Abstract
Purpose
Mutations in the PIK3CA gene are common in breast cancer and represent a clinically useful therapeutic target. Several larger, population-based studies have shown a positive prognostic significance associated with these mutations. This study aims to further identify characteristics of patients harboring PIK3CA mutations while evaluating the clinical impact of genomic testing for these mutations.
Methods
Tumors from 312 patients at Vanderbilt-Ingram Cancer Center were analyzed for PIK3CA mutations using a multiplex screening assay (SNaPshot). Mutation rates, receptor status, histopathologic characteristics, and time to recurrence were assessed. The number of patients participating in clinical trials, specifically trials relating to the PIK3CA mutation, was examined. Statistically significant differences between wild type and mutated tumors were determined using the Wilcoxon, Pearson, and Fischer exact tests.
Results
The PIK3CA mutation was found in 25% of tumors tested. Patients with PIK3CA mutations were significantly more likely to express hormone receptors, be of lower combined histological grade, and have a reduced time to recurrence. Patients found to have a PIK3CA mutation were significantly more likely to enter a PIK3CA specific clinical trial.
Conclusions
In addition to confirming previously established positive prognostic characteristics of tumors harboring PIK3CA mutations, this study demonstrates the feasibility and utility of mutation profiling in a clinical setting. PIK3CA mutation testing impacted treatment and resulted in more patients entering mutation specific clinical trials.
Keywords: PIK3CA mutation, PI3K, breast cancer, SNaPshot
Background
PIK3CA (3q26.3) encodes the p110α catalytic subunit of class IA phosphatidylinositol-3 kinase (PI3K), which phosphorylates phosphatidylinositol-4,5-bisphosphate (PIP2) to produce the second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3)[1] at the plasma membrane. Several proteins critical for cell survival, proliferation, migration, metabolism and angiogenesis, including AKT, SGK and PDK1, bind to PIP3 and become activated at the plasma membrane. The PI3K/AKT pathway is the most frequently mutated pathway in breast cancer and activating mutations in PIK3CA are the most common of these somatic alterations, occurring at a frequency of 20–40%[2–9]. The expression of mutant PIK3CA in human mammary epithelial cells results in constitutive activation of the PI3K/AKT pathway and induces multiple phenotypic alterations, including anchorage- and growth factor-independent proliferation, protection from apoptosis and drug resistance[10]. Greater than 80% of these mutations occur at two “hotspots” in exon 9 and exon 20, which encode the helical (E542K and E545K) and kinase (H1047R) domains, respectively[11].
Literature regarding the prognostic significance of PIK3CA mutations is conflicting. Specifically, González-Angulo et al. found an association between PIK3CA and poor prognostic features. Li et al. also showed a negative outcome for patients with tumors harboring PIK3CA mutations[3, 12]. In contrast, Maruyama et al., Perez-Tenorio et al., and Kalinsky et al. have shown an association between PIK3CA mutations and improved recurrence-free survival[4, 7, 13]. Loi et al. showed increased relapse free survival, but no overall survival benefit in patients with PIK3CA mutant tumors[14]. Cizkova et al. and Dupont et al. demonstrated a significant improvement in metastasis-free survival in patients with PIK3CA mutant breast cancers[5, 9].
In addition to conflicting prognostic significance, data surrounding the association of PIK3CA mutations with estrogen/progesterone receptor (ER/PR) expression or human epidermal growth factor receptor 2 (HER2) overexpression have also been variable. Several larger, population based studies show a significant association between PIK3CA mutations and ER/PR positive, HER2 negative tumors[4, 7, 15, 16]. Cizkova et al. showed a statistically significant improvement in metastatic free survival in PIK3CA mutants that were PR positive or HER2 negative and a trend toward increased survival in ER positive tumors, compared to ER+ tumors with wild-type PIK3CA [5]. Somewhat counterintuitively, the presence of a PIK3CA mutation has been associated with resistance to antiestrogen therapy and suggests a role for combination therapy with antiestrogens and PI3K inhibitors[17, 18]. In addition, HER2 amplified tumors that also harbor PIK3CA mutations are less responsive to combinations of HER2 inhibitors (trastuzumab/lapatinib and trastuzumab/pertuzumab), adding to the prognostic and therapeutic significance of PIK3CA mutation testing[5, 19–24].
Given the high frequency of these mutations and their prognostic implications, an increasing focus has been placed on the development of inhibitors PI3K pathway. Janku et al. showed a favorable response to PI3K/AKT/mTOR inhibitors in patients with PIK3CA mutant tumors who had failed conventional therapy[8]. Pan-PI3K, p110-isoform specific and dual PI3K/mTOR inhibitors are currently in various stages of clinical development (phases I – III) and offer promising examples of ways that tumor genomic information can inform cancer care[25, 26].
Herein, we describe the clinical and pathologic characteristics of breast cancers harboring a PIK3CA mutation detected by mutation profiling the SNaPshot method in an academic practice setting. This assay utilizes multiplex PCR, primer extension with fluorescently tagged dideoxy-nucleotides with capillary electrophoresis for detection, and can be performed rapidly with high sensitivity, requiring only 5–10% mutant allele frequency and minimal amounts of DNA (5–20 ng) from either fresh-frozen or formalin-fixed paraffin embedded tissues[27–29]. Briefly, the breast cancer DNA mutation panel screened using the SNaPshot assay includes 18 different somatic mutations within three genes in the PI3K pathway (PIK3CA, PTEN, AKT1; see Table 1). The aim of our study was to characterize the clinico-pathologic profile of our population of breast cancer patients with these alterations in their tumors, while also investigating whether the detection of these mutations would affect patient care. We hypothesized that the detection of PIK3CA mutations would result in a significant increase in the number of patients entering clinical trials, specifically trials of PI3K inhibitors and, as such, impact clinical decision making.
Table 1. PIK3CA mutations detected by SNaPshot assay.
