. To oxidize PCA, a graphite rod (Alfa Aesar #14738) working electrode was poised at a potential of +207 mV vs. NHE against a homemade platinum mesh counter electrode, which was placed in buffer within an attached small chamber separated from the bulk solution by a glass frit. Electrochemical parameters were maintained by a potentiostat (Gamry). An Ag/AgCl electrode (BaSi #RE5B) was used as the reference electrode. Anaerobic cultures of cells were stirred vigorously and maintained at 31°C for the duration of the experiment, with daily sampling for colony forming units. Phenazine turnover was confirmed by the accumulated charge over the duration of the experiment as previously described (Wang et al., 2010).
Pyruvate and arginine survival
For measuring anaerobic survival using pyruvate or arginine as a sole carbon source, cultures were grown overnight at 37 °C in a minimal medium containing 14.15 mM KH2PO4, 35.85 mM K2HPO4, 42.8 mM NaCl, 9.3 mM NH4Cl, 1 mM MgSO4, 7.2 µM FeCl2 and trace elements (Widdel et al., 1983), and 40 mM sodium pyruvate or 40 mM arginine (pH adjusted to 7.2 with NaOH or HCl). Overnight cultures were diluted at least 100-fold with fresh medium to an OD500 of 0.01 and incubated at 37 °C in shaking flasks to an OD500 of 0.4. After centrifuging the culture for 15 min at 5000×g, the supernatant was removed and the pellet was transferred to a glove chamber (Coy) containing an atmosphere of 15% CO2, 80% N2, and 5% H2. The pellet was washed twice with fresh anoxic medium and then resuspended to a final OD500 of approximately 0.4 in rubber-stoppered tubes. The sealed tubes were incubated at 37 °C in the anaerobic chamber without shaking.
Viability counting
For measurements of colony forming units (CFU), 20 µL of each culture was sampled anaerobically and then serially diluted in aerobic phosphate-buffered saline. Relevant dilutions were plated in drips of 10 µL on LB agar plates and incubated aerobically for 24 hours at 37°C. The average of at least 4 pipetting replicates was taken as the CFU count for a given CFU measurement.
Measurement of NAD+, NADH, and ATP
The method for measuring NAD+ and NADH in this study was based on established protocols (San et al., 2002; Price-Whelan et al., 2007). For each condition, two 1-mL samples in 1.7 ml tubes were centrifuged for 1 min at maximum speed (16,000×g). The supernatant was removed and 200 µL of either 0.2 M HCl (for NAD+ extraction) or 0.2 M NaOH (for NADH extraction) was added and mixed by vortexing. The samples were then incubated for 10 minutes at 55 °C, cooled on ice, and partially neutralized drop-wise with 200 µL of 0.1 M NaOH (for NAD+) or 0.1 M HCl (for NADH). Cell debris was pelleted by centrifugation (5 minutes at maximum speed) and 150 µL of the supernatant was transferred to a fresh tube for immediate analysis. To assay the amount of NAD+ or NADH in each sample, 5 µL of sample was added to 90 µL of the reagent mix in a 96-well microtiter plate. The reagent mix contained 2× bicine buffer (1.0 M, pH 8.0), 1× water, 1× 40 mM EDTA, 1× 100% ethanol, 1× 4.2 mM thiazolyl blue, and 2× 16.6mM phenazine ethosulfate. The 96-well plate was then heated to 30 °C. To initiate the colorimetric assay, 5 µL of a 1 mg mL−1 solution of alcohol dehydrogenase II (Sigma A3263) in 0.1 M bicine pH 8.0 was added to each well. A BioTek Synergy 4 plate reader was used to incubate the plate at 30 °C and record the absorbance of reduced thiazolyl blue at 570 nm every minute for 30 minutes. The rate of increase in absorbance at 570 nm over time was used as the measure of NAD(H) concentration. Unknown concentrations of NAD+ and NADH were determined from the slope and intercept of a standard curve derived from known concentrations of NADH.
For ATP measurement, 20 µL of culture was added to 180 µL of dimethyl sulfoxide (DMSO) to quench and dissolve the cells. The sample was then diluted with 800 µL of 0.1 M HEPES (pH 7.5) and stored at −80 °C for up to 7 days. ATP was quantified by mixing 25 µL of sample with 25 µL of Promega BacTiter-Glo reagent in a 96-well opaque white microtiter plate. The BacTiter-Glo reagent uses luciferase to produce luminescence in an ATP-dependent manner. Total luminescence was measured at 30 °C in a BioTek Synergy 4 plate reader. In control experiments using pure ATP, DMSO had no effect on luminescence at a final concentration of 9% (data not shown). Unknown concentrations of ATP were determined from the slope and intercept of a standard curve derived from known concentrations of ATP.