The incidence of each mutation is described as a percent of the total number of tumors harboring a PIK3CA mutation.
| Mutation | Incidence N (%) |
|---|---|
| PIK3CA c.3140A>G (H1047R)* | 31 (39%) |
| PIK3CA c.3140A>T (H1047L) | 4 (0.5%) |
| PIK3CA c.1624G>A (E542K)* | 15 (19%) |
| PIK3CA c.1633G>A (E545K)* | 23 (29%) |
| PIK3CA c.1633G>C (E545Q) | 0 (0%) |
| PIK3CA c.1634A>C (E545A) | 0 (0%) |
| PIK3CA c.1634A>G (E545G) | 3 (0.4%) |
| PIK3CA c.1634A>T (E545V) | 0 (0%) |
| PIK3CA c.1636C>A (Q546K) | 2 (0.3%) |
| PIK3CA c.1636C>G (Q546E) | 0 (0%) |
| PIK3CA c.1637A>C (Q546P) | 1 (0.1%) |
| PIK3CA c.1637A>G (Q546R) | 1 (0.1%) |
| PIK3CA c.1637A>T (Q546L) | 0 (0%) |
| PIK3CA c.1645G>A (D549N) | 0 (0%) |
Methods
Patient Database
The study population included 312 female patients with breast cancer whose tumor tissue underwent mutation profiling using the SNaPshot assay at the Vanderbilt University Medical Center CLIA certified Molecular Diagnostics Laboratory from July 2011 to January 2013. Written consent was obtained from all patients and the study was approved by the Institutional Review Board. All patient information was kept in a password protected database and reported in a manner to protect confidentiality. Patient characteristics, including age, menopausal status and race, were recorded at the time of initial diagnosis. Primary tumor characteristics recorded include Modified Nottingham combined histological grade, (3 to 5 low, 6 and 7 intermediate, and 8 and 9 high), pathology type (lobular or ductal), number of positive lymph nodes at diagnosis, and hormone receptor and HER2 expression, which is defined in detail below. These characteristics were based on re-review of H&E slides for all tested tissues (by two expert breast pathologists M.E.S. and M.G.K) and data from reports of the patient’s primary breast cancer. Information regarding subsequent locoregional and distant metastases was obtained from pathology reports and the patient’s medical records. The primary end point of interest included time to recurrent disease, which was defined as weeks from initial biopsy to repeat biopsy confirming locoregional/metastatic disease by site or radiologic evidence of recurrent disease. Histologic slides from procedures documenting recurrent/metastatic disease were requested and reviewed by the study pathologists in greater than 90% of cases (M.E.S and M.G.K.). A second endpoint was whether or not the patient enrolled in a clinical trial and whether the trial involved a PI3K inhibitor.
Hormone receptor and HER2 status
Estrogen and progesterone receptor detection was performed on formalin fixed, paraffin embedded tumor tissue using an enzyme immunoassay provided by Ventana Medical Systems (Tucson, Arizona). These tests were performed and reported according to the previously published Guidelines for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer[30]. HER2 status was determined by either immunohistochemistry (IHC) (Ventana Medical Systems) or FISH. HER2 gene amplification status by FISH was determined using the PathVysion HER-2 DNA probe kit (Abbott Molecular). All samples were deemed adequate for testing based on ASCO and CAP guidelines[31]. Amplification status was determined by calculating the ratio of HER2 to chromosome 17 centromere probe signals. A ratio less than 1.8 represented a normal result, while a ratio greater than 2.2 represented a positive gene amplification. A ratio between 1.8 and 2.2 was considered equivocal. We considered a patient to be HER2 positive if the result by IHC was 3+ and FISH was either not done or equivocal, or if FISH was positive. A patient was considered to be HER2 negative if the result by IHC was 0 or 1+ and FISH was equivocal or not done, or if FISH was negative with any IHC result.
Mutational Profiling
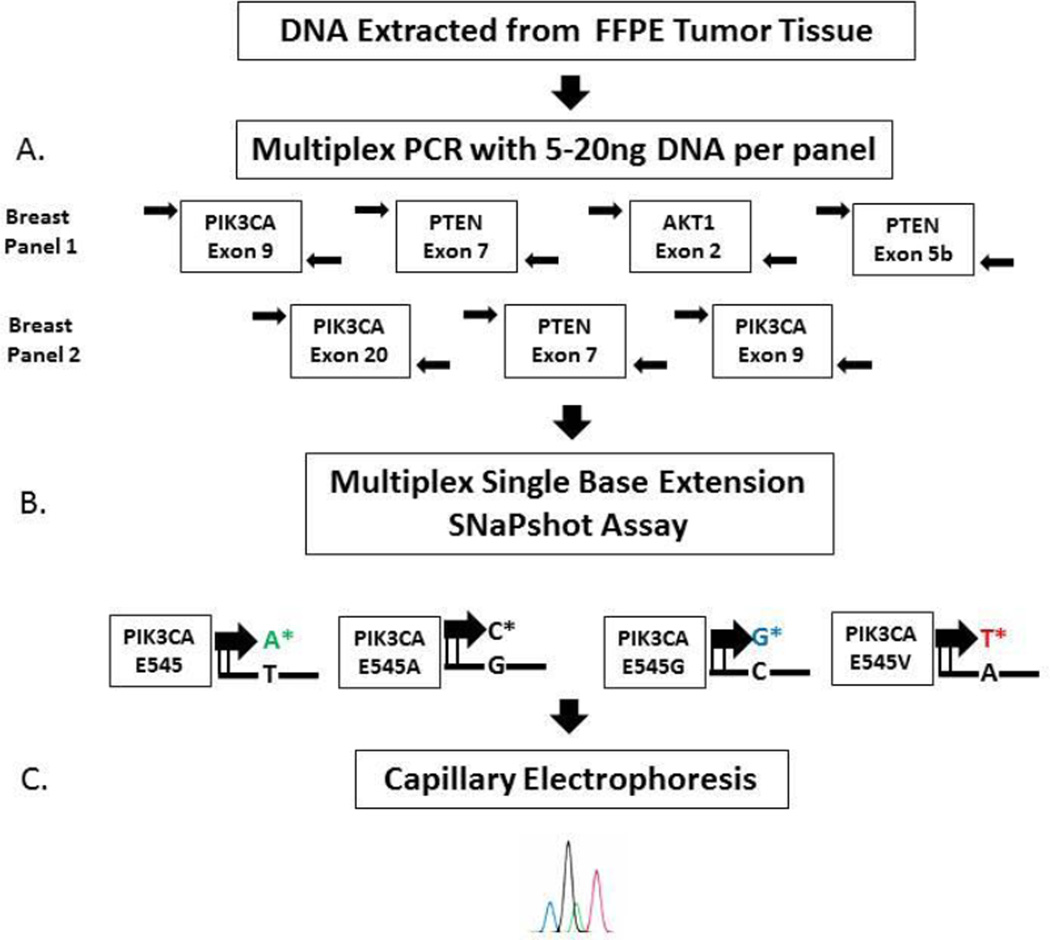
Appropriate formalin-fixed paraffin embedded tumor blocks containing invasive breast carcinoma were selected by an expert breast pathologist (M.E.S.). For specimens with >50% tumor cellularity, three to five 10-µm sections were utilized for testing, while for samples with <50% tumor cellularity, unstained slides with a corresponding H&E stained slide were submitted with the tumor area circled for macrodissection to permit enrichment for tumor DNA. Specimens with a tumor burden of <10% were rejected for analysis since this is below the sensitivity of the assay. DNA was eluted using QIAquick spin columns (Qiagen, Valencia CA, USA) following cell lysis and treatment with proteinase K. Tumor DNA (5–20 ng) was screened for 18 mutations in 3 genes (see Table 1) using multiplex base extension with the SNaPshot reaction mixture (Life Technologies, Carlsbad, CA, USA), and detection of mutations achieved utilizing capillary electrophoresis on an ABI Genetic Analyzer 3130XL as depicted in Figure 1.
Figure 1.
SNaPshot testing methodology.
Statistical Methods
Potential relationships between PIK3CA mutation status, the domain of the mutation [helical domain (exon 9 HD) versus kinase domain (exon 20, KD)] and the clinical and histopathological parameters outlined above were assessed. Associations were evaluated using the non-parametric Wilcoxon Rank Sum test, Pearson chi-squared test, or Fisher’s exact test. Differences between mutated and wild type tumors were deemed to be statistically significant at confidence levels of greater than 95% (p < 0.05). The Kaplan-Meier method for the time to recurrence by PIK3CA mutation and receptor status among patients who have recurrent disease was used, and the differences were compared with log-rank tests. All statistical analyses were performed using R 3.0.2.
Results
Mutational profiling
We assessed 18 mutations in the PI3K pathway in a cohort of 312 primary breast tumors using the SNaPshot assay. The incidence of PIK3CA mutations was 25% (79 tumors), with a total of 80 mutations detected, including one tumor with two different PIK3CA mutations. The most frequent mutation was H1047R in the kinase domain, consistent with previous reports[4, 5]. AKT1 and PTEN mutations were detected at frequencies of 3% (9 tumors) and 1% (2 tumors), respectively. Based on limited numbers of AKT1 and PTEN mutant tumors, genotype and phenotype correlations of these patients are not included in this study. Table 1 lists the frequency of each mutation in our cohort.
Patient characteristics
Patient characteristics were compared between tumors having a PIK3CA mutation and those with wild type PIK3CA (Table 2). The patient population had a median age at diagnosis of 51.8 years (95% CI, 34.1 – 71.2 years). Compared to patients with wild type tumors, those with PIK3CA mutations were significantly older at diagnosis (p=0.014) and tended to be post-menopausal (p = 0.079). The total population was 86% Caucasian, with no significant difference in ethnic makeup between the mutated and wild type cohorts (p = 0.178).
Table 2.
Patient and tumor characteristics of the total cohort and by PIK3CA mutation status.
| Characteristic | Total cohort (N = 312) |
Wild type (N= 233) |
PIK3CA Mutated (N = 79) |
P value* | |
|---|---|---|---|---|---|
| N (%) | N (%) | N (%) | |||
| Age | |||||
| ≤ 50 | 130 (42%) | 106 (45%) | 24 (30%) | ||
| > 50 | 164 (53%) | 113 (48%) | 51 (65%) | 0.014 | |
| Unknown | 18 (6%) | 14 (1%) | 4 (1%) | ||
| Menopausal status | |||||
| Pre | 84 (27%) | 66 (28%) | 18 (23%) | ||
| Post | 77 (25%) | 51 (22%) | 26 (33%) | 0.079 | |
| Unknown | 151 (49%) | 116 (50%) | 35 (44%) | ||
| Race | |||||
| Caucasian | 268 (86%) | 197 (85%) | 71 (90%) | ||
| Black | 27 (9%) | 24 (10%) | 3 (4%) | ||
| Asian/Pacific Islander | 2 (1%) | 2 (0.1%) | 0 (0%) | 0.178 | |
| Unknown or other | 15 (5%) | 10 (4%) | 4 (5%) | ||
| Tumor size | |||||
| 0 – 2 cm | 76 (24%) | 58 (25%) | 18 (23%) | ||
| 2.1 – 5 cm | 75 (24%) | 56 (24%) | 19 (24%) | ||
| > 5 cm | 19 (6%) | 16 (7%) | 3 (4%) | 0.74 | |
| Unknown or NA | 142 (46%) | 103 (44%) | 39 (49%) | ||
| Positive lymph nodes | |||||
| 0 | 107 (34%) | 75 (32%) | 32 (41%) | ||
| 1 – 3 | 91 (29%) | 71 (30%) | 20 (25%) | ||
| > 3 | 65 (21%) | 50 (21%) | 15 (19%) | 0.39 | |
| NA | 49 (16%) | 37 (16%) | 12 (15%) | ||
| Histologic subtype | |||||
| Ductal | 283 (91%) | 211 (93%) | 72 (92%) | ||
| Lobular | 12 (4%) | 8 (4%) | 4 (5%) | ||
| Other | 17 (5%) | 8 (4%) | 2 (3%) | 0.74 | |
| Histologic Grade | |||||
| Low | 40 (13%) | 23 (10%) | 17 (22%) | ||
| Intermediate | 122 (39%) | 80 (34%) | 42 (53%) | ||
| High | 143 (46%) | 124 (53%) | 19 (24%) | < 0.001 | |
| Unknown or NA | 7 (2%) | 6 (3%) | 1 (1%) | ||
| ER | |||||
| Postive | 208 (67%) | 142 (61%) | 66 (84%) | ||
| Negative | 93 (30%) | 85 (36%) | 8 (8%) | < 0.001 | |
| Unknown | 11 (4%) | 6 (3%) | 5 (6%) | ||
| PR | |||||
| Positive | 175 (56%) | 119 (51%) | 56 (71%) | ||
| Negative | 122 (39%) | 105 (45%) | 17 (22%) | <0.001 | |