Assessment of membrane potential
The membrane potential was measured qualitatively using the dyes 3,3′-diethyloxacarbocyanine iodide (DiOC2(3)) and TO-PRO-3. Cultures were diluted at least 100-fold into a permeabilization buffer containing 100 mM Tris, 1 mM EDTA, and 80 mM NaCl (pH adjusted to 7.4 using HCl). DiOC2(3) and TO-PRO-3 (both dissolved in DMSO) were then added to a final concentration of 30 µM and 100 nM, respectively. For depolarized controls, CCCP (dissolved in DMSO) was added to a final concentration of 15 µM. The final DMSO concentration did not exceed 2%. The samples were incubated for 2-5 minutes at room temperature and then analyzed on an Accuri C6 flow cytometer. DiOC2(3) fluorescence was measured using excitation at 488 nm and emission at 530 ± 15 nm (FL1) and 610 ± 10 nm (FL3). TO-PRO-3 fluorescence was measured using excitation at 640 nm and emission at 675 ± 12.5 nm (FL4). Intact cells were gated on a log-scale scatter plot of FL4 vs. FL2 (585 ± 20 nm) for further analysis. Cells with a membrane potential were distinguished by an increased red/green (FL3/FL1) fluorescence ratio relative to depolarized controls. For an example analysis, see Supplemental Figure 5.
High performance liquid chromatography
Metabolite samples were collected by centrifuging 550 µL of culture and passing the supernatant through a 0.2 µm nylon centrifugal filter. Samples were stored at −80 °C until analysis. Pyruvate, acetate, and succinate were quantified using an Agilent 1100 Series HPLC. A G1312A binary pump was used to a draw a continuous flow of 8 mN H2SO4 at 0.6 mL/min through a G1322A degasser. Samples were injected using a G1313A autosampler, and separation was effected using an Aminex HPX-87H column (Bio-Rad). Each sample was analyzed using a G1315A diode array detector to measure the UV absorbance at 206 nm referenced to 260 nm (bandwidth 16 nm) with a total run time of 25 min. Retention times for analytes were validated with single species standards. For each analyte, standards ranging from 0 to 40 mM were used to calibrate peak area against a known concentration. Data were analyzed using the ChemStation software (Agilent).
Supplementary Material
Acknowledgements
This work was supported by the Howard Hughes Medical Institute (HHMI). DKN is an HHMI Investigator, and NRG and SEK were both supported by NSF graduate research fellowships. This work was supported in part by the National Research Service Award (T32GM07676) from the National Institute of General Medical Sciences. We thank Nathan Dalleska and the Environmental Analysis Center (Caltech) for help with metabolite analyses, Ron Grimm for help with electrodes, and Ian Booth and members of the Newman lab for helpful discussions. The authors declare no conflicts of interest.