| Unknown | 15 (5%) | 9 (4%) | 6 (8%) | ||
| HER2 | |||||
| Positive | 52 (17%) | 47 (20%) | 6 (8%) | ||
| Negative | 233 (75%) | 171 (73%) | 62 (78%) | 0.008 | |
| Intedeterminate or unknown | 26 (8%) | 15 (6%) | 11 (14%) | ||
| Receptor status | |||||
| ER+ or PR+/HER2− | 169 (54%) | 113 (48%) | 56 (71%) | ||
| ER+ or PR+/HER2+ | 30 (10%) | 27 (12%) | 3 (4%) | ||
| ER−and PR−/HER2+ | 21 (7%) | 19 (8%) | 2 (3%) | ||
| ER−and PR−/HER2− | 64 (21%) | 58 (25%) | 6 (8%) | ||
| Other | 28 (9%) | 16 (7%) | 12 (15%) | < 0.001 | |
| Clinical trial participant | |||||
| yes | 82 (26%) | 57 (24%) | 25 (32%) | ||
| no | 227 (73%) | 173 (74%) | 54 (68%) | 0.23 | |
| Unknown | 3 (1%) | 3 (1%) | 0 (0%) | ||
| PI3K specific trial | |||||
| yes | 40 (13%) | 17 (7%) | 23 (29%) | ||
| no | 42 (13%) | 40 (17%) | 2 (3%) | <0.001 | |
| NA | 230 (74%) | 176 (76%) | 54 (68%) | ||
| Recurrent disease | |||||
| yes | 195 (63%) | 134 (58%) | 61 (77%) | ||
| no | 117 (38%) | 99 (42%) | 18 (23%) | 0.002 | |
| Metastatic at diagnosis | |||||
| yes | 44 (14%) | 29 (12%) | 15 (19%) | ||
| no | 266 (85%) | 202 (87%) | 64 (81%) | 0.16 | |
| Unknown | 2 (1%) | 2 (0.1%) | 0 (0%) | ||
Note: "Unknown," "other," or "NA" not included in p value calculation.
Tumor features
Tumor size and nodal status were not significantly different between tumors with and without PIK3CA mutations, although mutant tumors were numerically more likely to be lymph node negative (p=0.39). Further, PIK3CA mutant tumors were more likely to be of intermediate versus high combined histological grade in the wild type cohort (p<0.001). In the total study population, 91% of tumors were of invasive ductal morphology, and there was no significant change in this percentage when comparing mutant vs. wild type tumors (92% vs. 93%, p = 0.74). The majority of tumors in the cohort expressed hormone receptors, with 67% showing ER positivity and 56%, PR positivity. PIK3CA mutated tumors were significantly more likely to express ER and PR compared with wild type tumors (ER, 84% vs. 61%; PR, 71% vs. 51%, p<0.001). Overall, 17% of tumors examined were HER2 positive by IHC or FISH. The most common receptor status was ER or PR positive/HER2 negative, making up 54% of the study population, or 169 tumors. PIK3CA mutations were highly associated with HER2 negativity (20% vs. 8%, p = 0.008). In addition, mutated tumors were significantly more likely to be ER or PR positive/HER2 negative and significantly less likely to be triple negative (p<0.001). Table 2 summarizes these findings.
Metastatic and recurrent disease
A total of 197 patients (63%) had metastatic/recurrent disease at the time of this study, reflecting referral patterns for clinical trials in our tertiary center. The patient and tumor characteristics for this cohort are displayed in Table 3. These patients had a median age at diagnosis of 51.5 years (95% CI, 34.4 – 71.1 years). Frequency of PIK3CA mutations was 31% (62 tumors). Mutant PIK3CA cancers were most likely to be of intermediate combined histologic grade, versus high grade (p = 0.01). The majority of the primary tumors were hormone receptor positive, with 70% expressing ER and 58% expressing PR. ER positivity was significantly associated with PIK3CA mutations (p = 0.011) but PR expression was not (p = 0.17). The majority of metastatic or recurrent tumors were HER2 negative (73%), and the most common overall receptor status of the metastases was ER or PR positive/HER2 negative (56%), as seen with the overall patient population. In contrast to the population as a whole, this subgroup did not show a statistically significant difference in HER2 status between wild type and mutated tumors (p = 0.18). Mutated tumors were more likely to be ER or PR positive/HER2 negative and less likely to be triple negative (p = 0.081).
Table 3.
Patient and tumor characteristics of patients who were metastatic at primary diagnosis or who developed recurrent disease stratified by PIK3CA mutation status.
| Characteristic | Total cohort (N = 197) |
Wild type (N= 135) |
PIK3CA Mutated (N = 62) |
P value* | |
|---|---|---|---|---|---|
| N (%) | N (%) | N (%) | |||
| Age | |||||
| ≤ 50 | 84 (43%) | 67 (50%) | 17 (27%) | ||
| > 50 | 96 (49%) | 54 (40%) | 42 (68%) | < 0.001 | |
| Unknown | 17 (9%) | 14 (10%) | 3 (5%) | ||
| Menopausal status | |||||
| Pre | 43 (46%) | 31 (23%) | 12 (19%) | ||
| Post | 48 (51%) | 27 (20%) | 21 (34%) | 0.12 | |
| Unknown | 3 (3%) | 77 (57%) | 29 (47%) | ||
| Race | |||||
| Caucasian | 170 (86%) | 115 (85%) | 55 (89%) | ||
| Black | 15 (8%) | 12 (9%) | 3 (5%) | ||
| Unknown or other | 12 (6%) | 8 (6%) | 4 (6%) | ||
| Tumor size | |||||
| 0 –2 cm | 46 (23%) | 34 (25%) | 12 (19%) | ||
| 2.1 –5 cm | 54 (27%) | 36 (27%) | 18 (29%) | ||
| > 5 cm | 14 (7%) | 11 (8%) | 3 (5%) | 0.65 | |
| Unknown or NA | 83 (42%) | 54 (40%) | 29 (47%) | ||
| Positive lymph nodes | |||||
| 0 | 56 (28%) | 35 (26%) | 21 (34%) | ||
| 1 –3 | 53 (27%) | 35 (26%) | 18 (29%) | ||
| > 3 | 50 (25%) | 38 (28%) | 12 (19%) | 0.31 | |
| Unknown or NA | 38 (19%) | 27 (20%) | 11 (18%) | ||
| Histologic subtype | |||||
| Ductal | 176 (93%) | 120 (93%) | 56 (92%) | ||
| Lobular | 10 (5%) | 7 (5%) | 3 (5%) | ||
| Other | 4 (2%) | 2 (2%) | 2 (3%) | 0.715 | |
| Histologic Grade | |||||
| Low | 19 (10%) | 10 (7%) | 9 (15%) | ||
| Intermediate | 82 (42%) | 49 (36%) | 33 (53%) | ||
| High | 89 (45%) | 70 (52%) | 19 (31%) | 0.01 | |
| Unknown or NA | 7 (4%) | 6 (4%) | 1 (2%) | ||
| ER | |||||
| Postive | 137 (70%) | 88 (65%) | 49 (79%) | ||
| Negative | 49 (25%) | 41 (30%) | 8 (13%) | 0.011 | |
| Unknown | 11 (6%) | 68 (50%) | 5 (8%) | ||
| PR | |||||
| Positive | 114 (58%) | 75 (56%) | 39 (63%) | ||
| Negative | 69 (35%) | 52 (39%) | 17 (27%) | 0.17 | |
| Unknown | 14 (7%) | 4 (3%) | 6 (10%) | ||
| HER2 | |||||
| positive | 29 (15%) | 23 (17%) | 6 (10%) | ||
| negative | 143 (73%) | 98 (73%) | 45 (73%) | 0.18 | |
| Intermediate or Unknown | 25 (13%) | 14 (10%) | 11 (18%) | ||
| Receptor status | |||||
| ER+ or PR+/HER2− | 110 (56%) | 71 (53%) | 39 (63%) | ||
| ER+ or PR+/HER2+ | 17 (9%) | 14 (10%) | 3 (5%) | ||
| ER−and PR−/HER2+ | 11 (6%) | 9 (7%) | 2 (3%) | ||
| ER−and PR−/HER2− | 33 (17%) | 27 (20%) | 6 (10%) | ||
| Other | 26 (13%) | 14 (10%) | 12 (19%) | 0.081 | |
| Clinical trial participant | |||||
| yes | 62 (31%) | 38 (28%) | 24 (39%) | ||
| No | 133 (68%) | 95 (70%) | 38 (61%) | 0.16 | |
| Unknown | 2 (1%) | 2 (1%) | 0 (0%) | ||
| PI3K specific trial | |||||
| yes | 40 (20%) | 17 (13%) | 23 (37%) | ||
| No | 22 (11%) | 21 (16%) | 1 (2%) | < 0.001 | |
| NA | 135 (69%) | 97 (72%) | 38 (62%) | ||
| Metastatic at diagnosis | |||||
| yes | 44 (22%) | ||||
| No | 151 (77%) | ||||
| Unknown | 2 (1%) | ||||
Note: "Unknown," "other," or "NA" not included in p value calculation.
Time to recurrence
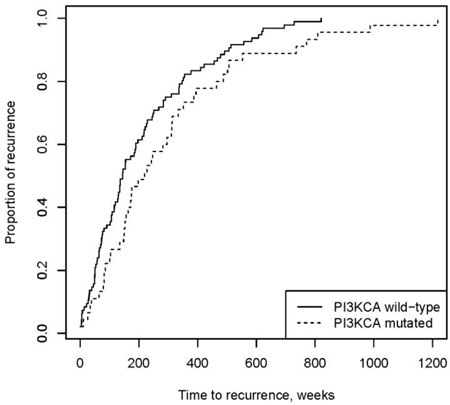
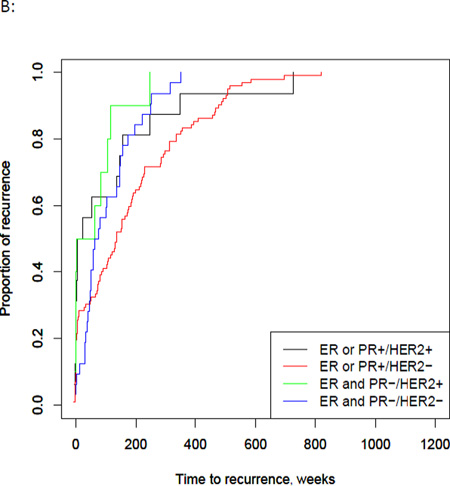
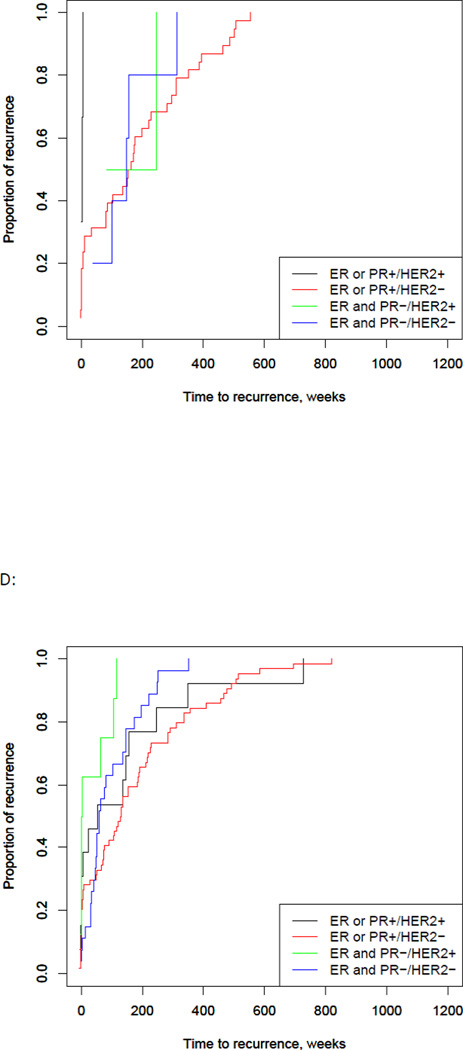
Excluding patients who had metastatic disease at diagnosis, 48% of the total patient population developed recurrent disease (151 patients). Time to recurrence data was available for 141 of these patients, with a median time to recurrence of 163 weeks (Interquartile range (IQR), 75 – 337 weeks). Patients with a PIK3CA mutations had a significantly longer median time to recurrence of 220 weeks (95% CI, 152 – 310 weeks), compared to 145 weeks in wild type (95% CI, 116 – 190 weeks, p = 0.04) (Fig. 2A). When tumors were stratified based on receptor status, median time to recurrence was longest for the ER or PR positive/HER2 negative group (median 135 weeks with 95% CI 86 – 174 weeks p = 0.01) (Fig. 2B). In the PIK3CA mutated group, the median time to recurrence was similar across the groups, except for the ER or PR positive/HER2 positive group, which was much shorter than the other groups (p = 0.01). However, there were only a small number of patients in this group (Fig. 2C). The wild type group correlated with the population as a whole, with the longest time to recurrence in the ER or PR positive/HER2 negative group (p = 0.005). Overall, mutated tumors had a longer time to recurrence compared with wild type across all receptor groups, except the ER or PR positive/HER2 positive group. These results were not significant, likely due to the small sample size of these groups (Figure 2).