References
- Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Ortega C, Harwood CS. Responses of seudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol. 2007;65:153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnishan J, Ayers LW. Rapid identification of nonfermentative gram-negative rods by the Corning N/F system. J Clin Microbiol. 1979;9:239–243. doi: 10.1128/jcm.9.2.239-243.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron SS, Rowe JJ. Antibiotic action of pyocyanin. Antimicrob Agents Chemother. 1981;20:814–820. doi: 10.1128/aac.20.6.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S, Schink B. Acetate oxidation through a modified citric acid cycle in Propionibacterium freundenreichii. Arch Microbiol. 1995;163:182–187. [Google Scholar]
- Behrends V, Ebbels TMD, Williams HD, Bundy JG. Time-resolved metabolic footprinting for nonlinear modeling of bacterial substrate utilization. Appl Environ Microbiol. 2009;75:2453–2463. doi: 10.1128/AEM.01742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz M, Schink B, Brune A. Humic acid reduction by Propionibacterium freudenreichii and other fermenting bacteria. Appl Environ Microbiol. 1998;64:4507–4512. doi: 10.1128/aem.64.11.4507-4512.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britigan BE, Roeder TL, Rasmussen GT. Interaction of the Pseudomonas aeruginosa secretory products pyocyanin and pyochelin generates hydroxyl radical and causes synergistic damage to endothelial cells. J Clin Invest. 1992;90:2187–2196. doi: 10.1172/JCI116104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamita HG, Ehringer WD, Koch AL, Doyle RJ. Evidence that the cell wall of Bacillus subtilis is protonated during respiration. Proc Nat Acad USA. 2001;98:15260–15263. doi: 10.1073/pnas.261483798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CA, Ingraham JL. Comparison of denitrification by Pseudomonas stutzeri Pseudomonas aeruginosa and Paracoccus denitrificans. Appl Environ Microbiol. 1983;45:1247–1253. doi: 10.1128/aem.45.4.1247-1253.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D-E, Shin S, Rhee J-S, Pan J-G. Acetate Metabolism in a pta Mutant of Escherichia coli W3110: Importance of Maintaining Acetyl Coenzyme A Flux for Growth and Survival. J Bacteriol. 1999;181:6656–6663. doi: 10.1128/jb.181.21.6656-6663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-Y, Cronan JE. Genetic and Biochemical Analyses of Escherichia coli Strains having a Mutation in the Structural Gene ( poxB) for Pyruvate Oxidase. J Bacteriol. 1983;154:756–672. doi: 10.1128/jb.154.2.756-762.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CD. Role of pyocyanin in the acquisition of iron from transferrin. Infect Immun. 1986;52:263–270. doi: 10.1128/iai.52.1.263-270.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LEP, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. Bacterial Community Morphogenesis Is Intimately Linked to the Intracellular Redox State. J Bacteriol. 2013;195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LEP, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- Dietrich LEP, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emde R, Schink B. Oxidation of glycerol, lactate, and propionate by Propionibacterium freudenreichii in a poised-potential amperometric culture system. Arch Microbiol. 1990;153:506–512. doi: 10.1128/aem.56.9.2771-2776.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emde R, Swain A, Schink B. Anaerobic oxidation of glycerol by Escherichia coli in an amperometric poised-potential culture system. Appl Microbiol Biot. 1989;32:170–175. [Google Scholar]
- Eschbach M, Schreiber K, Trunk K, Buer J, Jahn D, Schobert M. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J Bacteriol. 2004;186:4596–4604. doi: 10.1128/JB.186.14.4596-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Imlay JA. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Molecular Microbiology. 2011;79:1136–1150. doi: 10.1111/j.1365-2958.2010.07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer U, Lehninger AL, Thompson TE. Protonic conductance across phospholipid bilayer membranes induced by uncoupling agents for oxidative phosphorylation. Proc Natl Acad Sci USA. 1968;59:484–490. doi: 10.1073/pnas.59.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Coates AR. Transcription of the stationary-phase-associated hspX gene of Mycobacterium tuberculosis is inversely related to synthesis of the 16-kilodalton protein. J Bacteriol. 1999;181:1380–1387. doi: 10.1128/jb.181.5.1380-1387.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RC, Klepac-Ceraj V, Lorenzi MM, Grotzinger H, Martin TR, Newman DK. Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am J Respir Cell Mol Biol. 2012;47:738–745. doi: 10.1165/rcmb.2012-0088OC. [DOI] [PubMed] [Google Scholar]
- Jolliffe LK, Doyle RJ, Streips UN. The Energized Membrane and Cellular Autolysis in Bacillus subtilis. Cell. 1981;25:753–763. doi: 10.1016/0092-8674(81)90183-5. [DOI] [PubMed] [Google Scholar]
- Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. The Intracellular Concentration of Acetyl Phosphate in Escherichia coli Is Sufficient for Direct Phosphorylation of Two-Component Response Regulators. J Bacteriol. 2007;189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpen M, Kühl M, Bjarnsholt T, Moser C, Hansen CR, Liengaard L, et al. Nitrous Oxide Production in Sputum from Cystic Fibrosis Patiens with Chronic Pseudomonas aeruginosa Lung Infection. PLOS ONE. 2014;9:e84353. doi: 10.1371/journal.pone.0084353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nature Rev Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau GW, Hassett DJ, Britigan BE. Modulation of lung epithelial functions by Pseudomonas aeruginosa. Trends Microbiol. 2005;13:389–397. doi: 10.1016/j.tim.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol. 2006;44:417–445. doi: 10.1146/annurev.phyto.44.013106.145710. [DOI] [PubMed] [Google Scholar]
- Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi DV, Parejko JA, Mavrodi OV, Kwak Y-S, Weller DM, Blankenfeldt W, et al. Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp. Environ Microbiol. 2013;15:675–686. doi: 10.1111/j.1462-2920.2012.02846.x. [DOI] [PubMed] [Google Scholar]
- Möker N, Dean CR, Tao J. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J Bacteriol. 2010;192:1946–1955. doi: 10.1128/JB.01231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo D, Perlmutter NG, Hunt RH, Shapiro HM. Accurate flow cytometric membrane potential measurement in bacteria using diethyloxacarbocyanine and a ratiometric technique. Cytometry. 1999;35:55–63. doi: 10.1002/(sici)1097-0320(19990101)35:1<55::aid-cyto8>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Palmer K, Mashburn L, Singh P. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol. 2005;187:5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price-Whelan A, Dietrich LEP, Newman DK. Rethinking 'secondary' metabolism: physiological roles for phenazine antibiotics. Nature Chem Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- Price-Whelan A, Dietrich LEP, Newman DK. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:6372–6381. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve CA, Amy PS, Matin A. Role of protein synthesis in the survival of carbon-starved Escherichia coli K-12. J Bacteriol. 1984;160:1041–1046. doi: 10.1128/jb.160.3.1041-1046.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San K-Y, Bennett GN, Berríos-Rivera SJ, Vadali RV, Yang Y-T, Horton E, et al. Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metab Eng. 2002;4:182–192. doi: 10.1006/mben.2001.0220. [DOI] [PubMed] [Google Scholar]
- Schreiber K, Boes N, Eschbach M, Jaensch L, Wehland J, Bjarnsholt T, et al. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J Bacteriol. 2006;188:659–668. doi: 10.1128/JB.188.2.659-668.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebald W, Machleidt W, Wachter E. N,N'-dicyclohexylcarbodiimide binds specifically to a single glutamyl residue of the proteolipid subunit of the mitochondrial adenosinetriphosphatases from Neurospora crassa and Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1980;77:785–789. doi: 10.1073/pnas.77.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. Saccharomyces cerevisiae -based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol. 2006;72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro HM, Nebe-von-Caron G. Multiparameter flow cytometry of bacteria. 2004:33–44. doi: 10.1385/1-59259-773-4:033. [DOI] [PubMed] [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Nat Acad USA. 2010;107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NL, Tzeranis DS, Wang Y, So PTC, Newman DK. Quantifying the dynamics of bacterial secondary metabolites by spectral multiphoton microscopy. ACS Chem Biol. 2011;6:893–899. doi: 10.1021/cb200094w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alst NE, Sherrill LA, Iglewski BH, Haidaris CG. Compensatory periplasmic nitrate reductase activity supports anaerobic growth of Pseudomonas aeruginosa PAO1 in the absence of membrane nitrate reductase. Can J Microbiol. 2009;55:1133–1144. doi: 10.1139/w09-065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Wauven C, Piérard A, Kley-Raymann M, Haas D. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J Bacteriol. 1984;160:928–934. doi: 10.1128/jb.160.3.928-934.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kern SE, Newman DK. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J Bacteriol. 2010;192:365–369. doi: 10.1128/JB.01188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Newman DK. Redox Reactions of Phenazine Antibiotics with Ferric (Hydr)oxides and Molecular Oxygen. Environ Sci Technol. 2008;42:2380–2386. doi: 10.1021/es702290a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol. 2011;193:3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichart D, Querfurth N, Dreger M, Hengge-Aronis R. Global role for ClpP-containing proteases in stationary-phase adaptation of Escherichia coli. J Bacteriol. 2003;185:115–125. doi: 10.1128/JB.185.1.115-125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdel F, Kohring G-W, Mayer F. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. Arch Microbiol. 1983;134:286–294. doi: 10.1007/BF00406470. [DOI] [PubMed] [Google Scholar]
- Williamson KS, Richards LA, Perez-Osorio AC, Pitts B, McInnerney K, Stewart PS, et al. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol. 2012;194:2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Sykes DA, Watson D, Rutman A, Taylor GW, Cole PJ. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect Immun. 1988;56:2515–2517. doi: 10.1128/iai.56.9.2515-2517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.