Fig 2. PIK3CA, ER/PR, and HER2 status and time to recurrence.
A) PIK3CA mutations were significantly associated with a longer time to first recurrence compared to wild type (p=0.04). B) Time to recurrence by ER/PR and HER2 receptor status showed the longest recurrence –free survival in the ER/PR positive, HER2 negative group at 134.9 weeks (p = <0.001). C) In PIK3CA mutated tumors, the longest recurrence-free survival occurred in the ER/PR negative, HER2 positive group (n=2) with no significant difference between the groups. D) In wild type tumors, the longest recurrence-free survival occurred in the ER/PR positive, HER2 negative group at 128.9 weeks (p = <0.001).
Characteristics associated with helical and kinase domain mutations
Previous research has noted different biological behaviors between PIK3CA mutated tumors containing a helical domain (HD) mutation and those containing a kinase domain (KD) mutation[32]. Therefore, it is possible that these mutations have differences that affect clinical behavior. In our study, we found no significant difference in patient characteristics in those with HD versus KD mutations, including age at diagnosis, menopausal status, histological grade, or nodal status (Table 4). HD mutated tumors were less likely to express PR (61% vs. 83%, p = 0.04), but there was no difference in ER or HER2 expression between the two mutations. The overall receptor status was more likely to be ER or PR positive/HER2 negative in KD mutated tumors, compared with HD mutated tumors (p = 0.008). Interestingly, HD mutant tumors were significantly more likely to recur compared to KD mutated tumors (89% vs. 63%, p = 0.007) (Table 4).
Table 4.
Clinicopathologic characteristics of PIK3CA mutated tumors by mutation subtype.
| Characteristic | KD mutation (N = 35) |
HD mutation (N= 44) |
P value* | |
|---|---|---|---|---|
| N (%) | N (%) | |||
| Age | ||||
| ≤ 50 | 11 (31%) | 13 (30%) | ||
| > 50 | 24 (69%) | 27 (61%) | 0.92 | |
| Unknown | 0 (0%) | 4 (10%) | ||
| Menopausal status | ||||
| Pre | 10 (29%) | 8 (18%) | ||
| Post | 11 (31%) | 15 (34%) | 0.39 | |
| Unknown | 14 (40%) | 21 (47%) | ||
| Race | ||||
| Caucasian | 30 (86%) | 41 (93%) | ||
| Black | 2 (6%) | 1 (2%) | ||
| Asian/Pacific Islander | 0 (0%) | 0 (0%) | 0.58 | |
| Unknown or other | 3 (9%) | 2 (5%) | ||
| Tumor size | ||||
| 0 –2 cm | 8 (23%) | 10 (23%) | ||
| 2.1 –5 cm | 9 (26%) | 10 (23%) | ||
| > 5 cm | 2 (6%) | 1 (5%) | 0.9 | |
| Unknown or NA | 16 (46%) | 23 (53%) | ||
| Histologic subtype | ||||
| Ductal | 32 (91%) | 40 (93%) | ||
| Lobular | 2 (6%) | 2 (5%) | ||
| Other | 1 (3%) | 1 (2%) | 1 | |
| Grade | ||||
| Low | 10 (29%) | 7 (16%) | ||
| Intermediate | 18 (51%) | 24 (55%) | ||
| High | 7 (20%) | 12 (27%) | 0.39 | |
| Unknown or NA | 0 (0%) | 1 (2%) | ||
| Positive lymph nodes | ||||
| 0 | 16 (46%) | 16 (36%) | ||
| 1 –3 | 6 (17%) | 14 (32%) | ||
| > 3 | 9 (26%) | 6 (14%) | 0.18 | |
| Unknown or NA | 4 (11%) | 8 (18%) | ||
| ER | ||||
| Postive | 30 (86%) | 36 (82%) | ||
| Negative | 3 (9%) | 5 (11%) | 0.73 | |
| Unknown | 2 (6%) | 4 (9%) | ||
| PR | ||||
| Positive | 29 (83%) | 27 (61%) | ||
| Negative | 4 (11%) | 13 (30%) | 0.04 | |
| Unknown | 2 (6%) | 4 (9%) | ||
| HER2 | ||||
| Positive | 2 (6%) | 4 (9%) | ||
| Negative | 31 (89%) | 31 (70%) | 0.133 | |
| Unknown | 2 (6%) | 9 (20%) | ||
| Receptor status | ||||
| ER+ or PR+/HER2− | 30 (86%) | 26 (59%) | ||
| ER+ or PR+/HER2+ | 0 (0%) | 3 (7%) | ||
| ER−and PR−/HER2+ | 2 (6%) | 0 (0%) | ||
| ER−and PR−/HER2− | 1 (3%) | 5 (11%) | ||
| Other | 2 (6%) | 10 (23%) | 0.008 | |
| Recurrent disease | ||||
| yes | 22 (63%) | 39 (89%) | ||
| no | 12 (37%) | 5 (11%) | 0.007 | |
| Metastatic at diagnosis | ||||
| yes | 6 (17%) | 9 (20%) | ||
| no | 29 (83%) | 35 (80%) | 0.71 | |
| Unknown | 0 (0%) | 0 (0%) | ||
Note: "Unknown," "other," or "NA" not included in p value calculation.
KD = kinase domain, HD = helical domain
Clinical trial enrollment
Because of the high number of trials with PI3K inhibitors at our center and elsewhere, we asked whether the detection of PIK3CA mutations in a clinical care setting led to a clinical decision and whether these patients were more likely to enter trials of PI3K inhibitors. In the total population, 82 patients (26%) enrolled in a clinical trial. Of these, 50% entered a study of a PI3K inhibitor. Patients with PI3K mutant breast cancer were more likely to enter a trial of a PI3K inhibitor than those with wild type tumors (29%, versus 7%, p = <0.001) (Table 2). This was consistent when studying only the group of patients with recurrent or metastatic PI3K mutant cancer (37% vs. 13%, p = <0.001) (Table 3). As evidence of the impact of the knowledge of PIK3CA status on decisions regarding treatment, we present here two examples of patients whose treatment was changed due to mutation status.
The first patient was a 65 year old female with a history of a stage III invasive mammary carcinoma in 2011. The tumor was ER positive, PR positive, and HER2 negative. She received neoadjuvant chemotherapy with dose dense doxorubicin and cyclophosphamide followed by paclitaxel and then underwent a modified radical mastectomy. She began adjuvant anastrazole, and one year later, she developed back pain and was found to have metastatic disease to her bones. She began treatment with fulvestrant, which she continued for one year, until she was found to have multiple new metastases in her liver. Her oncologist recommended cytotoxic chemotherapy. She was seen at VICC and her tumor was submitted for mutation profiling by the SNaPshot assay. She was found to have a PIK3CA mutation at H1047R, a hotspot in the kinase domain. Despite a four hour commute to VICC, she initiated a clinical trial of an alpha specific PI3K inhibitor plus letrozole. Her decision to enroll was based on the fact that she would be able to avoid cytotoxic chemotherapy at the present time, that she would be able to take oral medications, and that she would be able to target a mutation in her tumor. At the time of publication, she has received ten months of therapy and is tolerating it well, with a marked decrease in the size of her tumor burden.
The second patient is a 42 year old female who presented with a Stage IV invasive mammary carcinoma that was ER/PR positive, and HER2 positive. Her staging PET scan at baseline revealed metastatic lesions in the manubrium and iliac wings. She was initiated on docetaxel, carboplatin, and trastuzumab with monthly denosumab, and due to side effects from treatment, switched to weekly paclitaxel and herceptin. Eight months later, she developed progression of her previously noted metastatic lesions and became interested in pursuing clinical trials. Her original tumor tissue was sent for mutation profiling with SNaPshot and showed a mutation at E542K in the helical domain of PIK3. Based on this information and despite the fact that she had previously received paclitaxel/trastuzumab, she decided to enter a clinical trial of paclitaxel, trastuzumab, and a PI3K inhibitor. Radiologic studies showed a partial response and then stable disease for 14 months, at which point she developed minimal progression in her bone disease. She has therefore recently switched therapy to ado-trastuzumab emtansine (Kadcyla).
Discussion
This study demonstrates the utility of tumor mutation profiling to inform and potentially enhance patient and physician decision making regarding clinical care. Previous data has been conflicting regarding the characteristics and behavior of PIK3CA mutated tumors. This study detected hot spot PIK3CA mutations in 25% of tumors tested. It is likely that these percentages underrepresent the total number of mutations, as less common mutations in PIK3CA are not detected by this assay. Interestingly, 14% of the mutations detected in this study would be considered outside of the commonly reported hot spots in the literature. Further, this assay has a sensitivity of 5–10%, making it more sensitive than previously described methods using Sanger dideoxy sequencing which have a detection level of 20–25%. To our knowledge, this represents the first application of this method for an expanded PIK3CA panel exclusively for breast cancer in a clinical care setting[27, 33–35].
Conflicting data have been published regarding the prognostic significance, hormone receptor association, and treatment implications of PIK3CA mutations. Consistent with the majority of prior publications, our study found a significant association between PIK3CA mutations and estrogen receptor positivity and HER2 negativity. In addition, we found a positive association between PIK3CA mutations and time to recurrent disease. This may be partially explained by the association between PIK3CA mutations and ER expression, given the natural history of these tumors[36, 37]. There was no statistically significant difference in time to recurrence in the PIK3CA mutated group when stratified by receptor status. Overall, the ER or PR positive/HER2 negative group had the longest time to recurrence, consistent the established prognostic implications of hormone receptor status. The recurrence free survival time for this receptor group was longest in the PIK3CA mutated group at 158 weeks, compared with 129 weeks in the wild type group and 135 weeks in the overall population; however, these differences were not statistically significant.
Previous studies have shown improved outcomes for KD mutations over HD mutations. Our study found patients with KD mutated tumors were less likely to develop recurrence and more likely to be associated with hormone receptor positivity than those with HD mutations; however, we found a shorter time to recurrence for patients with KD mutations. This is likely due to the small sample size of KD or HD mutants who developed recurrent disease in our study.
PIK3CA mutations occur frequently in breast cancer. These mutations are associated with good prognostic factors, including lower grade, hormone receptor positivity, and increased time to recurrent disease. Because of their frequency and availability of PI3K pathway inhibitors, we anticipate assays measuring these mutations will be common practice in the near future, particularly once PI3K-targeted drugs are FDA-approved. Objective and robust assays, including the SNaPshot assay, offer a quick, sensitive, and informative way to assess for these mutations for substantially lesser cost than next-generation sequencing assays. While previous publications have established links between PIK3CA mutations, ER positivity, and improved time to recurrent disease, none have evaluated the impact that this knowledge has made on patient and physician joint decision-making regarding treatment. The patients in early stage were tested because they agreed to donate tumor tissue to research. The patients with metastatic disease were tested because, in conjunction with their physicians, they wanted to better understand characteristics of their tumors in order to guide further, tumor-specific treatment. Our research shows that the objective knowledge of a PIK3CA mutation in the metastatic setting impacts clinical decision making and results in increased enrollment in specific PI3K inhibitor trials.
Acknowledgements
This study was supported by grant UL1 TR000445 from NCATS/NIH. The support of Ms. Jamie Farley in consenting patients for this study is acknowledged.
Footnotes
Ethical Standards
The experiments in this manuscript comply with the current laws of the United States.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Dillon RL, White DE, Muller WJ. The phosphatidyl inositol 3-kinase signaling network: implications for human breast cancer. Oncogene. 2007;26(9):1338–1345. doi: 10.1038/sj.onc.1210202. [DOI] [PubMed] [Google Scholar]
- 2.Campbell IG, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64(21):7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 3.Li SY, et al. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat. 2006;96(1):91–95. doi: 10.1007/s10549-005-9048-0. [DOI] [PubMed] [Google Scholar]
- 4.Kalinsky K, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15(16):5049–5059. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 5.Cizkova M, et al. PIK3CA mutation impact on survival in breast cancer patients and in ERalpha, PR and ERBB2-based subgroups. Breast Cancer Res. 2012;14(1):R28. doi: 10.1186/bcr3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller TW, et al. Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 2011;13(6):224. doi: 10.1186/bcr3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maruyama N, et al. Clinicopathologic analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Res. 2007;13(2 Pt 1):408–414. doi: 10.1158/1078-0432.CCR-06-0267. [DOI] [PubMed] [Google Scholar]
- 8.Janku F, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;30(8):777–782. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont Jensen J, et al. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res. 2011;17(4):667–677. doi: 10.1158/1078-0432.CCR-10-1133. [DOI] [PubMed] [Google Scholar]
- 10.Isakoff SJ, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65(23):10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 11.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27(41):5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Angulo AM, et al. Frequency of mesenchymal-epithelial transition factor gene (MET) and the catalytic subunit of phosphoinositide-3-kinase (PIK3CA) copy number elevation and correlation with outcome in patients with early stage breast cancer. Cancer. 2013;119(1):7–15. doi: 10.1002/cncr.27608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Tenorio G, et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res. 2007;13(12):3577–3584. doi: 10.1158/1078-0432.CCR-06-1609. [DOI] [PubMed] [Google Scholar]
- 14.Loi S, et al. Somatic mutation profiling and associations with prognosis and trastuzumab benefit in early breast cancer. J Natl Cancer Inst. 2013;105(13):960–967. doi: 10.1093/jnci/djt121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saal LH, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65(7):2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 16.Dumont AG, Dumont SN, Trent JC. The favorable impact of PIK3CA mutations on survival: an analysis of 2587 patients with breast cancer. Chin J Cancer. 2012;31(7):327–334. doi: 10.5732/cjc.012.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller TW, Balko JM, Arteaga CL. Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. J Clin Oncol. 2011;29(33):4452–4461. doi: 10.1200/JCO.2010.34.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell RA, et al. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J Biol Chem. 2001;276(13):9817–9824. doi: 10.1074/jbc.M010840200. [DOI] [PubMed] [Google Scholar]
- 19.Berns K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12(4):395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 20.Eichhorn PJ, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68(22):9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cizkova M, et al. Outcome impact of PIK3CA mutations in HER2-positive breast cancer patients treated with trastuzumab. Br J Cancer. 2013;108(9):1807–1809. doi: 10.1038/bjc.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen JD, et al. PIK3CA mutations, PTEN, and pHER2 expression and impact on outcome in HER2-positive early-stage breast cancer patients treated with adjuvant chemotherapy and trastuzumab. Ann Oncol. 2012;23(8):2034–2042. doi: 10.1093/annonc/mdr546. [DOI] [PubMed] [Google Scholar]
- 23.Baselga J, et al. Biomarker analyses in CLEOPATRA: A phase III, placebo-controlled study of pertuzumab in HER2-positive, first-line metastatic breast cancer (MBC). Cancer Res; Thirty-Fifth Annual CTRC-AACR San Antonio Breast Cancer Symposium; Dec 4–8, 2012; San Antonio, TX. 2012. Dec 15, pp. S5–S1. 2012. [Google Scholar]
- 24.Baselga JMI, Nuciforo PG, et al. PI3KCA mutations and correlation with pCR in the NeoALTTO trial (BIG 01–06). Presented at: European Cancer Congress 2013; September 27-October 1, 2013; Amsterdam, The Netherlands. 2013. Abstract 1859. [Google Scholar]
- 25.Janku F, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73(1):276–284. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bendell JC, et al. Phase I, dose-escalation study of BKM120, an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2012;30(3):282–290. doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
- 27.Hurst CD, et al. A SNaPshot assay for the rapid and simple detection of four common hotspot codon mutations in the PIK3CA gene. BMC Res Notes. 2009;2:66. doi: 10.1186/1756-0500-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su Z, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13(1):74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovly CM, et al. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic trials. PLoS One. 2012;7(4):e35309. doi: 10.1371/journal.pone.0035309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammond ME, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version) Arch Pathol Lab Med. 2010;134(7):e48–e72. doi: 10.5858/134.7.e48. [DOI] [PubMed] [Google Scholar]
- 31.Wolff AC, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131(1):18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 32.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105(7):2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Board RE, et al. Multiplexed assays for detection of mutations in PIK3CA. Clin Chem. 2008;54(4):757–760. doi: 10.1373/clinchem.2007.098376. [DOI] [PubMed] [Google Scholar]
- 34.Lurkin I, et al. Two multiplex assays that simultaneously identify 22 possible mutation sites in the KRAS, BRAF, NRAS and PIK3CA genes. PLoS One. 2010;5(1):e8802. doi: 10.1371/journal.pone.0008802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramirez-Ardila DE, et al. Hotspot mutations in PIK3CA associate with first-line treatment outcome for aromatase inhibitors but not for tamoxifen. Breast Cancer Res Treat. 2013;139(1):39–49. doi: 10.1007/s10549-013-2529-7. [DOI] [PubMed] [Google Scholar]
- 36.Knight WA, et al. Estrogen receptor as an independent prognostic factor for early recurrence in breast cancer. Cancer Res. 1977;37(12):4669–4671. [PubMed] [Google Scholar]
- 37.Kinne DW, et al. Estrogen receptor protein in breast cancer as a predictor of recurrence. Cancer. 1981;47(10):2364–2367. doi: 10.1002/1097-0142(19810515)47:10<2364::aid-cncr2820471007>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]